Abstract
Adenosine deaminases acting on RNA (ADARs) catalyze adenosine (A) to inosine (I) editing of RNA that possesses double-stranded (ds) structure. A-to-I RNA editing results in nucleotide substitution, because I is recognized as G instead of A both by ribosomes and by RNA polymerases. A-to-I substitution can also cause dsRNA destabilization, as I:U mismatch base pairs are less stable than A:U base pairs. Three mammalian ADAR genes are known, of which two encode active deaminases (ADAR1 and ADAR2). Alternative promoters together with alternative splicing give rise to two protein size forms of ADAR1: an interferon-inducible ADAR1-p150 deaminase that binds dsRNA and Z-DNA, and a constitutively expressed ADAR1-p110 deaminase. ADAR2, like ADAR1-p110, is constitutively expressed and binds dsRNA. A-to-I editing occurs with both viral and cellular RNAs, and affects a broad range of biological processes. These include virus growth and persistence, apoptosis and embryogenesis, neurotransmitter receptor and ion channel function, pancreatic cell function, and post-transcriptional gene regulation by microRNAs. Biochemical processes that provide a framework for understanding the physiologic changes following ADAR-catalyzed A-to-I ( = G) editing events include mRNA translation by changing codons and hence the amino acid sequence of proteins; pre-mRNA splicing by altering splice site recognition sequences; RNA stability by changing sequences involved in nuclease recognition; genetic stability in the case of RNA virus genomes by changing sequences during viral RNA replication; and RNA-structure-dependent activities such as microRNA production or targeting or protein–RNA interactions.
Introduction
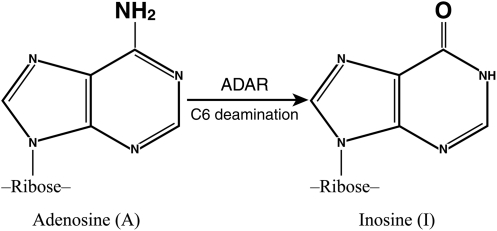
Adenosine deaminases acting on RNA (ADARs) catalyze the hydrolytic C6 deamination of adenosine (A) to produce inosine (I) in RNA substrates that possess regions of double-stranded (ds) character (Fig. 1). A-to-I editing is of increasingly broad physiologic significance (Samuel 2001; Bass 2002). Editing by ADARs results in nucleotide substitution in RNA, because the purine I generated as the result of the deamination reaction is recognized as G instead of A, both by ribosomes during translational decoding of mRNA and by RNA-dependent polymerases during RNA replication. In addition, A-to-I editing leads to destabilization of dsRNA structures, because stable A:U base pairs are changed to less stable I:U mismatch base pairs (Valente and Nishikura 2005; Toth and others 2006). Two mammalian genes (ADAR1 and ADAR2) are known that encode enzymatically active ADARs (Bass and others 1997; Toth and others 2006). A third gene, ADAR3, is known but has not yet been shown to encode a catalytically active deaminase (Chen and others 2000; Toth and others 2006). This review focuses on the organization and regulation of ADAR1 and ADAR2 gene expression, the biochemical properties of the encoded proteins, and the biological significance of A-to-I editing catalyzed by these ADARs.
FIG. 1.
RNA editing by adenosine deamination. Substitution RNA editing by hydrolytic C6 deamination of adenosine (A) to generate inosine (I) in RNA with double-stranded (ds) character catalyzed by adenosine deaminase acting on RNA (ADAR) enzymes. I is recognized as G instead of A during ribosome decoding of mRNA and during RNA replication by RNA-dependent polymerases. Conversion of an A:U base pair to an I:U mismatch destabilizes duplex RNA and alters RNA structures.
RNA Adenosine Deaminase (ADAR) Genes and Their Regulation
ADAR1 gene
Chromosome assignment of ADAR1
The human ADAR1 gene maps to a single locus on chromosome 1 band q21.1–21.2 by fluorescence in situ hybridization (Wang and others 1995; Weier and others 1995). Mouse Adar1 maps by fluorescence in situ hybridization to a single locus on chromosome 3 band F2 (Weier and others 2000). These assignments for the human and mouse ADAR1 genes are consistent with human–mouse homology maps that localize other genes from human chromosome 1q to the mouse chromosome 3F region. DNA sequence analyses of ADAR1 genomic and cDNA clones are consistent with a single mammalian ADAR1 gene (Wang and others 1995; Liu and others 1997; George and Samuel 1999b; George and others 2005).
ADAR1 exon–intron organization
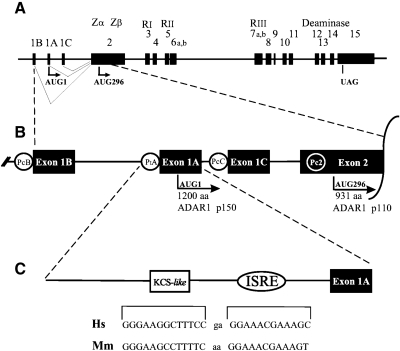
The human ADAR1 gene spans ∼40 kb and includes 17 exons (Wang and others 1995; Liu and others 1997; George and Samuel 1999a; Kawakubo and Samuel 2000) as summarized in Fig. 2. The consensus sequence for the human ADAR1 cDNA is 6,474 nt and predicts an open-reading frame (ORF) of 3,678 nt with the capacity to encode a 1,226 amino acid protein (Kim and others 1994b; Patterson and Samuel 1995), but it is now known that ADAR1 transcription initiates from multiple promoters, one interferon (IFN) inducible and the others constitutively active, and that the transcripts undergo alternative splicing to encode two differently sized ADAR1 proteins, an IFN-inducible (p150) and a constitutively expressed (p110) form of ADAR1 (Patterson and Samuel 1995; Liu and others 1997; George and Samuel 1999a, 1999b).
FIG. 2.
Promoter and exon–intron organization of the human ADAR1 gene. (A) Organization of introns and exons of the ADAR1 gene that spans about ∼40 kb on human chromosome 1q21. Exons are indicated by the numbers 1–15 (according to Liu and others 1997; George and Samuel 1999a, 1999b) and are represented by the filled boxes; introns and the 5′- and 3′-flanking regions are shown by the solid lines; and the alternative exons by outlined shaded boxes. (B) Alternative promoters drive expression of the human ADAR1 gene. An interferon (IFN)-inducible promoter (PIA) drives expression of inducible transcripts that possess the exon 1A; exons 1B and 1C are found in transcripts derived from constitutively active alternative promoters (PCB, PCC). In addition, a constitutively active promoter (PC2) is found in exon 2. The AUG1 translation initiation site of the IFN-inducible p150 protein (1,200 amino acids) is in exon 1A; the constitutively expressed p110 protein (931 amino acids) initiates at AUG296 in exon 2. (C) The IFN-inducible promoter possesses a 12-bp IFN-stimulated response element (ISRE) and an adjacent 13-bp kinase conserved sequence (KCS)-like element. Human, Hs, Homo sapiens; mouse, Mm, Mus musculus (adapted from Toth and others 2006, with permission).
Exon 1 occurs in three alternative forms, designated as 1A, 1B, and 1C (George and Samuel 1999a, 1999b; Kawakubo and Samuel 2000) (Fig. 2A). Only the IFN-inducible 1A form of exon 1 contains a translation initiation codon, designated as AUG1 (George and Samuel 1999a). The constitutively expressed forms of exon 1, 1B and 1C, both lack a translation initiation codon and therefore translation begins at the downstream AUG296 codon present in exon 2 (Patterson and Samuel 1995; Liu and others 1997). The junction of the different exon 1 structures (1A, 1B, or 1C) with exon 2 is exactly conserved, in both human (George and Samuel 1999a, 1999b) and mouse (George and others 2005) ADAR1 transcripts. In addition to the 3 different forms of exon 1, tissue and cell line differences are observed in the occurrence of alternative splice variants of exons 6 and 7 (Liu and others 1997; Shtrichman and others 2002; George and others 2005). Alternative exon 7b has been linked predominantly to IFN-inducible transcripts that contain exon 1A (the p150-coding RNA), whereas exon 7a is found in constitutively expressed transcripts containing exon 1B (the p110-coding RNA) (George and others 2005). Exon 7b is 26 amino acids shorter than exon 7a. With this exon arrangement, the IFN-inducible transcripts possessing exons 1A and 7b encode an inducible ADAR1 protein (p150) of 1,200 amino acids. By contrast, the constitutively expressed transcripts beginning at the 5′-end with either exon 1B or 1C (George and Samuel 1999a, 1999b) or within exon 2 (Kawakubo and Samuel 2000) encode a constitutive ADAR1 protein (p110) predicted to be 931 amino acids (Fig. 2B). The two AUG codons used for translation initiation, AUG1 in exon 1A and AUG296 in exon 2, both have a strong translation initiation site based on their −3/+4 flanking nucleotides (Kozak 1989; Kim and others 1994b; Patterson and Samuel 1995; Liu and others 1997).
Exon 2 is unusually large (1,586 nt) for a mammalian exon that is entirely an ORF. However, this exon is only completely decoded when present in the inducible transcripts (Liu and others 1997; George and Samuel 1999b). In the constitutive transcripts, the 5′-region of exon 2 upstream of AUG296 as well as all of exon 1B (or 1C) together account for the 5′-untranslated region (UTR) (George and Samuel 1999a, 1999b). Further alternative splicing of ADAR1 transcripts in the 5′-region that functions either as the 5′-UTR or as part of the ORF also has been described (Lykke-Andersen and others 2007). The 3′-UTR of human ADAR1 transcripts is 2,749 nt and is positioned entirely within exon 15, the largest ADAR1 exon (2,984 nt) that has an UAG for translation termination. The 3′-UTR also includes the canonical hexameric AAUAAA polyadenylation signal present 17 nt upstream of the poly(A) tail. Curiously, a repeated sequence of 227 nt (87% identity) of unknown function, as well as three copies of the AUUUA sequence associated with mRNA instability, also are present in the 3′-UTR (Patterson and Samuel 1995).
The mouse Adar1 gene consists of 15 exons with an organization similar to that of the human ADAR1 gene (Liu and others 1997; Hartner and others 2004; Wang and others 2004; George and others 2005). The ORF of the rat ADAR1 cDNA (O'Connell and others 1995) is similar to that of both the mouse and human ADAR1 cDNAs (Kim and others 1994b; Patterson and Samuel 1995; Kumar and Carmichael 1997). The homologies as deduced from the cDNA sequence data are extensive: between human and mouse there is >85% identity at the nucleotide level, and between mouse and rat the identity is >90% at the nucleotide level.
ADAR1 promoters and transcriptional regulation
ADAR1 transcript levels are increased by IFN treatment and by pathogen infection as measured by either northern hybridization or reverse transcription–polymerase chain reaction. Hybridization probes derived from exons 2 to 15 of the human cDNA clone detect a single major transcript of ∼6.7 kb, the steady-state level of which is increased by IFN treatment (Patterson and others 1995; Patterson and Samuel 1995; George and Samuel 1999b). A similarly sized RNA is seen in the absence of IFN treatment. When exon 1A- or exon 1B-specific hybridization probes are used, both detect an RNA of about 6.7 kb. However, the transcript detected in human cells with an exon 1A-specific probe is IFN-inducible, whereas the abundant transcript detected with the exon 1B-specific probe is not inducible (George and Samuel 1999a).
The organization of the multiple promoters of the human ADAR1 gene (George and Samuel 1999a, 1999b) is shown in Fig. 2B. One promoter is IFN inducible. The others are constitutively active. The IFN-inducible promoter (PIA) is TATA-less and possesses a consensus 12-bp IFN-stimulated response element (ISRE) (Fig. 2C) characteristic of type I IFN-regulated genes (Stark and others 1998). The PIA promoter drives expression of the exon 1A-containing ADAR1 transcripts (George and Samuel 1999a, 1999b; George and others 2005). The region immediately upstream of the ISRE of the inducible promoter is designated as a kinase conserved sequence (KCS)-like element (Ward and others 2002; Markle and others 2003) because of homology with the KCS identified in the IFN-inducible human and mouse protein kinase regulated by RNA (PKR) gene promoters (Kuhen and Samuel 1997) (Fig. 2C). Mouse Adar1 gene expression, like human ADAR1 gene expression, is driven by alternative promoters (Shtrichman and others 2002; George and others 2005). The 12-bp ISREs of the human and mouse inducible promoters differ in only one nucleotide position (Fig. 2C).
According to the canonical JAK-STAT signaling pathway for transcriptional activation of gene expression by type I IFN, following binding of IFN-α/β to the interferon alpha receptor (IFNAR), the TYK2 and JAK1 kinases mediate activation of STAT1 and STAT2 factors that, together with IRF9, form the heterotrimeric ISGF3 protein complex that binds at the ISRE to enhance transcription (Stark and others 1998). Induction by IFN of the p150 ADAR1 protein is impaired as expected in mouse embryo fibroblasts (MEFs) genetically deficient in either the JAK1 kinase or STAT2 factor, but surprisingly not in MEFs deficient in STAT1 (George and others 2008). ADAR1 p150 induction also is independent of Sp3 (George and others 2008), a component of the KCS binding complex found at the PKR promoter (Das and others 2006). The STAT1-independent, STAT2-dependent property of induction by IFN seen for the ADAR1 p150 form is intriguing. A second nucleic acid editing enzyme, APOBEC3G, which catalyzes C-to-U deamination of DNA, is likewise reportedly induced by type I IFN in a STAT1-independent, STAT2-dependent manner (Sarkis and others 2006). The ADAR1 gene promoters that drive constitutive expression of exon 1B- and 1C-containing transcripts lack ISREs and are not inducible by IFN (George and Samuel 1999a; Kawakubo and Samuel 2000; George and others 2008). While the gene organization involving multiple promoters and alternative exon 1 structures is preserved between the human and mouse ADAR1 genes, the actual sequences of the promoters and alternative exon 1 structures are not well conserved between species outside of the ISRE and KCS-like elements of the inducible promoter (George and Samuel 1999a, 1999b; George and others 2005, 2008).
The ADAR1 gene is ubiquitously expressed. However, tissue-selective expression of the exon 1A- and 1B-containing transcripts is observed in mouse organs (Shtrichman and others 2002; George and others 2005). Mouse Adar1 cDNA probes hybridize to a major IFN-inducible mRNA of ∼6.5 kb, comparable in size to the human transcript. The ∼6.5 kb RNA is abundantly expressed in the liver of mice following oral infection with Salmonella and in cultured mouse fibroblasts treated with IFN. Transcripts possessing 7b, the short form of exon 7 that is found in exon 1A-containing transcripts, are apparently most abundantly expressed in response to inflammation or IFN treatment (Yang and others 2003a, 2003b; George and others 2005). Exon 1A-containing transcripts are detected in mouse organs such as liver, spleen, Peyer's patches, kidney, heart, lung, and cecum, and are increased following Salmonella infection. However, in the brain, only very low levels of the exon 1A-containing transcripts are detectable and no increase is seen after infection. By contrast, exon 1B-containing transcripts while found in all tissues are highest in the brain and are not increased after infection (Shtrichman and others 2002; George and others 2005). Analysis of the spatiotemporal expression patterns for ADAR1 and ADAR2 in the mouse forebrain shows that both proteins are broadly distributed in most regions of the forebrain by P0, with high expression levels maintained in adulthood (Jacobs and others 2009).
ADAR2 gene
Chromosome assignment of ADAR2
The human ADAR2 gene maps through a polymerase chain reaction-based strategy to chromosome 21 band q22.3 (Mittaz and others 1997), a region that has been implicated in several genetic disorders.
ADAR2 exon–intron organization
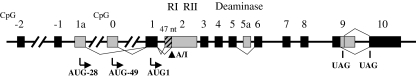
The human ADAR2 gene spans ∼150 kb. While human and mouse ADAR2 genes have generally similar exon/intron organizations (Slavov and Gardiner 2002), the complexity of their exon structures has not been appreciated until recently. Both the human and mouse ADAR2 genes are now known to possess 15 exons (Slavov and Gardiner 2002; Maas and Gommans 2009) as summarized in Fig. 3. Two exons, denoted as exons -2 and -1, specify the 5′-UTR. Exons 0–9 represent the ORF that encodes ADAR2. Similar to ADAR1, various ADAR2 transcript isoforms exist as a result of multiple alternative splicing events (Gerber and others 1997; Lai and others 1997; Mittaz and others 1997; Slavov and Gardiner 2002; Kawahara and others 2005; Maas and Gommans 2009).
FIG. 3.
Exon–intron organization of the human ADAR2 gene. Organization of introns and exons of the ADAR2 gene that spans ∼150 kb on human chromosome 21q22. Exons are numbered from −2 to 10 (according to Slavov and Gardiner 2002; and Maas and Gommans 2009) and represented by the filled boxes; introns and the 5′- and 3′-flanking regions are shown by the solid lines. The shaded boxes indicate alternatively utilized exons or exons containing alternative splice sites. Autoediting by ADAR2 within intron 1 creates a 3′ splice site (from AA to AI) that leads to an insertion of 47 nucleotides (nt). CpG, CpG islands present in the putative promoter regions.
Comparison of human and mouse genomic sequences with the 5′-terminal sequences of ADAR2 cDNAs identifies 3 exons, denoted exon 1a, 0, and 1 for human and exon 1b, 0, and 1 for mouse, respectively (Slavov and Gardiner 2002; Maas and Gommans 2009). Each of these exons contains a potential AUG codon for initiation of ADAR2 translation. For human ADAR2, inclusion of exon 1a between exons −1 and 1 results in an N-terminal extension of 28 amino acids that includes 7 Arg (R)/Lys (K) residues. By contrast, in mouse ADAR2 the inclusion of exon 1b between exons 1 and 2 causes a frame shift that disrupts the original ORF; use of the next AUG codon within exon 1b leads to the replacement of the original negatively charged N-terminal sequence MDIEDEENS with MPLG. Therefore, for both human and mouse ADAR2, use of an alternative exon 1 can produce ADAR2 protein isoforms that possess altered charge properties in their N-terminal regions (Slavov and Gardiner 2002). In all brain regions and tissues tested, ∼40% of human ADAR2 transcripts are found to contain exon 1a; however, exon 1b of mouse Adar2 is transcriptionally utilized at a much lower frequency (Slavov and Gardiner 2002). In addition, the AUG-28 codon in human exon 1a (or mouse exon 1b) does not occur in the −3/+4 context of a consensus Kozak sequence (A/GCCatgG) (Kozak 1989), raising the possibility that these transcripts are poorly translated. Recently, an additional exon designated as exon 0 positioned between exon −1 and exon 1 has been characterized for both human and mouse ADAR2 (Maas and Gommans 2009). Inclusion of exon 0 with initiation at AUG-49 results in the addition of 49 amino acids N-terminal of the previously identified initiation methionine (AUG1) in exon 1; the added sequence exhibits a high similarity to the Arg-rich domain present in the N-terminal region of ADAR3. However, selective use of exon 0 also occurs at a very low frequency, with the highest levels observed in <10% of total human ADAR2 transcripts in the hippocampus (Maas and Gommans 2009). Neither the relative protein abundance nor the functional properties of the ADAR2 isoforms possessing the altered N-terminal residues have been determined.
ADAR2 isoforms are also produced as a result of alternative splicing in the coding exon 5. Human ADAR2 possesses exon 5 and 5a, with exon 5a encoding an in-frame Alu-J cassette in the catalytic domain, inclusion of which appears to reduce its enzyme activity in vitro (Gerber and others 1997). In mouse Adar2, however, an alternative splicing event within intron 5 extends exon 5 by 30 nt, generating an ADAR2 protein isoform with relatively higher editing activity for GluR-B RNA in vitro (Rueter and others 1999).
Alternative splicing events between exons 9 and 10 of both human and mouse ADAR2 contribute to producing multiple isoforms with differing C-terminal sequences or 3′-UTR regions. In human ADAR2, use of an alternative splice site within exon 9 leads to replacement of the C-terminal 29 residues by 2 amino acids encoded by exon 10, which generates an inactive protein for editing the GluR-B pre-mRNA (Lai and others 1997). A recent characterization of human brain ADAR2 transcripts in different developmental stages (fetal, neonatal, and adult) reveals 4 additional C-terminal variants resulting from alternative splicing that leads to the inclusion of intron 9 as well as an alternative splice site in exon 9 at position 83 nt downstream of the stop codon (Kawahara and others 2005). Mouse Adar2 likewise contains 2 forms of exon 9 and exon 10 denoted as short (s) and long (l). Out of the 4 possible splicing combinations, however, only 2 variant forms, 9s + 10l and 9l + 10s, are detectable (Slavov and Gardiner 2002). Despite the lack of experimental evidence, it is tempting to speculate that the variations within the 3′ UTR may affect the stability, transport, or translational efficiency of ADAR2 mRNA.
The most unique alternative splicing event known thus far for ADAR2 is the one mediated by A-to-I autoediting to create a new 3′-splice acceptor site (AA to AI) in the intron 1 sequence of ADAR2 (Rueter and others 1999). This editing-dependent splicing results in the addition of 47 nts to the 5′-end of exon 2 of ADAR2, causing a frame-shift in the coding region that is predicted to generate a 9-kDa protein due to premature translational termination. Further, studies of knockin mice suggest that this autoediting event may act as a negative feedback mechanism for modulating ADAR2 expression (Feng and others 2006).
Thus, similar to ADAR1, the exceptional diversity of ADAR2 transcripts and, consequently, ADAR2 protein isoforms presumably reflect the physiological requirement for regulation of the functional activities of ADAR2 enzyme.
ADAR2 transcriptional regulation
In contrast to the occurrence of multiple promoters that drive expression of ADAR1, the promoter regions that direct ADAR2 transcription have not been fully characterized. Although a putative promoter region has been described upstream of exon 0 (Maas and Gommans 2009), it has yet to be established whether multiple promoters are utilized together with alternative splicing to produce exon 1a (AUG-28), exon 0 (AUG-49), or exon 1 (AUG1) containing transcripts to control expression of ADAR2.
Similar to ADAR1, ADAR2 is ubiquitously expressed in most tissues but most abundantly in the brain (Melcher and others 1996b; Gan and others 2006). Northern blot analyses have revealed multiple ADAR2 mRNA species in a broad size range from ∼1.9 to ∼8.8 kb, indicating the multiplicity of ADAR2 transcripts (Gerber and others 1997; Lai and others 1997; Mittaz and others 1997; Kawahara and others 2005). The human ADAR2 transcript of 8.5–8.8 kb appears to be the predominant RNA form in the brain. ADAR2 displays distinct spatio expression patterns in the adult rat brain as shown by in situ hybridization analysis, with the highest expression level found in the thalamic nuclei (Melcher and others 1996b). During embryonic development, ADAR2 mRNA can be detected as early as embryonic day E19, thereafter increasing gradually in several brain regions (Paupard and others 2000; Hang and others 2008; Jacobs and others 2009). This is in agreement with the finding that RNA editing by ADAR2 increases progressively during brain development (Lomeli and others 1994). However, the molecular details with respect to how ADAR2 transcription is regulated in specific neurons or at different developmental stages remain poorly understood.
Significant levels of transcription of both Adar1 and Adar2 also occur in murine pancreatic islets (Gan and others 2006). Adar2, but not Adar1, transcription is selectively upregulated in islets of obese mice induced by feeding a high-fat diet. Further, in rat insulinoma insulin-secreting INS-1 β-cell cultures, glucose can stimulate Adar2 transcription, and thus ADAR2-mediated RNA editing (Gan and others 2006). The detailed mechanism(s) by which Adar2 transcription is metabolically regulated in pancreatic β-cells is yet to be elucidated. It remains unclear what regulatory elements may be present in the Adar2 promoter that are important in sensing metabolic stimuli such as glucose.
RNA Adenosine Deaminase (ADAR) Proteins and Their Biochemical Activities
ADAR1 proteins
Size isoforms of ADAR1 proteins
The IFN-inducible ADAR1 protein (p150) predicted from the ORF is 1,200 (human) or 1,152 (mouse) amino acids, whereas the constitutively expressed protein (p110) is predicted to be 931 (human) or 903 (mouse) amino acids (Patterson and Samuel 1995; George and Samuel 1999a; George and others 2005). The sizes of the ADAR1 proteins, deduced from cDNA sequence ORFs, are somewhat smaller than the sizes estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Patterson and Samuel 1995; George and others 2005): ∼133 kDa for the human p150 protein, and ∼103 kDa for the human p110 protein (Patterson and Samuel 1995). ADAR1 also has been known variously as dsRAD, DRADA, or K88 in the earlier literature (Patterson and Samuel 1995; Bass and others 1997). Of the two size forms of ADAR1, p150 is sometimes referred to as the long form and p110 as the short form.
Functional domains of ADAR1 proteins
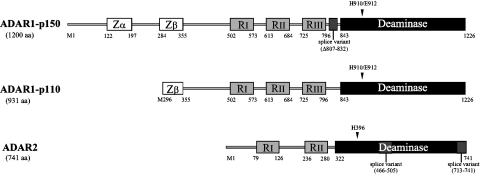
The IFN-inducible p150 ADAR1 protein is an N-terminally extended form of the p110 constitutive protein and includes within the N-terminal region two copies of a Z-DNA binding motif (Zα and Zβ). Both p150 and p110 ADAR1 are active deaminases, and both possess the catalytic domain in the C-terminal region and three copies of the dsRNA-binding motif (RI, RII, and RIII) in the central region of the proteins. The location of the catalytic, RNA-binding and Z-DNA-binding domains of ADAR1 are shown schematically in Fig. 4. Biochemical and mutational studies have established the functional importance of each of these domains, as well as sequences important for sumoylation, nuclear localization, and nuclear export of ADAR1.
FIG. 4.
Domain organization of human ADAR1 and ADAR2 proteins. The 2 size forms of ADAR1, the IFN-inducible (p150, long form) and constitutively expressed (p110, short form), and the one known size form of ADAR2, are illustrated. The numbers immediately below each schematic refer to the amino acid (aa) position of each domain or splice variant. Arrowheads identify specific amino acid positions that, when mutated, impair function of the catalytic domain. For ADAR1, the 2 Z-DNA-binding domains (Zα and Zβ), 3 dsRNA-binding domains (RI, RII, and RIII), and the deaminase catalytic domain are shown; an alternatively spliced exon 7 is 26 amino acids shorter in p150 ADAR1. For ADAR2, the 2 dsRNA-binding domains, the catalytic domain, and splice site variants are shown. Additional splice variants are known for both ADAR1 and ADAR2 (adapted from Toth and others 2006, with permission).
The C-terminal ∼380 amino acids of ADAR1 that specifies the catalytic domain was initially recognized to have extensive homology with proteins known to possess cytosine deaminase activity (Kim and others 1994b; Lai and others 1995; O'Connell and others 1995; Patterson and Samuel 1995; Maas and others 1996; Melcher and others 1996b). His910 and Glu912 of ADAR1 are key amino acids of the CHAE motif of the catalytic core characteristic of deaminases. Substitution of His910 with Gln, and Glu912 with Ala, completely abolishes A-to-I editing activity (Lai and others 1995; Liu and Samuel 1996). Atomic level structural studies of other deaminase proteins, including ADAR2 (Macbeth and others 2005), established that the His of the CHAE motif is involved in binding of zinc and that Gln is important in proton transfer. Further, Cys966 and Cys1036, which are believed to play a role in zinc coordination along with His910, are necessary for ADAR1 deaminase activity (Lai and others 1995). O-phenanthroline, a chelator of zinc ions, inhibits the enzymatic activity of purified natural ADAR1 (Kim and others 1994a). The catalytically active form of ADAR1 is a dimer (Cho and others 2003; Gallo and others 2003; Valente and Nishikura 2007). Fluorescence resonance energy transfer (FRET) and immunoprecipitation analyses have further demonstrated that both ADAR1 and ADAR2 form homodimers in human cells, that ADAR1 and ADAR2 may also form heterodimers, and that the dimerization likely is RNA-independent (Chilibeck and others 2006; Cenci and others 2008). Finally, while the crystal structure of the catalytic domain of human ADAR2 revealed inositol hexakisphosphate bound in the ADAR2 catalytic core and required for RNA editing (Macbeth and others 2005), there is no evidence for a similar cofactor requirement for ADAR1.
Three copies of the dsRNA-binding domain (dsRBD; designated as RI, RII, and RIII in Fig. 4) present in both p150 and p110 ADAR1 are homologous to the dsRBD repeats first discovered in the RNA-dependent protein kinase PKR (McCormack and others 1992; Kim and others 1994b; O'Connell and others 1995; Patterson and Samuel 1995; Liu and Samuel 1996). The RI, RII, and RIII dsRBDs are located in exons 3, 5, and 7, respectively, of human ADAR1 (Fig. 2). Mutagenesis of a conserved Lys present in the core sequence of each of the dsRBD domains (Lys554, Lys665, and Lys777) provided a test of their relative importance, both for RNA binding and for enzymatic activity (McCormack and others 1994; Liu and Samuel 1996; Fierro-Monti and Mathews 2000). The RI and RIII domains are the most important, and RII less so, for binding to synthetic dsRNA (Liu and Samuel 1996; Liu and others 1997). RIII emerges as the most important of the three dsRBDs, and RII the least important, for A-to-I editing activity, more or less correlating with their importance in dsRNA binding (Liu and Samuel 1996; George and Samuel 1999b). The RIII single mutant lacks detectable enzymatic activity, as do all of the double mutants and the triple RI, RII, RIII Lys substitution mutant (Liu and Samuel 1996). Similar conclusions were drawn from studies with mutants where dsRBDs are deleted (Lai and others 1995). Finally, when the three dsRBDs of ADAR1 are substituted with the two from PKR, appreciable editing activity is retained with the chimeric PKR:ADAR1 protein when measured with a synthetic dsRNA substrate, but not with natural dsRNA substrates (Liu and others 2000).
The additional N-terminal sequence present in the inducible p150 protein, but lacking in p110, originates because of the alternative exon 1 structures and different translation initiation sites (Fig. 4). The N-terminus of p150 ADAR1 includes two copies of a Z-DNA-binding domain (Herbert and others 1997), designated as Zα and Zβ, that was first identified as a region with homology to the poxvirus E3L protein (Patterson and Samuel 1995; Kim and others 2003). Zα and Zβ are encoded by exon 2 (Fig. 2A). The p110 protein possesses only Zβ. While the Zα domain is sufficient to bind Z-DNA, the Zβ domain does not bind Z-DNA as a separate entity (Schwartz and others 1999). The physiologic function of the Z-DNA binding by p150 has not been clearly defined. Although mutation of the Z-DNA-binding domains decreases the efficiency of ADAR1 editing of short (15-bp) dsRNA substrates (Herbert and Rich 2001), the Zα and Zβ domains together are not an obligate requirement either for dsRNA binding or for deaminase activity because ADAR1 p110 possesses only Zβ and ADAR2 has neither Zα or Zβ (Fig. 4). For the poxvirus E3L IFN-resistance protein, the Z-DNA-binding domain plays a role in viral pathogenesis in the mouse model, but again the biochemical basis of the observed biologic effect is not yet elucidated (Kim and others 2003). The A form of dsRNA with purine-pyrimidine repeats can be transformed to a left-handed helix Z-RNA, and the Zα domain of ADAR1 can bind Z-RNA (Placido and others 2007).
Finally, human ADAR1 is sumoylated at Lys418 (Fig. 4), a modification that alters ADAR1 A-to-I editing activity (Desterro and others 2005). Modification of proteins by the small ubiquitin-like modifier (SUMO) has emerged as an important regulatory mechanism for localization and function of proteins (Yeh 2009). Modification of ADAR1 by SUMO-1 reduces editing activity in vitro. The mutation Lys418Arg, which abolishes SUMO-1 conjugation, stimulates ADAR1 editing activity both in vivo and in vitro (Desterro and others 2005).
Subcellular localization of ADAR1
Immunofluorescence microscopy and biochemical fractionation approaches initially established that the p110 form of ADAR1 localizes predominantly, if not exclusively, to the nucleus, whereas the IFN-inducible p150 protein is found both in the cytoplasm and the nucleus (Patterson and Samuel 1995).
Nuclear localization signal (NLS) and nuclear export signal (NES) sequences have been identified in ADAR1. One NLS is present within the RIII copy of the dsRBD of human ADAR1 (Eckmann and others 2001; Strehblow and others 2002); however, nuclear localization does not require dsRNA binding (Eckmann and others 2001). Rather, RNA binding involving RI and RIII may conversely impede activity of the NLS positioned in the RIII dsRBD (Strehblow and others 2002). ADAR1 localizes to the nucleolus (Patterson and Samuel 1995; Desterro and others 2003; Yang and others 2003b). Further, the p150 form of ADAR1 shuttles between the cytoplasm and the nucleus (Eckmann and others 2001; Poulsen and others 2001). Inhibition of the nuclear export protein Crm1 by the pharmacologic agent leptomycin B (LMB) stimulates nuclear accumulation of ADAR1. Nuclear export of p150 is mediated by a Rev-like NES present in the Zα domain, a region of ADAR1 that interacts with Crm1 and RanGTP (Poulsen and others 2001). Transportin-1 is a nuclear import factor for ADAR1, and dsRNA binding modulates the interaction of transportin-1 and exportin-5 with the dsRBD to regulate nucleocytoplasmic shuttling of ADAR1 (Fritz and others 2009).
RNA substrates edited by ADAR1
A-to-I RNA editing occurs with both viral and cellular mRNAs. ADAR1 modifying activity is implicated in two types of processes. First, the deamination can be site-selective and occur at one or a few sites. Second, the editing can occur at multiple sites when RNA substrates possess high duplex character.
Site-selective A-to-I editing is exemplified by the editing of viral RNAs, including the hepatitis delta virus (HDV) antigenome RNA (Jayan and Casey 2002a) and the human herpes virus 8 (HHV8) kaposin K12 transcript RNA (Gandy and others 2007), and editing of cellular mRNA transcripts in the nervous system that encode neurotransmitter receptors for l-glutamate (GluR) and serotonin (5-HT2cR) (Higuchi and others 1993; Burns and others 1997; Liu and others 1999; Liu and Samuel 1999; Seeburg and Hartner 2003). In these cases, the high positional selectivity of the A-to-I RNA editing events leads to the synthesis of specific protein products with altered function. In the edited HDV, HHV8, GluR, and 5-HT2cR RNAs, the I generated by editing is recognized as G instead of A by the translational machinery, thereby producing selective amino acid substitutions (Bass 2002).
A-to-I editing of HDV RNA represents a well-characterized example of viral RNA editing. Editing of HDV RNA occurs on the highly structured antigenome at a site designated as “amber/W,” thereby changing an UAG amber stop codon to an UIG tryptophan (W) codon that permits synthesis of the large delta antigen HDAg-L (Luo and others 1990). Constitutively expressed p110 form of ADAR1, specifically the exon 7a splice variant, is the ADAR believed to be primarily responsible for editing the amber/W site (Jayan and Casey 2002b; Wong and Lazinski 2002).
In the cases of neurotransmitter receptor transcript substrates, highly selective A-to-I editing events are specified by imperfect duplex RNA structures formed between adjacent exonic and intronic sequences. The editing causes selective amino acid substitutions that generate receptor proteins with altered properties (Seeburg and Hartner 2003). The pre-mRNA substrate encoding the B subunit of the glutamate receptor (GluR-B) has two functionally important sites, designated as Q/R and R/G. The Q/R site occurs in exon 11, where a genome encoded glutamine (Q) codon (CAG) becomes an arginine (R) codon (CIG) following A-to-I editing. R/G editing involves the conversion of an arginine codon (AGA) to a glycine (G) codon (IGA) in exon 13. In addition, multiple sites of A-to-I editing are seen in intron 11, termed hotspots, that include +60. Both ADAR1 and ADAR2 edit the R/G site of GluR-B; by contrast, Q/R site editing is carried out primarily if not exclusively by ADAR2 (Melcher and others 1996a; Liu and Samuel 1999; Higuchi and others 2000; Hartner and others 2004; Wang and others 2004). The +60 intron hotspot is edited by ADAR1 (Liu and Samuel 1999). Exon 3 of 5-HT2CR pre-mRNA transcripts possesses 5 editing sites dictated by an imperfect inverted repeat sequence present in the adjacent intron 3 (Burns and others 1997; Fitzgerald and others 1999; Liu and others 1999; Niswender and others 1999; Wang and others 2000b). Editing of all 5 sites in 5-HT2CR pre-mRNA leads to 3 amino acid substitutions in exon 3 that specifies the second extracellular loop of the 7-transmembrane receptor. Biochemical and subsequent genetic analyses established that ADAR1 and ADAR2 both are necessary to achieve full editing (Burns and others 1997; Liu and others 1999; Niswender and others 1999; Higuchi and others 2000; Wang and others 2000b; Hartner and others 2004).
A-to-I editing of substrates such as GluR-B and 5-HT2CR RNAs must occur before splicing, and hence is nuclear. Both ADAR1 and ADAR2 proteins are found associated with spliceosomal Sm and SR proteins within 200S lnRNP complexes that constitute the natural pre-mRNA processing machinery (Raitskin and others 2001). Further, ADAR1 and the human Upf1 protein involved in RNA surveillance are found associated within nuclear RNA-splicing complexes. RNAi knockdown of ADAR1 is accompanied by upregulation of a number of genes previously shown to undergo A-to-I editing in Alu repeats and downregulated by human Upf1 (Agranat and others 2008). Analysis of a set of selectively edited substrates by large-scale 454 sequencing suggests that recognition and coupling of editing sites are determined by the tertiary structure of the RNA (Ensterö and others 2009). When the target preferences of 3 novel coding RNAs and 2 SINES were examined using embryos from either Adar1 or Adar2 knockout mice, some sites not unexpectedly were found to be edited selectively by ADAR1 and others by ADAR2 (Riedmann and others 2008). ADARs act on duplex RNA without obvious sequence-specific binding, but with a 3′-neighbor preference of G or a purine and, with the trinucleotide, UAG is favored (Lehmann and Bass 2000; Kim and others 2004; Riedmann and others 2008).
In contrast to the HDV, GluR-B, and 5HT-2CR examples of highly selective A-to-I editing, adenosine deamination by ADAR1 can occur at multiple sites when the RNA substrate possesses extensive duplex character. Indeed, dsRNA-specific adenosine deaminase activity was first described as a dsRNA duplex-unwinding activity in Xenopus oocytes during antisense RNA studies (Bass and Weintraub 1987; Rebagliati and Melton 1987). However, now it is recognized that, rather than unwinding of dsRNA duplexes, it is a destabilization of the edited dsRNA that results when more stable A:U base pairs become less stable I:U base pairs (Bass and Weintraub 1988; Wagner and others 1989). A-to-I editing of viral RNA genomes during certain lytic and persistent infections apparently can occur at multiple sites. Such hyperediting of viral RNAs has been implicated during replication and subsequent persistent infection with single-stranded RNA viruses, including measles virus, where biased A-to-I (G) hypermutations were first described (Cattaneo and others 1988), hepatitis C virus (Taylor and others 2005), and lymphocytic choriomeningitis virus (Zahn and others 2007), and also during infection with the dsDNA virus, mouse polyoma virus (Kumar and Carmichael 1997).
Possibly, the shuttling of ADAR1 between the cytoplasm and nucleus may impact in a temporal and spatial manner in which dsRNA substrates are edited. For example, editing of GluR-B RNA at the R/G site is efficiently catalyzed by p150 NES mutants that accumulate in the nucleus, whereas GluR-B RNA is poorly edited by wild-type ADAR1 localized in the cytoplasm (Poulsen and others 2001). Perhaps in the nucleus ADAR1 functions to edit pre-mRNAs before splicing, whereas in the cytoplasm ADAR1 may function to edit substrates such as viral RNAs or microRNAs (miRNAs) (Bass 2002; Seeburg and Hartner 2003; Valente and Nishikura 2005; Toth and others 2006). In some instances the function of ADAR1 may involve interactions with protein or RNA independent of, or in addition to, its RNA deaminase catalytic activity. For example, ADAR1 increases plasmid-based gene expression at the translational level by decreasing PKR-dependent eIF-2α phosphorylation independent of editing activity (Gommans and Maas 2008; Wang and Samuel 2009). In HIV-infected lymphocytes, ADAR1 interacts with PKR and enhances viral replication without the requirement for editing activity (Clerzius and others 2009; Doria and others 2009). ADAR1 also interacts with nuclear factor 90 (NF90) proteins and affects NF90-mediated gene expression independently of editing activity (Nie and others 2005); ADAR1 is found in complexes that include Vigilin that has a high affinity for I-containing RNA, RNA helicase A, and Ku86/70 (Wang and others 2005); finally, I-containing dsRNA binds a stress granule-like complex and downregulates expression of both reporter and endogenous genes (Scadden 2007).
Antagonists of ADAR1 activity
Two viral gene products, one RNA and the other a protein, have been shown to antagonize ADAR1. Adenovirus VAI RNA and the vaccinia virus E3L protein both antagonize ADAR1 deaminase activity, at least in vitro (Lei and others 1998; Liu and others 2001; George and others 2009). VAI RNA and E3L protein both mediate IFN resistance in the context of viral infections; VAI and E3L were the first antagonists identified of the IFN-inducible RNA-dependent protein kinase PKR (Kitajewski and others 1986; Chang and others 1992; Samuel 2001; George and others 2009).
VAI RNA, a product of RNA polymerase III and a highly structured small RNA, antagonizes PKR activity by binding to the dsRBDs and preventing subsequent autophosphorylation and kinase activation and eIF-2α phosphorylation (Kitajewski and others 1986; O'Malley and others 1986; Mathews and Shenk 1991; McCormack and Samuel 1995). VAI RNA inhibits the deaminase activity of both p110 and p150 ADAR1 measured with synthetic dsRNA in vitro (Lei and others 1998). Mutant adenovirus that does not express VAI RNA replicates poorly, and surprisingly the stable knockdown of PKR does not rescue the growth of VAI mutant virus (Zhang and Samuel 2007), consistent with the notion that VAI RNA has an additional physiologically important target such as ADAR1. The mechanism by which VAI RNA antagonizes ADAR1 is not yet known, nor is the biological significance in the context of adenovirus infection. However, VAI RNA has been used to assess the role of ADAR1 in hepatitis C virus (HCV) RNA replication. IFN-alpha treatment of HCV RNA replicon-containing cells causes an inhibition of HCV viral RNA. Inhibition of ADAR1 by VAI RNA or siRNA knockdown stimulates replicon activity and reduces the inosine detected in HCV replicon RNA (Taylor and others 2005).
Vaccinia virus E3L is a 190 amino acid protein expressed at early times after infection. E3L is an established antagonist of two IFN-inducible enzymes: the PKR kinase (Samuel 1979; Chang and others 1992) and OAS, the RNA-dependent oligoadenylate synthetases that produce 2′5′-oligoadenylate activators of RNase L (Rivas and others 1998; Rebouillat and Hovanessian 1999). Similar to p150 ADAR1, the poxvirus E3L protein also binds both Z-DNA and dsRNA and localizes to the cytoplasm and the nucleus (Chang and Jacobs 1993; Yuwen and others 1993; Patterson and Samuel 1995; Herbert and others 1997; Liu and others 1998). E3L inhibits deamination of synthetic dsRNA in vitro by both size forms of ADAR1, p150 and p110 (Liu and others 2001). Although sequestration of dsRNA would represent one possible and obvious mechanism for the inhibition of ADAR1 by E3L, neither reovirus σ3 protein nor some E3L mutants that retain dsRNA-binding activity inhibit ADAR1 activity, suggesting that the RNA binding activity alone of E3L is not sufficient for ADAR1 antagonism. No physical interaction between E3L and ADAR1 has yet been reported. Mutant vaccinia virus deleted for E3L replicates poorly in human cells. The stable knockdown of PKR complements to a significant extent, but albeit not completely, the E3L deletion mutant phenotype by restoration of both viral protein synthesis and virus growth and reduction of apoptosis (Zhang and others 2008). These results suggest that PKR, rather than ADAR1, is a primary E3L target in infected HeLa cells in culture (Zhang and others 2008).
ADAR2 proteins
Functional domains of ADAR2 proteins
Like ADAR1, ADAR2 possesses multiple copies of the highly conserved dsRNA-binding domain (dsRBD), but ADAR2 has two copies (RI and RII) of the dsRBD instead of three as found in ADAR1 proteins. While having a C-terminal catalytic deaminase domain (Bass 2002; Maas and others 2003) like ADAR1, ADAR2 does not contain a Z-DNA-binding domain as found in ADAR1. While the RII is essential for editing of most naturally occurring RNA substrates, RI shows substrate-dependent importance for its editing activity (Xu and others 2006), suggesting distinct substrate-binding properties of these 2 dsRBDs. Curiously, deletion of the N-terminal RI dsRBD of the human ADAR2 retains its editing activity in vitro on a short 15-bp substrate RNA harboring the GluR-B R/G site (Macbeth and others 2004). The N-terminal portion containing RI can act as an inhibitor in trans for the N-terminally truncated ADAR2 protein. This implies a possible regulatory role for RI of ADAR2 in recognizing its endogenous cellular RNA targets. Structural analyses have provided important insights into the functional features of the dsRBDs in substrate recognition and catalysis. Studies using directed hydroxyl radical cleavage data demonstrated that both RI and RII of human ADAR2 exhibit similar selective binding at locations adjacent to the GluR-B Q/R site of a 30-bp duplex RNA substrate, whereas different binding selectivity is found for the dsRBD of PKR (Stephens and others 2004). According to the NMR solution structures, the two dsRBDs of rat ADAR2 with 50% amino acid identity possess a common αβββα topology, differing slightly in the orientation of α-helix 1 and the conformation of the β1 − β2 loop (Stefl and others 2006). NMR chemical shift perturbation analysis of the two ADAR2 dsRBDs interacting with a 71-nt GluR-B RNA harboring the R/G site suggests that the RI recognizes a conserved penta-loop, while the RII recognizes two bulged bases adjacent to the editing site (Stefl and others 2006). The linker region bridging the two dsRBDs of ADAR2 is unstructured and does not appear to be involved in the interaction with the RNA stem-loop.
The crystal structure of the catalytic domain of human ADAR2 has been determined at 1.7 Å resolution (Macbeth and others 2005). A structurally conserved deamination motif is formed by 77 residues, consisting of 2 α-helices, 4 β-strands, and connecting loops. The deaminase catalytic domain contains 1 zinc ion and, unexpectedly, one molecule of inositol hexakisphosphate (IP6) buried in a cavity lined with several Arg and Lys residues. Whether IP6 is directly involved in deamination catalysis remains to be delineated, though IP6 is a required cofactor for enzymatic activity.
Subcellular localization of ADAR2
ADAR2 is localized exclusively in the nucleus, primarily residing in nucleoli (Desterro and others 2003; Sansam and others 2003). In addition to an NLS in the N-terminal domain of ADAR2 that is responsible for its nuclear localization, several lines of evidence indicate that the dsRBDs are also crucial for the translocation, if not retention, of ADAR2 to the nucleoplasm, likely through their ability to bind rRNA (Sansam and others 2003; Xu and others 2006). Photobleaching experiments in live cells indicate that both ADAR2 and the p110 form of ADAR1 constantly shuttle in and out of the nucleolus (Desterro and others 2003; Sansam and others 2003). In cells expressing the substrate GluR-B RNA, ADAR2 can preferentially localize to the nucleoplasm where the substrate transcripts are located (Desterro and others 2003); enhanced translocation of ADAR2 from the nucleolus to the nucleoplasm is associated with increased editing of endogenous RNA substrates (Sansam and others 2003). Therefore, the subcellular mobilization of ADAR2 may serve as a mechanism for modulating its editing activity. Similar to ADAR1, the ADAR2 protein is found to associate with the spliceosome complex (Raitskin and others 2001), likely facilitating its anchorage in the nucleoplasm and providing the cellular compartmentalization for its editing function.
RNA substrates edited by ADAR2
ADAR2 and ADAR1 can catalyze the site-specific editing of a number of common RNA substrates, including the aforementioned ionotropic glutamate receptors and G-protein-coupled serotonin-2C receptor (Sommer and others 1991; Lomeli and others 1994; Melcher and others 1996b; Burns and others 1997; Lai and others 1997; Seeburg and others 1998; Liu and others 1999; Liu and Samuel 1999; Aruscavage and Bass 2000; Bhalla and others 2004). Comparative genomics analysis also has identified the mRNA encoding human voltage-dependent K+ channel Kv1.1 (Hoopengardner and others 2003) that undergoes efficient editing by ADAR2, replacing an isoleucine (ATT) residue with valine (ITT) in the highly conserved ion-conducting pore-lining S6 domain and thereby controlling the channel inactivation (Bhalla and others 2004). Kv1.1 editing requires a hairpin structure formed within the mRNA coding region instead of an exonic complementary sequence within the adjacent intron to form a stem loop structure (Bhalla and others 2004). This is in contrast to the cases of GluR-B (Higuchi and others 1993), ADAR2 (Dawson and others 2004), and 5-HT2cR (Burns and others 1997) pre-mRNA substrates that contain exonic complementary sequence. Other newly identified mammalian substrates targeted for editing by ADARs in the coding regions include FLNA, BLCAP, CYFIP2, and IGFBP7 (Levanon and others 2005). Both ADAR1 and ADAR2 can edit BLCAP (Galeano and others 2009), whereas ADAR2 predominantly edits the K/E site of CYFIP2 RNA (Kwak and others 2008) and FLNA may be an ADAR2 substrate as indicated by the fact that FLNA mRNAs coimmunoprecipitate with ADAR2 (Kwak and others 2008). However, the functional consequences of these editing events have not been demonstrated.
Among the endogenous cellular RNA transcripts edited by ADAR2 is its own pre-mRNA, creating a 3′ splice site (AA to AI) for the subsequent alternative splicing (see Fig. 3) near the N-terminal portion (Rueter and others 1999). Viral RNA species that are targeted for A-to-I modifications by both ADAR1 and ADAR2 include the site-selective editing at the amber/W site of HDV RNA (Sato and others 2001; Jayan and Casey 2002a). However, the p110 form of ADAR1 appears to be the predominant enzyme responsible for the HDV RNA editing (Wong and Lazinski 2002), and the exact role of ADAR2 in editing viral RNAs is currently unclear.
Antagonists of ADAR2 activity
In contrast to ADAR1, which has been shown to be antagonized by viral proteins, viral RNA molecules, and synthetic RNA aptamers (Lei and others 1998; Liu and others 2000, 2001), it remains unclear whether there exist specific viral RNAs or viral proteins that act as antagonists against ADAR2. However, the N-terminal portion of ADAR2 that includes the dsRBD RI may act to inhibit its own deaminase activity (Macbeth and others 2004). ADAR3 protein has been shown also as an ADAR2 inhibitor when assayed in vitro (Chen and others 2000), possibly through competitive sequestration of the RNA substrates. The brain-specific small RNA MBII-52 that may function as a nucleolar C/D RNA can target the C-site of the 5-HT2C receptor pre-mRNA for 2′-O-methylation, consequently altering its editing by ADAR2 (Vitali and others 2005). However, whether this nucleolar editing antagonism is a general regulatory mechanism for ADAR2 is unknown.
Other RNA substrates of ADARs
Most cellular substrates of selective A-to-I editing by ADARs, including GluR and 5-HT2CR pre-mRNAs, were identified serendipitously. Computational searches of the human transcriptome using large numbers of expressed sequences revealed several thousand candidate A-to-I editing sites (Athanasiadis and others 2004; Kim and others 2004; Levanon and others 2004, 2005). These newly identified editing events occur primarily in noncoding regions of RNA, often in Alu repeats. Pairs of inverted Alu repeats in the 3′-UTR regions of mRNAs conceivably could form duplex structures that lead to A-to-I editing by ADARs, and hence altered gene expression (Chen and Carmichael 2008).
Biological Activities of RNA Adenosine Deaminases ADAR1 and ADAR2
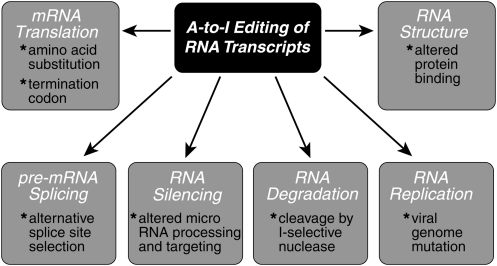
A broad range of biological processes are affected by the ADAR proteins. These include virus growth and persistence, apoptosis and embryogenesis, neurotransmitter receptor function, pancreatic cell function, and post-transcriptional gene regulation by miRNAs. Established biochemical mechanisms that provide a framework for understanding the physiologic changes following ADAR-mediated editing are summarized in Fig. 5. Because I is recognized as G, A-to-I editing may affect gene expression and function at a number of levels. These include mRNA translation by changing codons and hence the amino acid sequence of synthesized proteins; pre-mRNA splicing by altering splice site recognition sequences; RNA stability by changing sequences involved in nuclease recognition; genetic stability in the case of RNA virus genomes by changing template and thus product sequences during viral RNA replication; and RNA-structure-dependent activities such as miRNA production or targeting or protein–RNA interactions.
FIG. 5.
Effects that A-to-I RNA editing may exert on gene expression and function. Adenosine (A) deamination to inosine (I) in duplex RNA structures catalyzed by ADARs substitutes an I for an A in an RNA sequence. Because I is recognized as G, A-to-I editing may effect gene expression and function in a number of ways that include mRNA translation by changing codons and hence the amino acid sequence of expressed proteins; pre-mRNA splicing by changing a conserved A in splice site recognition sequences; RNA stability by altering sequences involved in nuclease recognition; RNA virus genome stability by changing template and thus product sequences during viral RNA replication; and, RNA-structure-dependent activities such as microRNA production or targeting or protein-RNA interactions (adapted from Samuel 2003 with permission).
Virus growth and persistence
While sequence changes consistent with A-to-I editing by ADAR deaminases are seen in some viral RNAs, including measles virus (Cattaneo and others 1988), hepatitis C virus (Taylor and others 2005), lymphocytic choriomeningitis virus (Zahn and others 2007), parainfluenza virus (Murphy and others 1991), vesicular stomatitis virus (O'Hara and others 1984), and polyoma virus (Kumar and Carmichael 1997), the precise role of ADARs in virus infection in most instances is not well defined. Because the ADAR1 cDNA was isolated as an IFN-inducible gene product (Patterson and others 1995) and the p150 form of ADAR1 is an interferon-stimulated gene (ISG) with an ISRE-containing promoter (George and Samuel 1999a, 1999b; George and others 2005), a reasonable consideration is that ADAR1 may function in an antiviral role. However, evidence of an antiviral activity for ADAR1 is limited (Samuel 2001; Toth and others 2006). Overexpression approaches failed to demonstrate an antiviral activity of either ADAR1 or ADAR2 in single-cycle infections with vesicular stomatitis virus (Li and others 2010). While ADAR1 antiviral activity is implicated in some studies with 2 hepatitis viruses, HCV (Taylor and others 2005) and HDV (Jayan and Casey 2002b; Sato and others 2004), recent work with other viruses is most consistent with a proviral role for ADAR1 as exemplified by measles virus (Toth and others 2009), vesicular stomatitis virus (Nie and others 2007; Li and others 2010), HHV8 (Gandy and others 2007), human immunodeficiency virus (Phuphuakrat and others 2008; Clerzius and others 2009; Doria and others 2009), and with HDV (Casey 2006; Linnstaedt and others 2009). The proviral effect of ADAR1 appears to result, at least in some instances, from the suppression of PKR activation and function (Nie and others 2007; Clerzius and others 2009; Toth and others 2009; Li and others 2010).
Under normal physiologic conditions, ADAR1 promotes rather than inhibits HDV replication (Casey 2006). HDV has a structured, circular RNA genome and depends on a helper virus, hepatitis B virus, for replication. HDV expresses 2 forms of delta antigen, small HDAg necessary for viral RNA replication and large HDAg that mediates packaging of the viral genome (Taylor 2003). ADAR1 is proviral for HDV, because RNA editing on the HDV antigenome converts the UAG stop codon that terminates small HDAg synthesis to an UIG tryptophan codon, thereby allowing synthesis of large HDAg. Without editing, genome packaging does not occur; ADAR1, therefore, is necessary for virus replication. However, high levels of ADAR1 protein inhibit HDV replication (Jayan and Casey 2002a; Hartwig and others 2004; Casey 2006). Overexpression of ADAR1 or ADAR2 in Huh-7 cells, or treatment with IFN that induces the p150 form of ADAR1, results in increased HDV RNA editing and decreased HDV replication (Jayan and Casey 2002a; Hartwig and others 2004). Likewise, a replication-competent HDV virus mutant that shows increased editing at the amber/W site produces increased amounts of large HDAg early in infection, and replication of the mutant virus terminates prematurely (Sato and others 2004; Casey 2006). It is not unreasonable to presume that an antiviral activity for ADAR1 against HDV might occur during natural infection, especially if the level of ADAR1 p150 is increased during IFN-α therapy for hepatitis B virus (HBV).
In the case of HCV, evidence for an antiviral role of ADAR1 comes from replicon studies and is based on reduction of ADAR1 activity either by adenovirus VAI RNA expression or siRNA knockdown of ADAR1. Such reduction in ADAR1 reportedly causes a reduction in I-containing viral RNA in replicon expressing cells and impairs the IFN-induced inhibition of HCV RNA replication (Taylor and others 2005). Whether this finding based on the HCV replicon system will extend to the cell culture model of HCV virus infection, eg, based on the HCV JFH-1 molecular clone virus and Huh-7-derived cell lines, is not yet clear. An intriguing counterintuitive possibility is that ADAR1 is antiviral in the context of HCV by mediating a reduction in PKR activation in HCV-infected cells; HCV infection has been found to block IFN-induced protein induction through activation of PKR phosphorylation, which causes inhibition of cellular (including ISG protein) but not viral (HCV) protein production (Garaigorta and Chisari 2009). Consistent with this notion, in cells infected with either measles virus (Toth and others 2009) or VSV (Li and others 2010), ADAR1 deficiency leads to enhanced PKR activation.
RNA editing of the HHV8 kaposin K12 transcript during lytic replication is believed to eliminate its transforming activity (Gandy and others 2007). The K12 RNA, an abundant transcript expressed in latent Kaposi's sarcoma-associated HHV8 infection, also is induced during lytic replication. The transcripts that possess an A at position 117990 are tumorigenic, whereas those with G (or I) are not. There is no detectable G in the viral DNA sequence at this position, only A. Purified human ADAR1 efficiently edits K12 RNA in vitro and the extent of A-to-I (G) editing in vivo correlates with the replicative state of the virus (Gandy and others 2007).
Mouse polyoma virus, a DNA virus that replicates in the nucleus, expresses early and late transcripts from opposite strands of the viral genome. Early strand RNA is downregulated by antisense late-strand RNA. Approximately half of the early strand RNAs in the nucleus at late times after infection possess G (or I) in place of A as determined by sequence analysis, and about 50% of the A's are replaced in these transcripts, consistent with hyperdeamination by an ADAR (Kumar and Carmichael 1997). However, it is not yet known whether ADAR1 or ADAR2, or both, are responsible for the downregulation of early polyoma RNA.
Persistent measles virus infection can occur in the brain, causing a rare but often fatal disease known as subacute sclerosing panencephalitis (SSPE) (Oldstone 2009). When viral RNA from SSPE patients was characterized, clustered A-to-I(G) mutations were found in the M gene and less frequently in other measles virus genes (Cattaneo and others 1988; Oldstone 2009). The M mRNA mutations seen in SSPE patients are presumed to prevent synthesis of functional M protein that is required for virion assembly and release, and thereby contribute to persistence. Parainfluenza (Murphy and others 1991) and vesicular stomatitis (O'Hara and others 1984) viruses likewise are implicated as targets of ADAR1 deaminase activity based on observed sequence changes. In the case of human parainfluenza virus 3 RNA isolated from persistently infected LLC-MK2 cells in culture, biased A-to-I (G) changes are observed toward the 3′-end of the viral RNA along with U-to-C transitions (Murphy and others 1991).
Somewhat surprisingly, ADAR1 has emerged as a proviral host factor in the context of measles virus (Toth and others 2009), vesicular stomatitis virus (Nie and others 2007; Li and others 2010), and HIV (Phuphuakrat and others 2008; Clerzius and others 2009; Doria and others 2009) infections. This proviral activity of ADAR1 correlates with the suppression of PKR activation and eIF-2α phosphorylation (Toth and others 2009; Li and others 2010) and the enhancement of synthesis of viral proteins (Phuphuakrat and others 2008; Doria and others 2009; Toth and others 2009). Among the mechanisms that might contribute to the proviral effects of ADAR1 that involve suppressive modulatory effects on PKR include inactivation of PKR activator RNAs and ADAR1-PKR protein interaction. ADAR1-mediated deamination of dsRNA through A-to-I editing would destabilize RNA structures, the very property by which ADARs were originally discovered (Bass and Weintraub 1987, 1988; Rebagliati and Melton 1987; Wagner and others 1989), which presumably could decrease the amount of PKR activator RNA. The other possibility is that ADAR interaction with PKR results in a multiprotein complex and subsequent alteration in protein localization that effectively neutralizes a host function such as that mediated by PKR which normally is antiviral. The enhanced apoptosis and reduced virus yields seen following infection of cells stably deficient in ADAR1 with measles virus C or V mutants correlates with enhanced activation of PKR and IRF3 (Toth and others 2009). Induction of IFN-β is amplified by PKR in cells infected with the C or V deletion mutant of measles virus (McAllister and others 2010), but the role of ADAR1 in the process is not yet known.
Apoptosis and embryogenesis
Mouse gene knockouts have been described for both Adar1 (Wang and others 2000a, 2004; Hartner and others 2004, XuFeng and others 2009) and Adar2 (Higuchi and others 2000). Their phenotypes are different. Homozygosity for Adar1 null leads to liver disintegration, defects in hematopoiesis, and embryonic death between E11.5 and E12.5 (Hartner and others 2004; Wang and others 2004). Homozygosity for Adar2 null is not embryonic lethal, but Adar2 null mice are shorter-lived than wild-type mice and display neurological abnormality, including episodes of epileptic seizures (Higuchi and others 2000).
Analysis of Adar1−/− dead embryos reveals apoptosis in many tissues, especially the liver (Wang and others 2004). Subsequent studies of inducible Adar1 gene disruption in mice suggest that ADAR1 is essential for maintenance of both fetal and adult hematopoietic stem cells. Loss of ADAR1 in hematopoietic stem cells leads to global upregulation of type I and II IFN-inducible transcripts and rapid apoptosis (Hartner and others 2009). ADAR1 appears to be essential for survival of differentiating hematopoietic progenitor cells as opposed to more primitive stem cells in adult mice. In the absence of ADAR1, transplanted ADAR1-deficient hematopoietic stem cells are incapable of reconstituting irradiated recipients although they are phenotypically present in the recipient bone marrow (XuFeng and others 2009). MEF cells derived from the Adar1−/− mice are prone to serum-deprivation-induced apoptosis (Wang and others 2004). Likewise, virus-induced apoptosis is enhanced in human cells in culture made stably deficient in ADAR1 p150 and p110 through short hairpin RNA-mediated knockdown (Toth and others 2009). Conceivably the antiapoptotic activity of ADAR1 arises in part through suppression of activation of proapoptotic RNA-dependent activities as exemplified by PKR and IRF3 (Toth and others 2009; Li and others 2010). Whether it is the absence of p110 or p150 or both ADAR1 protein isoforms that causes increased apoptosis and embryonic lethality is not yet established, as the Adar1 gene disruptions described to date eliminate both p110 and p150. In wild-type embryos, constitutively expressed exon 1B-containing transcripts are detectable at E10, whereas the IFN-inducible exon 1A-containing transcripts are not (George and others 2005). This suggests the possibility that p110, but not p150, is important for embryogenesis. However, another possibility is that the transcriptional activation of inducible p150 ADAR1 is necessary for embryonic development but simply not triggered to a detectable level at E10. Selective disruption of either the IFN-inducible PIA promoter and the corresponding exon 1A region, or the constitutive promoters and corresponding exon 1B and 1C regions, might resolve this question.
The ADAR1 requirement for embryo survival during development cannot be complemented by ADAR2 (Hartner and others 2004, 2009; Wang and others 2004); and, ADAR2 is not required for embryogenesis (Higuchi and others 2000). Adar2−/− mice, although prone to epileptic seizures, do survive several weeks after birth. Lack of editing of GluR-B RNA at the Q/R site appears to be the primary causal basis of the neurological phenotype of Adar2 null mice, because the behavioral dysfunction of Adar2−/− mice is rescued by replacing the unedited CAG codon of each GluR-B allele with a GluR-B allele encoding an edited (CGG) RNA codon (Higuchi and others 2000).
Neurotransmitter receptor function
Adar gene targeting studies in the mouse have established unequivocally that A-to-I RNA editing can modulate neuronal activity and have confirmed the site selective roles of ADAR1 and ADAR2 in the editing of the neurotransmitter receptor pre-mRNAs encoding the AMPA glutamate receptor GluR-B and the serotonin 5-HT2C receptor (Higuchi and others 2000; Hartner and others 2004; Wang and others 2004). The α3 subunit of the γ-aminobutyric acid type A (GABAA) receptor also is edited by ADAR1 and ADAR2 (Ohlson and others 2007; Rula and others 2008). Editing leads to highly selective amino acid substitutions in the GluR-B, 5-HT2CR, and α3 GABAA receptor proteins that affect their functional activities and hence neurophysiology.
The ionotropic glutamate receptors are a diverse group of receptors that are important in the mediation of excitatory synaptic neurotransmission. RNA editing by ADAR proteins at 2 sites contributes to the functional diversity of AMPA and kainate glutamate receptors (Seeburg and Hartner 2003). A-to-I editing by ADAR2 at the Q/R site in exon 11 of hydrophobic transmembrane domain 2 alters calcium permeability of the glutamate receptor channel; channels composed of the edited GluR-B subunits show reduced permeability to Ca++ ions, whereas channels composed of unedited subunits show higher divalent ion permeability (Burnashev and others 1992). Q/R editing also affects assembly and trafficking of the glutamate receptors (Greger and others 2003). A-to-I editing at the second GluR-B site, the R/G site in exon 13, affects the kinetic properties of the GluR-B receptor; editing leads to faster recovery rates from receptor desensitization (Lomeli and others 1994). A-to-I editing of the ionotropic GABAA receptor α3 subunit at the I/M site is regulated in a spatiotemporal manner and leads to the conversion of a genomic Ile codon to a Met codon that alters chloride ion flux (Ohlson and others 2007; Rula and others 2008). GABAA α3 RNA editing is low in the embryo at E15 but approaches 90% by P7 and persists in the adult mouse brain; GABAA receptors with the nonedited α3 subunit are activated more rapidly and deactivated more slowly than the edited receptors (Rula and others 2008).
The 5-HT2CR serotonin receptor is a 7-transmembrane G-protein-coupled receptor, linked to phospholipase C with the production of inositol phosphates and diacylglycerol (Baxter and others 1995; Burns and others 1997; Werry and others 2008). Pre-mRNA transcripts for 5-HT2CR undergo A-to-I editing at 5 different positions in exon 3 (Burns and others 1997; Liu and others 1999; Niswender and others 1999). Editing at all 5 of the sites results in 3 amino acid substitutions in the second extracellular loop of the receptor; however, several different isoforms of the 5-HT2CR protein can result from different combinations of edited or unedited adenosines at the 5 sites (Fitzgerald and others 1999). 5-HT2CR receptor isoform expression patterns vary in different regions of the brain (Burns and others 1997; Liu and others 1999; Niswender and others 1999; Wang and others 2000b). The fully edited 5-HT2CR isoform (with Val, Ser, and Val substituting for Ile, Asp and Ile, respectively) displays a ∼10–15-fold reduction in G-protein-mediated signaling compared to the fully unedited isoforms as assessed by dose–response analysis of inositol phosphate accumulation with serotonergic agonists (Burns and others 1997). The difference in agonist potency between edited and unedited isoforms is caused by both decreased receptor-G protein coupling (Burns and others 1997; Niswender and others 1999) and decreased agonist affinity (Fitzgerald and others 1999).
Metabolic and pancreatic cell function
Several lines of evidence have implicated a role of ADAR2 editing enzyme in metabolism. Mice expressing either the wild-type or deaminase-deficient ADAR2 transgenes exhibit hyperphagia and develop adult-onset obesity (Singh and others 2007). However, the exact mechanism that underlies the ADAR2 effects on the central control of energy balance is not yet known. In addition, a potential endocrine function is suggested by the findings that ADAR2 expression and ADAR2-mediated RNA editing are selectively upregulated in primary pancreatic islets of mice with diet-induced obesity and insulin resistance (Gan and others 2006). While ADAR2 and ADAR2-dependent RNA editing are increased in cultured INS-1 β-cells in response to glucose stimulation, whether ADAR2 is directly involved in the control of the pancreatic endocrine function is yet to be demonstrated. Thus, more detailed studies using tissue-specific gene-targeting strategies will be required to dissect the central or peripheral function of ADAR2 in metabolic homeostasis.
Post-transcriptional gene regulation by miRNAs
ADAR proteins can affect post-transcriptional gene regulation by miRNAs, small noncoding RNAs that function as regulatory RNAs. By base pairing to mRNAs, miRNAs can downregulate with high specificity the production of proteins encoded by targeted mRNAs, either by mediating mRNA degradation or translational suppression without degradation (Filipowicz and others 2008). miRNAs are produced from precursor transcripts, pri-miRNAs, that possess base-paired stem structures and undergo nuclear processing by the Drosha nuclease to generate ∼70 nt hairpin pre-miRNAs that are transported to the cytoplasm by exportin 5 and further processed by the Dicer nuclease to yield ∼20 bp miRNA duplexes. Drosha and Dicer are endonucleases that function in complexes with proteins that possess dsRBD of the nature first discovered in PKR and also present in the ADARs (Samuel 2001; Toth and others 2006; Filipowicz and others 2008). MiRNAs are assembled into RNAi-induced silencing complexes (RISCs) and base pair with nearly perfect complementarity to the targeted mRNA, typically in the 3′-UTR region, to silence expression. ADAR1 and ADAR2 have been shown to affect the miRNA gene silencing process at multiple steps, which retrospectively is not surprising, given that the miRNA silencing process depends upon dsRNA structures and dsRNA-binding proteins. Among the steps of the miRNA pathway altered by ADARs are the processing of miRNA precursors and the targeting of miRNAs (Habig and others 2007; Heale and others 2009b).
Among the first hints of A-to-I editing of miRNAs was the sequence changes seen for human and mouse pri-miRNA-22 transcripts (Luciano and others 2004) and then the subsequent demonstration that editing of pri-miRNA-142 leads to inhibition of processing by the Drosha nuclease (Yang and others 2006). A-to-I editing of two adenosines within the dsRNA structure of pri-miRNA-142 blocks the processing cleavage by the Drosha-DGCR8 complex, thereby decreasing the level of mature miRNA-142 produced. A-to-I editing of pri-miRNA-151 at 2 positions within its dsRNA structure inhibits cleavage by the Dicer-TAR RNA binding protein (TRBP) complex and leads to an accumulation of edited pre-miRNA-151 (Kawahara and others 2007a). The first evidence that A-to-I editing could alter the target specificity of miRNA was obtained with miRNA-376a, where editing in the seed sequence changes the targeting to silence expression of the phosphoribosyl pyrophosphate synthetase 1 enzyme in uric acid synthesis in a manner dependent upon ADAR2 (Kawahara and others 2007b). A significant correlation also has been observed between A-to-I editing events in 3′-UTR regions of mRNA and changes in miRNA complementarities, suggesting that creation of miRNA target sites by editing of mRNAs may also be a function of ADARs (Borchert and others 2009).
The frequency of miRNA editing appears significant. Initial sequence analysis suggested that ∼10% of the pri-miRNAs were edited in human tissues (Blow and others 2006). A large-scale survey of human brain pri-miRNAs by direct sequencing identified 86 editing sites in 47 pri-miRNAs, leading to the prediction that ∼16% of human pri-miRNAs are subject to A-to-I editing (Kawahara and others 2008). The majority of the pri-miRNA editing events seem to affect steps of miRNA processing rather than re-directing targeting through altered seed sequences. Expression of miRNAs has been described for various groups of viruses, including herpesviruses, retroviruses, and small DNA viruses, and cellular miRNAs have been identified that target viral sequences (Berkhout and Jeang 2007). Essentially nothing is known regarding whether the IFN-induced ADAR1 deaminase is able to edit any of these miRNAs in a manner that alters the virus–host interaction. However, it has been observed that the liver-specific miRNA-122, implicated in the control of HCV replication and the response to IFN in hepatoma cells (Pedersen and others 2007), is present at lower pretreatment levels in individuals with hepatitis C responding poorly to pegIFN alpha therapy than in responders (Sarasin-Filipowicz and others 2009).
ADAR proteins also can modulate gene silencing independent of their deaminase activity (Yang and others 2005; Heale and others 2009a). This was first suggested by the finding that ADAR can limit the efficacy of RNAi by synthetic siRNAs (Yang and others 2005). Both ADAR1 and ADAR2 bind to siRNAs, with the cytoplasmic p150 form of ADAR1 showing the highest affinity. Short siRNAs (<36 bp) while not edited, still bind with high affinity. Not unexpectedly, when such sequestration is circumvented, RNAi becomes significantly more potent as seen in MEF cells deficient in ADAR1 (Yang and others 2005). ADAR2 can modulate the processing of miRNA-376a independent of deaminase catalytic activity (Heale and others 2009a). Extensive editing of siRNAs is seen in Drosophila, and using a Drosophila-based assay for RNAi, human ADAR1 p150 inhibits RNAi through a mechanism that is partially independent of editing activity (Heale and otheres 2009a). ADAR1 p150 also inhibits siRNA silencing when tested in the background of Adar1 null MEFs (Yang and others 2005).
ADARs in human disease
Among the human genetic diseases associated with ADAR1 and A-to-I editing are dyschromatosis symmetrica hereditaria (DSH-1) and a variety of neurologic disorders (Maas and others 2006; Toth and others 2006). DSH-1 is an autosomal dominant pigmentary dermatological disorder that causes freckle-like pigmentation changes typically on the hands and feet. The disease locus for DSH-1 has been mapped to the ADAR1 locus on chromosome 1q21 (Weier and others 1995; Miyamura and others 2003; Zhang and others 2003; He and others 2004). Multiple ADAR1 mutations, including missense, nonsense, frameshift, and splicing mutations, have been identified in DSH-1 patients (Miyamura and others 2003; Gao and others 2005; Suzuki and others 2005; Liu and others 2006). While neurological effects are not usually seen, progressive mental changes are associated with some rare mutations. The effects of these ADAR1 mutations on ADAR1 protein function and the mechanistic basis by which the ADAR1 mutations cause the DSH pigment disorder are largely unknown.
Altered editing of neurotransmitter receptor RNAs has been observed in patients with several neurological disorders, including schizophrenia, Alzheimer's disease, Huntington's disease, and depression (Maas and others 2006; Toth and others 2006; Werry and others 2008) as well as astrocytomas (Cenci and others 2008). When A-to-I editing of the serotonin 5-HT2CR receptor was examined in the frontal cortex of schizophrenic patients, an overall decrease in editing was seen (Sodhi and others 2001). Reduction in 5-HT2CR editing of one of the ADAR1 sites, the B site, reached statistical significance. A statistically significant difference in editing at the other 5-HT2CR site edited by ADAR1, the A-site, also was reported in patients who had committed suicide (Niswender and others 2001). Decreased editing of GluR-B at the Q/R site, a site edited by ADAR2, has been reported in the prefrontal cortex of Alzheimer's disease and schizophrenic subjects and in the striatum of Huntington's disease patients (Akbarian and others 1995) as well as in neurons from patients with amyotrophic lateral sclerosis (Kwak and Kawahara 2005; Aizawa and others 2010) and astrocytomas, both adult (Maas and others 2001) and pediatric (Cenci and others 2008).
Acknowledgments
This work was supported in part by the U.S.P.H.S. National Institute of Allergy and Infectious Diseases, NIH (Research Grants AI-12520 and AI-20611), and by the National Natural Science Foundation of China (Grants 30830033 and 30988002). The authors thank the many investigators within the ADAR and RNA editing fields for their basic and clinical research contributions that made this review possible, and we apologize to those whose work may not have been cited because of the length constraints of the review.
Author Disclosure Statement
No competing financial interests exist.
References
- Agranat L. Raitskin O. Sperling J. Sperling R. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc Natl Acad Sci U S A. 2008;105(13):5028–5033. doi: 10.1073/pnas.0710576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H. Sawada J. Hideyama T. Yamashita T. Katayama T. Hasebe N. Kimura T. Yahara O. Kwak S. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010;120(1):75–84. doi: 10.1007/s00401-010-0678-x. [DOI] [PubMed] [Google Scholar]
- Akbarian S. Smith MA. Jones EG. Editing for an ampa receptor subunit RNA in prefrontal cortex and striatum in Alzheimer's disease, Huntington's disease and schizophrenia. Brain Res. 1995;699(2):297–304. doi: 10.1016/0006-8993(95)00922-d. [DOI] [PubMed] [Google Scholar]
- Aruscavage PJ. Bass BL. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA. 2000;6(2):257–269. doi: 10.1017/s1355838200991921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A. Rich A. Maas S. Widespread A-to-I RNA editing of alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. Nishikura K. Keller W. Seeburg PH. Emeson RB. O'Connell MA. Samuel CE. Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3(9):947–949. [PMC free article] [PubMed] [Google Scholar]
- Bass BL. Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48(4):607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- Bass BL. Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Baxter G. Kennett G. Blaney F. Blackburn T. 5-HT2 receptor subtypes: a family re-united? Trends Pharmacol Sci. 1995;16(3):105–110. doi: 10.1016/s0165-6147(00)88991-9. [DOI] [PubMed] [Google Scholar]
- Berkhout B. Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem. 2007;282(37):26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- Bhalla T. Rosenthal JJ. Holmgren M. Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11(10):950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Blow MJ. Grocock RJ. van Dongen S. Enright AJ. Dicks E. Futreal PA. Wooster R. Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7(4):R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM. Gilmore BL. Spengler RM. Xing Y. Lanier W. Bhattacharya D. Davidson BL. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 2009;18(24):4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N. Monyer H. Seeburg PH. Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8(1):189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Burns CM. Chu H. Rueter SM. Hutchinson LK. Canton H. Sanders-Bush E. Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387(6630):303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Casey JL. RNA editing in hepatitis delta virus. Curr Top Microbiol Immunol. 2006;307:67–89. doi: 10.1007/3-540-29802-9_4. [DOI] [PubMed] [Google Scholar]
- Cattaneo R. Schmid A. Eschle D. Baczko L. ter Meulen V. Billeter MA. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55(2):255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci C. Barzotti R. Galeano F. Corbelli S. Rota R. Massimi L. Di Rocco C. O'Connell MA. Gallo A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283(11):7251–7260. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- Chang HW. Jacobs BL. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene-products to double-stranded-RNA. Virology. 1993;194(2):537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- Chang HW. Watson JC. Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein-kinase. Proc Natl Acad Sci U S A. 1992;89(11):4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CX. Cho DS. Wang Q. Lai F. Carter KC. Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL. Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7(21):3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- Chilibeck KA. Wu T. Liang C. Schellenberg MJ. Gesner EM. Lynch JM. MacMillan AM. FRET analysis of in vivo dimerization by RNA-editing enzymes. J Biol Chem. 2006;281(24):16530–16535. doi: 10.1074/jbc.M511831200. [DOI] [PubMed] [Google Scholar]
- Cho DS. Yang W. Lee JT. Shiekhattar R. Murray JM. Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278(19):17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- Clerzius G. Gélinas JF. Daher A. Bonnet M. Meurs EF. Gatignol A. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol. 2009;83(19):10119–10128. doi: 10.1128/JVI.02457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S. Ward SV. Tacke RS. Suske G. Samuel CE. Activation of the RNA-dependent protein kinase PKR promoter in the absence of interferon is dependent upon Sp proteins. J Biol Chem. 2006;281(6):3244–3253. doi: 10.1074/jbc.M510612200. [DOI] [PubMed] [Google Scholar]
- Dawson TR. Sansam CL. Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem. 2004;279(6):4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- Desterro JM. Keegan LP. Lafarga M. Berciano MT. O'Connell M. Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116(pt9):1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Desterro JMP. Keegan LP. Jaffray E. Hay RT. O'Connell MA. Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16(11):5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria M. Neri F. Gallo A. Farace MG. Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37(17):5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR. Neunteufl A. Pfaffstetter L. Jantsch MF. The human but not the xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell. 2001;12(7):1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensterö M. Daniel C. Wahlstedt H. Major F. Ohman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37(20):6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Sansam CL. Singh M. Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol Cell Biol. 2006;26(2):480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-Monti I. Mathews MB. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem Sci. 2000;25(5):241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- Filipowicz W. Bhattacharyya SN. Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fitzgerald LW. Iyer G. Conklin DS. Krause CM. Marshall A. Patterson JP. Tran DP. Jonak GJ. Hartig PR. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology. 1999;21(suppl):82S–90S. doi: 10.1016/S0893-133X(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Fritz J. Strehblow A. Taschner A. Schopoff S. Pasierbek P. Jantsch MF. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol. 2009;29(6):1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano F. Leroy A. Rossetti C. Gromova I. Gautier P. Keegan LP. Massimi L. DiRocco C. O'Connell MA. Gallo A. Human BLCAP transcript: new editing events in normal and cancerous tissues. Int J Cancer. 2009;127(1):127–137. doi: 10.1002/ijc.25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A. Keegan LP. Ring GM. O'Connell MA. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 2003;22(13):3421–3430. doi: 10.1093/emboj/cdg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z. Zhao L. Yang L. Huang P. Zhao F. Li W. Liu Y. RNA editing by ADAR2 is metabolically regulated in pancreatic islets and beta-cells. J Biol Chem. 2006;281(44):33386–33394. doi: 10.1074/jbc.M604484200. [DOI] [PubMed] [Google Scholar]
- Gandy SZ. Linnstaedt SD. Muralidhar S. Cashman KA. Rosenthal LJ. Casey JL. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J Virol. 2007;81(24):13544–13551. doi: 10.1128/JVI.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M. Wang PG. Yang S. Hu SL. Zhang KY. Zhu YG. Ren YQ. Du WH. Zhang GL. Cui Y. Chen JJ. Yan KL. Xiao FL. Xu SJ. Huang W. Zhang XJ. Two frameshift mutations in the RNA-specific adenosine deaminase gene associated with dyschromatosis symmetrica hereditaria. Arch Dermatol. 2005;141(2):193–196. doi: 10.1001/archderm.141.2.193. [DOI] [PubMed] [Google Scholar]
- Garaigorta U. Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6(6):513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX. Das S. Samuel CE. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology. 2008;380(2):338–343. doi: 10.1016/j.virol.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX. Li Z. Okonski KM. Toth AM. Wang Y. Samuel CE. Tipping the balance: antagonism of PKR kinase and ADAR1 deaminase functions by virus gene products. J Interferon Cytokine Res. 2009;29(9):477–487. doi: 10.1089/jir.2009.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX. Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 1999a;96(8):4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX. Samuel CE. Characterization of the 5′-flanking region of the human RNA- specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene. 1999b;229(1–2):203–213. doi: 10.1016/s0378-1119(99)00017-7. [DOI] [PubMed] [Google Scholar]
- George CX. Wagner MV. Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem. 2005;280(15):15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- Gerber A. O'Connell MA. Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3(5):453–463. [PMC free article] [PubMed] [Google Scholar]
- Gommans WM. Maas S. Characterization of ADAR1-mediated modulation of gene expression. Biochem Biophys Res Commun. 2008;377(1):170–175. doi: 10.1016/j.bbrc.2008.09.109. [DOI] [PubMed] [Google Scholar]
- Greger IH. Khatri L. Kong X. Ziff EB. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40(4):763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- Habig JW. Dale T. Bass BL. miRNA editing—we should have inosine this coming. Mol Cell. 2007;25(6):792–793. doi: 10.1016/j.molcel.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang PN. Tohda M. Matsumoto K. Developmental changes in expression and self-editing of adenosine deaminase type 2 pre-mRNA and mRNA in rat brain and cultured cortical neurons. Neurosci Res. 2008;61(4):398–403. doi: 10.1016/j.neures.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Hartner JC. Schmittwolf C. Kispert A. Muller AM. Higuchi M. Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279(6):4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Hartner JC. Walkley CR. Lu J. Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10(1):109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig D. Schoeneich L. Greeve J. Schutte C. Dorn I. Kirchner H. Hennig H. Interferon-alpha stimulation of liver cells enhances hepatitis delta virus RNA editing in early infection. J Hepatol. 2004;41(4):667–672. doi: 10.1016/j.jhep.2004.06.025. [DOI] [PubMed] [Google Scholar]
- He PP. He CD. Cui Y. Yang S. Xu HH. Li M. Yuan WT. Gao M. Liang YH. Li CR. Xu SJ. Chen JJ. Chen HD. Huang W. Zhang XJ. Refined localization of dyschromatosis symmetrica hereditaria gene to a 9.4-cM region at 1q21-22 and a literature review of 136 cases reported in China. Br J Dermatol. 2004;150(4):633–639. doi: 10.1111/j.0007-0963.2004.05861.x. [DOI] [PubMed] [Google Scholar]
- Heale BS. Keegan LP. McGurk L. Michlewski G. Brindle J. Stanton CM. Caceres JF. O'Connell MA. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009a;28(20):3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale BS. Keegan LP. O'Connell MA. ADARs have effects beyond RNA editing. Cell Cycle. 2009b;8(24):4011–4012. doi: 10.4161/cc.8.24.10214. [DOI] [PubMed] [Google Scholar]
- Herbert A. Alfken J. Kim YG. Mian IS. Nishikura K. Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci U S A. 1997;94(16):8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. Rich A. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc Natl Acad Sci U S A. 2001;98(21):12132–12137. doi: 10.1073/pnas.211419898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M. Single FN. Köhler M. Sommer B. Sprengel R. Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75(7):1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Higuchi M. Stefan M. Single FN. Hartner J. Rozov A. Burnashev N. Feldmeyer D. Sprengel R. Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406(6791):78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hoopengardner B. Bhalla T. Staber C. Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301(5634):832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Jacobs MM. Fogg RL. Emeson RB. Stanwood GD. ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev Neurosci. 2009;31(3):223–237. doi: 10.1159/000210185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayan GC. Casey JL. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J Virol. 2002a;76(8):3819–3827. doi: 10.1128/JVI.76.8.3819-3827.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayan GC. Casey JL. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J Virol. 2002b;76(23):12399–12404. doi: 10.1128/JVI.76.23.12399-12404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. Ito K. Ito M. Tsuji S. Kwak S. Novel splice variants of human ADAR2 mRNA: skipping of the exon encoding the dsRNA-binding domains, and multiple c-terminal splice sites. Gene. 2005;363:193–201. doi: 10.1016/j.gene.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Kawahara Y. Megraw M. Kreider E. Iizasa H. Valente L. Hatzigeorgiou AG. Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36(16):5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. Zinshteyn B. Chendrimada TP. Shiekhattar R. Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007a;8(8):763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. Zinshteyn B. Sethupathy P. Iizasa H. Hatzigeorgiou AG. Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007b;315(5815):1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakubo K. Samuel CE. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene. 2000;258(1–2):165–172. doi: 10.1016/s0378-1119(00)00368-1. [DOI] [PubMed] [Google Scholar]
- Kim DD. Kim TT. Walsh T. Kobayashi Y. Matise TC. Buyske S. Gabriel A. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14(9):1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U. Garner TL. Sanford T. Speicher D. Murray JM. Nishikura K. Purification and characterization of double-stranded RNA adenosine deaminase from bovine nuclear extracts. J Biol Chem. 1994a;269(18):13480–13489. [PubMed] [Google Scholar]
- Kim U. Wang Y. Sanford T. Zeng Y. Nishikura K. Molecular cloning of cDNA for double-stranded-RNA adenosine-deaminase, a candidate enzyme for nuclear-RNA editing. Proc Natl Acad Sci U S A. 1994b;91(24):11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG. Muralinath M. Brandt T. Pearcy M. Hauns K. Lowenhaupt K. Jacobs BL. Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A. 2003;100(12):6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajewski J. Schneider RJ. Safer B. Munemitsu SM. Samuel CE. Thimmappaya B. Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2-alpha kinase. Cell. 1986;45(2):195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation—an update. J Cell Biol. 1989;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhen KL. Samuel CE. Isolation of the interferon-inducible RNA-dependent protein kinase PKR promoter and identification of a novel DNA element within the 5′-flanking region of human and mouse Pkr genes. Virology. 1997;227(1):119–130. doi: 10.1006/viro.1996.8306. [DOI] [PubMed] [Google Scholar]
- Kumar M. Carmichael GG. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci U S A. 1997;94(8):3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S. Kawahara Y. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J Mol Med. 2005;83(2):110–120. doi: 10.1007/s00109-004-0599-z. [DOI] [PubMed] [Google Scholar]
- Kwak S. Nishimoto Y. Yamashita T. Newly identified ADAR-mediated A-to-I editing positions as a tool for ALS research. RNA Biol. 2008;5(4):193–197. doi: 10.4161/rna.6925. [DOI] [PubMed] [Google Scholar]
- Lai F. Chen CX. Carter KC. Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17(5):2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F. Drakas R. Nishikura K. Mutagenic analysis of double-stranded-RNA adenosine-deaminase, a candidate enzyme for RNA editing of glutamate-gated ion-channel transcripts. J Biol Chem. 1995;270(29):17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- Lehmann KA. Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39(42):12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- Lei M. Liu Y. Samuel CE. Adenovirus VAI RNA antagonizes the RNA-editing activity of the ADAR adenosine deaminase. Virology. 1998;245(2):188–196. doi: 10.1006/viro.1998.9162. [DOI] [PubMed] [Google Scholar]
- Levanon EY. Eisenberg E. Yelin R. Nemzer S. Hallegger M. Shemesh R. Fligelman ZY. Shoshan A. Pollock SR. Sztybel D. Olshansky M. Rechavi G. Jantsch MF. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22(8):1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Levanon EY. Hallegger M. Kinar Y. Shemesh R. Djinovic-Carugo K. Rechavi G. Jantsch MF. Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33(4):1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Wolff KC. Samuel CE. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology. 2010;396(2):316–322. doi: 10.1016/j.virol.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD. Kasprzak WK. Shapiro BA. Casey JL. The fraction of RNA that folds into the correct branched secondary structure determines hepatitis delta virus type 3 RNA editing levels. RNA. 2009;15(6):1177–1187. doi: 10.1261/rna.1504009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. Jiang L. Liu WL. Kang XJ. Ao Y. Sun M. Luo Y. Song Y. Lo WH. Zhang X. Two novel mutations and evidence for haploinsufficiency of the ADAR gene in dyschromatosis symmetrica hereditaria. Br J Dermatol. 2006;154(4):636–642. doi: 10.1111/j.1365-2133.2006.07133.x. [DOI] [PubMed] [Google Scholar]
- Liu Y. Emeson RB. Samuel CE. Serotonin-2c receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274(26):18351–18358. doi: 10.1074/jbc.274.26.18351. [DOI] [PubMed] [Google Scholar]
- Liu Y. George CX. Patterson JB. Samuel CE. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem. 1997;272(7):4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- Liu Y. Herbert A. Rich A. Samuel CE. Double-stranded RNA-specific adenosine deaminase: nucleic acid binding properties. Methods. 1998;15(3):199–205. doi: 10.1006/meth.1998.0624. [DOI] [PubMed] [Google Scholar]
- Liu Y. Lei M. Samuel CE. Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA- binding domains from ADAR1 and protein kinase PKR. Proc Natl Acad Sci U S A. 2000;97(23):12541–12546. doi: 10.1073/pnas.97.23.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Samuel CE. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA- specific adenosine deaminase. J Virol. 1996;70(3):1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Samuel CE. Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274(8):5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- Liu Y. Wolff KC. Jacobs BL. Samuel CE. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology. 2001;289(2):378–387. doi: 10.1006/viro.2001.1154. [DOI] [PubMed] [Google Scholar]
- Lomeli H. Mosbacher J. Melcher T. Hoger T. Geiger JR. Kuner T. Monyer H. Higuchi M. Bach A. Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266(5191):1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Luciano DJ. Mirsky H. Vendetti NJ. Maas S. RNA editing of a miRNA precursor. RNA. 2004;10(8):1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GX. Chao M. Hsieh SY. Sureau C. Nishikura K. Taylor J. A specific base transition occurs on replicating hepatitis delta virus-RNA. J Virol. 1990;64(3):1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S. Piñol-Roma S. Kjems J. Alternative splicing of the ADAR1 transcript in a region that functions either as a 5′-UTR or an ORF. RNA. 2007;13(10):1732–1744. doi: 10.1261/rna.567807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. Gommans WM. Novel exon of mammalian ADAR2 extends open reading frame. PLoS One. 2009;4(1):e4225. doi: 10.1371/journal.pone.0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. Kawahara Y. Tamburro KM. Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3(1):1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. Melcher T. Herb A. Seeburg PH. Keller W. Krause S. Higuchi M. O'Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271(21):12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- Maas S. Patt S. Schrey M. Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98(25):14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. Rich A. Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278(3):1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- Macbeth MR. Lingam AT. Bass BL. Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. RNA. 2004;10(10):1563–1571. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR. Schubert HL. Vandemark AP. Lingam AT. Hill CP. Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309(5740):1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle D. Das S. Ward SV. Samuel CE. Functional analysis of the KCS-like element of the interferon-inducible RNA-specific adenosine deaminase ADAR1 promoter. Gene. 2003;304:143–149. doi: 10.1016/s0378-1119(02)01200-3. [DOI] [PubMed] [Google Scholar]
- Mathews MB. Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65(11):5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS. Toth AM. Zhang P. Devaux P. Cattaneo R. Samuel CE. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J Virol. 2010;84(1):380–386. doi: 10.1128/JVI.02630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack SJ. Ortega LG. Doohan JP. Samuel CE. Mechanism of interferon action: motif I of the interferon-induced, RNA-dependent protein kinase (PKR) is sufficient to mediate RNA-binding activity. Virology. 1994;198(1):92–99. doi: 10.1006/viro.1994.1011. [DOI] [PubMed] [Google Scholar]
- McCormack SJ. Samuel CE. Mechanism of interferon action: RNA-binding activity of full-length and R-domain forms of the RNA-dependent protein kinase PKR—determination of KD values for VAI and TAR RNAs. Virology. 1995;206(1):511–519. doi: 10.1016/s0042-6822(95)80067-0. [DOI] [PubMed] [Google Scholar]
- McCormack SJ. Thomis DC. Samuel CE. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology. 1992;188(1):47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- Melcher T. Maas S. Herb A. Sprengel R. Higuchi M. Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996a;271(50):31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- Melcher T. Maas S. Herb A. Sprengel R. Seeburg PH. Higuchi M. A mammalian RNA editing enzyme. Nature. 1996b;379(6564):460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- Mittaz L. Scott HS. Rossier C. Seeburg PH. Higuchi M. Antonarakis SE. Cloning of a human RNA editing deaminase (ADARB1) of glutamate receptors that maps to chromosome 21q22.3. Genomics. 1997;41(2):210–217. doi: 10.1006/geno.1997.4655. [DOI] [PubMed] [Google Scholar]
- Miyamura Y. Suzuki T. Kono M. Inagaki K. Ito S. Suzuki N. Tomita Y. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet. 2003;73(3):693–699. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DF. Dimock K. Kang CY. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology. 1991;181(2):760–763. doi: 10.1016/0042-6822(91)90913-v. [DOI] [PubMed] [Google Scholar]
- Nie Y. Ding L. Kao PN. Braun R. Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25(16):6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y. Hammond GL. Yang JH. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol. 2007;81(2):917–923. doi: 10.1128/JVI.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM. Copeland SC. Herrick-Davis K. Emeson RB. Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2c receptor silences constitutive activity. J Biol Chem. 1999;274(14):9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- Niswender CM. Herrick-Davis K. Dilley GE. Meltzer HY. Overholser JC. Stockmeier CA. Emeson RB. Sanders-Bush E. RNA editing of the human serotonin 5-HT2C receptor: Alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24(5):478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- O'Connell MA. Krause S. Higuchi M. Hsuan JJ. Totty NF. Jenny A. Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15(3):1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P. Nichol S. Horodyski F. Holland J. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell. 1984;36(4):915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- Ohlson J. Pedersen JS. Haussler D. Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13(5):698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB. Modeling subacute sclerosing panencephalitis in a transgenic mouse system: uncoding pathogenesis of disease and illuminating components of immune control. Curr Top Microbiol Immunol. 2009;330:31–54. doi: 10.1007/978-3-540-70617-5_2. [DOI] [PubMed] [Google Scholar]
- O'Malley RP. Mariano TM. Siekierka J. Mathews MB. A mechanism for the control of protein-synthesis by adenovirus VA RNA1. Cell. 1986;44(3):391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- Patterson JB. Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15(10):5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB. Thomis DC. Hans SL. Samuel CE. Mechanism of interferon action —double-stranded RNA-specific adenosine-deaminase from human-cells is inducible by alpha-interferon and gamma-interferon. Virology. 1995;210(2):508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- Paupard M-C. O'Connell MA. Gerber AP. Zukin RS. Patterns of developmental expression of the RNA editing enzyme rADAR2. Neuroscience. 2000;95(3):869–879. doi: 10.1016/s0306-4522(99)00431-5. [DOI] [PubMed] [Google Scholar]
- Pedersen IM. Cheng G. Wieland S. Volinia S. Croce CM. Chisari FV. David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–822. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuphuakrat A. Kraiwong R. Boonarkart C. Lauhakirti D. Lee TH. Auewarakul P. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol. 2008;82(21):10864–10872. doi: 10.1128/JVI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placido D. Brown BA., 2nd Lowenhaupt K. Rich A. Athanasiadis A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15(4):395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen H. Nilsson J. Damgaard CK. Egebjerg J. Kjems J. Crm1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol Cell Biol. 2001;21(22):7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitskin O. Cho DS. Sperling J. Nishikura K. Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc Natl Acad Sci U S A. 2001;98(12):6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati MR. Melton DA. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- Rebouillat D. Hovanessian AG. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J Interferon Cytokine Res. 1999;19(4):295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- Riedmann EM. Schopoff S. Hartner JC. Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14(6):1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C. Gil J. Melkova Z. Esteban M. Diaz-Guerra M. Vaccinia virus E3L protein is an inhibitor of the interferon-induced 2-5A synthetase enzyme. Virology. 1998;243(2):406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- Rueter SM. Dawson RT. Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399(6731):75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Rula EY. Lagrange AH. Jacobs MM. Hu N. Macdonald RL. Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J Neurosci. 2008;28(24):6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc Natl Acad Sci U S A. 1979;76(2):600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. RNA editing minireview series. J Biol Chem. 2003;278(3):1389–1390. doi: 10.1074/jbc.R200032200. [DOI] [PubMed] [Google Scholar]
- Sansam CL. Wells KS. Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci U S A. 2003;100(24):14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M. Krol J. Markiewicz I. Heim MH. Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15(1):31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- Sarkis PT. Ying S. Xu R. Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177(7):4530–4540. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- Sato S. Cornillez-Ty C. Lazinski DW. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J Virol. 2004;78(15):8120–8134. doi: 10.1128/JVI.78.15.8120-8134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S. Wong SK. Lazinski DW. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J Virol. 2001;75(18):8547–8555. doi: 10.1128/JVI.75.18.8547-8555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol Cell. 2007;28(3):491–500. doi: 10.1016/j.molcel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T. Rould MA. Lowenhaupt K. Herbert A. Rich A. Crystal structure of the Z domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284(5421):1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- Seeburg PH. Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol. 2003;13(3):279–283. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- Seeburg PH. Higuchi M. Sprengel R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res Brain Res Rev. 1998;26(2–3):217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- Shtrichman R. Heithoff DM. Mahan MJ. Samuel CE. Tissue selectivity of interferon-stimulated gene expression in mice infected with dam(+) versus dam(-) Salmonella enterica Serovar typhimurium strains. Infect Immun. 2002;70(10):5579–5588. doi: 10.1128/IAI.70.10.5579-5588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. Kesterson RA. Jacobs MM. Joers JM. Gore JC. Emeson RB. Hyperphagia-mediated obesity in transgenic mice misexpressing the RNA-editing enzyme ADAR2. J Biol Chem. 2007;282(31):22448–22459. doi: 10.1074/jbc.M700265200. [DOI] [PubMed] [Google Scholar]
- Slavov D. Gardiner K. Phylogenetic comparison of the pre-mRNA adenosine deaminase ADAR2 genes and transcripts: conservation and diversity in editing site sequence and alternative splicing patterns. Gene. 2002;299(1–2):83–94. doi: 10.1016/s0378-1119(02)01016-8. [DOI] [PubMed] [Google Scholar]
- Sodhi MS. Burnet PW. Makoff AJ. Kerwin RW. Harrison PJ. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol Psychiatry. 2001;6(4):373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- Sommer B. Köhler M. Sprengel R. Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67. 1991;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Stark GR. Kerr IM. Williams BR. Silverman RH. Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Stefl R. Xu M. Skrisovska L. Emeson RB. Allain FH. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure. 2006;14(2):345–355. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Stephens OM. Haudenschild BL. Beal PA. The binding selectivity of ADAR2's dsRBMs contributes to RNA-editing selectivity. Chem Biol. 2004;11(9):1239–1250. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Strehblow A. Hallegger M. Jantsch MF. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell. 2002;13(11):3822–3835. doi: 10.1091/mbc.E02-03-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N. Suzuki T. Inagaki K. Ito S. Kono M. Fukai K. Takama H. Sato K. Ishikawa O. Abe M. Shimizu H. Kawai M. Horikawa T. Yoshida K. Matsumoto K. Terui T. Tsujioka K. Tomita Y. Mutation analysis of the ADAR1 gene in dyschromatosis symmetrica hereditaria and genetic differentiation from both dyschromatosis universalis hereditaria and acropigmentatio reticularis. J Invest Dermatol. 2005;124(6):1186–1192. doi: 10.1111/j.0022-202X.2005.23732.x. [DOI] [PubMed] [Google Scholar]
- Taylor DR. Puig M. Darnell MER. Mihalik K. Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79(10):6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JM. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 2003;11(4):185–190. doi: 10.1016/s0966-842x(03)00045-3. [DOI] [PubMed] [Google Scholar]
- Toth AM. Li Z. Cattaneo R. Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284(43):29350–29356. doi: 10.1074/jbc.M109.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM. Zhang P. Das S. George CX. Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- Valente L. Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Valente L. Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem. 2007;282(22):16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P. Basyuk E. Le Meur E. Bertrand E. Muscatelli F. Cavaillé J. Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169(5):745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RW. Smith JE. Cooperman BS. Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A. 1989;86(8):2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Khillan J. Gadue P. Nishikura K. Requirement of the RNA editing deaminase Adar1 gene for embryonic erythropoiesis. Science. 2000a;290(5497):1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Wang Q. Miyakoda M. Yang W. Khillan J. Stachura DL. Weiss MJ. Nishikura K. Stress-induced apoptosis associated with null mutation of Adar1 RNA editing deaminase gene. J Biol Chem. 2004;279(6):4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Wang Q. O'Brien PJ. Chen C-X. Cho D-SC. Murray JM. Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2c receptors. J Neurochem. 2000b;74(3):1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- Wang Q. Zhang Z. Blackwell KL. Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 2005;15(4):384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Wang Y. Samuel CE. Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2alpha phosphorylation. J Mol Biol. 2009;393(4):777–787. doi: 10.1016/j.jmb.2009.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Zeng Y. Murray JM. Nishikura K. Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene: the enzyme for glutamate-activated ion channel RNA editing. J Mol Biol. 1995;254(2):184–195. doi: 10.1006/jmbi.1995.0610. [DOI] [PubMed] [Google Scholar]
- Ward SV. Markle D. Das S. Samuel CE. The promoter-proximal KCS element of the PKR kinase gene enhances transcription irrespective of orientation and position relative to the ISRE element and is functionally distinct from the KCS-like element of the ADAR deaminase promoter. J Interferon Cytokine Res. 2002;22(8):891–898. doi: 10.1089/107999002760274917. [DOI] [PubMed] [Google Scholar]
- Weier HUG. George CX. Greulich KM. Samuel CE. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1–21.2. Genomics. 1995;30(2):372–375. doi: 10.1006/geno.1995.0034. [DOI] [PubMed] [Google Scholar]
- Weier HUG. George CX. Lersch RA. Breitweser S. Cheng JF. Samuel CE. Assignment of the RNA-specific adenosine deaminase gene (Adar) to mouse chromosome 3f2 by in situ hybridization. Cytogenet Cell Genet. 2000;89(3–4):214–215. doi: 10.1159/000015615. [DOI] [PubMed] [Google Scholar]
- Werry TD. Loiacono R. Sexton PM. Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signaling, pharmacology and brain function. Pharmacol Ther. 2008;119(1):7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Wong SK. Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci U S A. 2002;99(23):15118–15123. doi: 10.1073/pnas.232416799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. Wells KS. Emeson RB. Substrate-dependent contribution of double-stranded RNA-binding motifs to ADAR2 function. Mol Biol Cell. 2006;17(7):3211–3220. doi: 10.1091/mbc.E06-02-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XuFeng R. Boyer MJ. Shen H. Li Y. Yu H. Gao Y. Yang Q. Wang Q. Cheng T. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A. 2009;106(42):17763–17768. doi: 10.1073/pnas.0903324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH. Luo X. Nie Y. Su Y. Zhao Q. Kabir K. Zhang D. Rabinovici R. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003a;109(1):15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH. Nie Y. Zhao Q. Su Y. Pypaert M. Su H. Rabinovici R. Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J Biol Chem. 2003b;278(46):45833–45842. doi: 10.1074/jbc.M308612200. [DOI] [PubMed] [Google Scholar]
- Yang W. Chendrimada TP. Wang Q. Higuchi M. Seeburg PH. Shiekhattar R. Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13(1):13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WD. Wang QD. Howell KL. Lee JT. Cho DSC. Murray JM. Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280(5):3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;284(13):8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuwen H. Cox JH. Yewdell JW. Bennink JR. Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195(2):732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- Zahn RC. Schelp I. Utermöhlen O. von Laer D. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J Virol. 2007;81(2):457–464. doi: 10.1128/JVI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. Jacobs BL. Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J Virol. 2008;82(2):840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. Samuel CE. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J Virol. 2007;81(15):8192–8200. doi: 10.1128/JVI.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ. Gao M. Li M. Li CR. Cui Y. He PP. Xu SJ. Xiong XY. Wang ZX. Yuan WT. Yang S. Huang W. Identification of a locus for dyschromatosis symmetrica hereditaria at chromosome 1q11-1q21. J Invest Dermatol. 2003;120(5):776–780. doi: 10.1046/j.1523-1747.2003.12130.x. [DOI] [PubMed] [Google Scholar]