Abstract
E2F transcription factors and their target genes have been known to play an important role in cell growth control. We found that curcumin, a polyphenolic phytochemical isolated from the plant Curcuma longa, markedly suppressed E2F4 expression in HCT116 colon cancer cells. Hydrogen peroxide was also found to decrease E2F4 protein level, indicating the involvement of reactive oxygen species (ROS) in curucmin-induced downregulation of E2F4 expression. Involvement of ROS in E2F4 downregulation in response to curcumin was confirmed by the result that pretreatment of cells with N-acetylcystein (NAC) before exposure of curcumin almost completely blocked the reduction of E2F4 expression at the protein as well as mRNA level. Anti-proliferative effect of curcumin was also suppressed by NAC which is consistent to previous reports showing curcumin-superoxide production and induction of poly (ADP-ribose) polymerase (PARP) cleavage as well as apoptosis. Expression of several genes, cyclin A, p21, and p27, which has been shown to be regulated in E2F4-dependent manner and involved in the cell cycle progression was also affected by curcumin. Moreover, decreased (cyclin A) and increased (p21 and p27) expression of these E2F4 downstream genes by curcumin was restored by pretreatment of cells with NAC and E2F4 overexpression which is induced by doxycycline. In addition, E2F4 overexpression was observed to partially ameliorate curcumin-induced growth inhibition by cell viability assay. Taken together, we found curcumin-induced ROS down-regulation of E2F4 expression and modulation of E2F4 target genes which finally lead to the apoptotic cell death in HCT116 colon cancer cells, suggesting that E2F4 appears to be a novel determinant of curcumin-induced cytotoxicity.
Keywords: Curcumin, Cell proliferation, E2F4, Reactive oxygen species
INTRODUCTION
E2F transcription factor was originally discovered as a cellular factor which binds to adenovirus E2 promoter [1]. Since E2F was found to associate with and to be regulated by retinoblastoma protein (pRb), considerable attention has been paid to E2F as a cell growth regulator [1,2]. The E2F family members consist of eight genes: E2F1 - E2F8 and nine proteins including two E2F3 proteins, E2F3a and E2F3b, driven by alternative promoters [3,4]. In the amino acid terminus of each E2F proteins, there are highly conserved DNA-binding domain and adjacent hydrophobic domain required for dimerization with either DP1 or DP2, with exception of E2F7 and E2F8 which have two DNA-binding domains. In the carboxy terminus, transcriptional activation domain is located and overlapped with a region involved in binding to pRb and related pocket proteins, p107 and p130, while E2F6, E2F7 and E2F8 lack transactivation domain and/or Rb family protein binding region [5].
Among E2F family members, E2F1, E2F2, and E2F3a are categorized into activator E2Fs because they induce transcriptional activation of a set of genes, which results in DNA replication and cell cycle progression [6]. In contrast, E2F3b-E2F8, which are categorized into repressor E2Fs, generally function in the repression of E2F target gene expression through Rb-dependent (E2F3b, E2F4, and E2F5) and -independent (E2F6, E2F7, and E2F8) mechanism, respectively [7-9]. Accumulating evidence indicates that this conventional classification of E2Fs as either activators or repressors, correlating with activator or inhibitor of cell cycle progression, respectively, is oversimplified and unsuitable to explain distinct role of individual E2Fs in the regulation of target gene expression and/or cell cycle progression. For example, E2F1-E2F3, but not E2F4 and E2F5, overexpressed in WI-38 human primary fibroblasts was found to act as a transcription repressors of the plasminogen activator (PA) inhibitor 1 and urokinase-type PA genes which plays a significant role in angiogenesis, inflammation, and tumor metastasis and of which expression is regulated in a cell cycle-dependent manner [10]. In addition, preferential E2F4 expression was found in human intestinal crypt cells, colorectal cancer cells, and proliferative region of mouse embryonic intestine [11], suggesting E2F4 is highly expressed in proliferating cells and its expression might be required for cell cycle progression.
Curcumin, diferuloylmethane, is the yellow pigment with polyphenolic structure. This natural product is an active ingredient of tumeric extracted from the roots of the plant Curcuma longa (Linn) and used as a common coloring and flavoring agent in food. It is interesting that in China and India, curcumin has been used for thousands of years as a folk medicine to treat diverse diseases, long before the recent extensive studies revealed various biological activities of curcumin. It has been reported that curcumin has anti-inflammatory and antioxidant activities as well as a potent anticancer effect on numerous cancer cell types [12-16]. Moreover, in phase I and phase II clinical trials, it was proved that oral administration of curcumin (up to 12 g/day) was safe to human and conferred chemopreventive benefits on patients with high-risk and precancerous legions [17-19]. In this study, we examined the effect of curcumin on E2F4 expression and molecular mechanism involved in the regulation of its expression.
METHODS
Reagents and antibodies
HCT116 cells were obtained from the American Type Culture Collection. Curcumin and N-acetylcystein were from Sigma (St. Louis, MO). Cell Counting-8 Kit was from Dojindo Laboratories (Kumamoto, Japen). Primers for RT-PCR were synthesized from domestic company, Bioneer Inc (Daejoun). TRI reagent was from Solgent (Daejoun). AmpliTaq DNA polymerase was purchased from Roche Inc (Indianapolis, IN). Antibodies against E2F4, cyclin A, cyclin D1, p21, and p27 and horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG antibodies were from Santa Cruz (Santa Cruz, CA). Lipofectamin™ 2000 was from Invitrogen (Carlsbad, CA) and Blastacidin and Zeocin™ were obtained from Gibco BRL (Gaithersburg, MD).
Cell culture
Cells were grown in RPMI 1640 (Gibco BRL, Gaithersburg, MD) containing 10% FBS, 2 mM L-glutamine, 10 U/ml penicillin, and 10 g/ml streptomycin at 37℃ in 5% CO2 in a water-saturated atmosphere.
RT-PCR
Cells (1×106) were seeded in 100 mm culture dish and then stimulated with the indicated concentrations of curcumin for various periods. Total RNA was extracted using TRI reagent and subjected to the cDNA synthesis and PCR. The specific primers were as follows:
E2F1, forward 5'-CCCAGGGAAAGGTGTGAAAT-3' reverse 5'-CTTCTTGGCAATGAGCTGGA-3';
E2F4, forward 5'-GGAACGTCACAGAGGACGTG-3' reverse 5'-GGGCTCTGGAGCAAATCTTC-3';
c-Myc, forward 5'-CCTCGGATTCTCTGCTCTCC-3' reverse 5'-ACCTCTTGAGGACCAGTGGG-3';
CDK2, forward 5'-GGTCCTCCACCGAGACCTTA-3' reverse 5'-GAAAGATCCGGAAGAGCTGG-3';
cyclin A, forward 5'-GATGAGCATGTCACCGTTCC-3' reverse 5'-AGGGACCAATGGTTTTCTGG-3';
cyclin D1, forward 5'-TGCTCCTGGTGAACAAGCTC-3' reverse 5'-AGGACAGGAAGTTGTTGGGG-3';
p21, forward 5'-ATGTCAGAACCGGCTGGGGATGTC-3' reverse 5'-AAGGCCCCGCACACGCTCCCAGGC-3';
p27, forward 5'-GGCAAGTACGAGTGGCAAGA-3' reverse 5'-TTCCTTATTCCTGCGCATTG-3';
GAPDH, forward 5'-GGAGCCAAAAGGGTCATCAT-3' reverse 5'-GTGATGGCATGGACTGTGGT-3'.
Western blot analysis
Total cell lysates were prepared with lysis buffer (1% Triton X-100, 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM PMSF, 1 mM Na3VO4, and protease inhibitor cocktail). Protein concentrations were determined using Bio-Rad protein assays. Proteins from cell lysates (50 µg) were separated on 12% SDS-PAGE, and electrotransferred to nitrocellulose membranes. Membranes were blocked for 30 minutes at room temperature in Tris buffered saline-0.05% Tween-20 (TTBS) containing 5% non-fat dry milk, and then incubated with TTBS containing a primary antibody for 4 h at room temperature. After 5×10 min washes in TTBS, membranes were incubated with peroxidase-conjugated secondary antibody for 1 hr. Following 5 additional 10 min washes with TTBS, protein bands of interest were visualized using an enhanced chemiluminescence detection system (Amersham).
FACS analysis
To detect apoptotic cells, Annexin V-FITC staining (BD Biosciences, San Jose, CA) was performed according to the manufacturer's instructions. After treatment of cells with 10 mM NAC for 2 h and subsequent exposure to 20 µM curcumin for 18 h, cells were then stained with Annexin V-FITC in the dark and subjected to Flow cytometry using a Bacton-Dickinson FACS Caliber and analyzed by Cell Quest software (Becton-Dickinson, San Jose, CA).
Cell viability assay
The number of viable cells was measured using Cell Counting Kit-8 assay kit (Dojindo Laboratories, Kumamoto, Japen). Cells were seeded at a density of 1×104 per well on 96-well plates and treated with curcumin in the absence or presence of NAC for 20 h. After treatment, 10 µl of CCK-8 solution was added to each well and further incubated for 4 h. The absorbance of each well was measured at 450 nm using a microplate reader (Model 680 microplate reader, Bio-Rad Laboratories). Values are the mean±S.D. for triplicate wells and normalized to the value of control group to determine the % of viability.
Establishment of doxycyclin-inducible E2F4 overexpressing cell line
The recombinant plasmid pcDNA4/TO/Myc/His-B-E2F4 which expresses E2F4 in the presence of doxycycline was constructed by cloning of the E2F4 cDNA into the BamHI and XhoI sites of pcDNA4/TO/Myc/His-B.
HCT116 cells (1×106 cells in 100 mm dish) were transfected with 2 µg of purified plasmids using Lipofectamin™ 2000 (Invitrogen, Carlsbad, CA). To select clones capable of doxycycline-inducible E2F4 expression, RPMI 1640 medium with Blastacidin (5 µg/ml) and Zeocin (100 µg/ml) was added to the cells and the medium was refreshed every 3 days. After 3 weeks, cells that formed colonies were isolated and doxycycline-regulated E2F4 expression of these cells was analyzed by immunoblotting.
Statistical analysis
Data are expressed as the mean±SD values from at least three independent experiments. To determine statistical significance as compared with controls, Student's t and ANOVA test were used with a threshold of p values less than 0.05.
RESULTS
Curcumin suppresses E2F4 expression in HCT116 cells
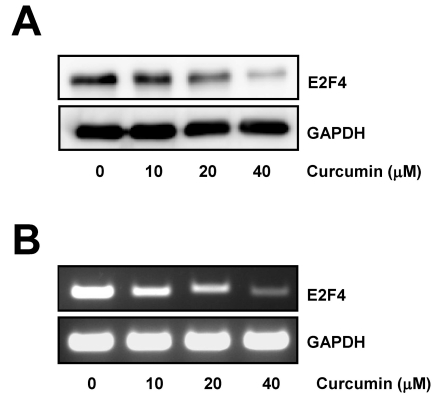
To assess the effect of curcumin on E2F4 expression in colon cancer cells, HCT116 cells were treated with various concentrations of curcumin and the level of E2F4 protein was observed. As shown in Fig. 1A, curcumin markedly suppressed E2F4 expression in a dose-dependent manner.
Fig. 1.
Curcumin induces downregulation of E2F4 expression. (A) HCT116 cells were treated with the indicated concentrations of curcumin for 20 h. Whole cell lysates were prepared and resolved by SDS-PAGE and the level of E2F4 protein was determined with E2F4 specific antibody. Equal amounts of proteins were loaded and immunoblot with anti-GAPDH antibody was used as the loading control. (B) Cells were treated as in (A) for 12 h. Total RNAs from those cells were isolated and used for analysis of E2F4 mRNA expression by RT-PCR. The level of E2F4 mRNA was normalized to that of GAPDH. The data shown are representative of three independent experiments.
Next, we performed RT-PCR with total RNAs isolated from cells which were incubated with a range of concentrations of curcumin for 12 h to examine if the level of E2F4 mRNA was downregulated by curcumin. RT-PCR result showed that E2F4 mRNA level was significantly reduced by curcumin treatment, indicating that curcumin caused downregulation of E2F4 expression at transcriptional level (Fig. 1B).
ROS produced by curcumin is responsible for the cell growth inhibition and the downregulation of E2F4 expression
Since curcumin has been known to generate ROS, which plays a role as a key mediator to exert its biological activities including cell growth control [20-22], we confirmed the effect of cucumin-induced ROS on cell proliferation and examined its effect on E2F4 expression.
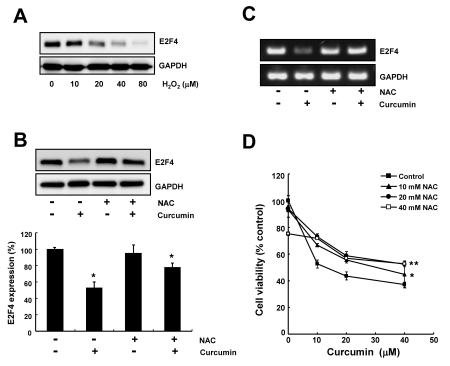
We first examined the effect of hydrogen peroxide on E2F4 expression. As shown in Fig. 2A, the level of E2F4 protein was found to be decreased by hydrogen peroxide treatment (10~80 µM) in a dose-dependent manner.
Fig. 2.
Curcumin-induced ROS is involved in the cell growth inhibition and the downregulation of E2F4 expression. (A) HCT116 cells were treated with indicated concentrations of hydrogen peroxide for 24 h and total cell lysate were analyzed by immunoblotting. GAPDH was used as a loading control. (B) Cells were pre-treated with 10 mM NAC for 2 h and subsequently exposed to 20 µM curcumin for 20 h. Total cell lysate were subjected to immunoblotting. GAPDH was used as a loading control. Relative amount of E2F4 protein was calculated using the software Image Gauge 3.01 (Fujifilm). Each bar represents mean±SD of three independent experiments (*p<0.05, compared with controls). (C) Cells were treated as described in (B). RT-PCR was conducted to determine E2F4 mRNA level. GAPDH was used as an internal control. (D) Cells were pre-treated with 10 mM NAC for 2 h and then exposed to the indicated concentrations of curcumin for 20 h. Cell viability was measured by WST-8 assay. Values are expressed as the mean±SD of three independent experiments performed in triplicate (*p<0.05; control vs 10 mM NAC, **p<0.04; control vs 20 mM NAC).
We next investigated the effect of a potent antioxidant agent, NAC on curcumin-regulated E2F4 expression. We found that downregulation of E2F4 expression by curcumin was markedly recovered by preincubation of cells with NAC (Fig. 2B, 2C). RT-PCR results also showed that curcumin-mediated downregulation of E2F4 expression was abolished by pretreatment of cells with NAC. These data indicate that curcumin-induced ROS is responsible for the suppression of E2F4 expression (Fig. 2B, 2C).
Consistent with Western blot analysis, cell viability assay was also revealed that anti-proliferative activity of curcumin was diminished by preincubation of cells with NAC, since pretreatment of cells with NAC and subsequent exposure of a range of curcumin was shown to interfer curcumin-mediated cell death (Fig. 2D).
Pretreatment of antioxidant blocks curcumin-induced downregulation of E2F4 target gene expression
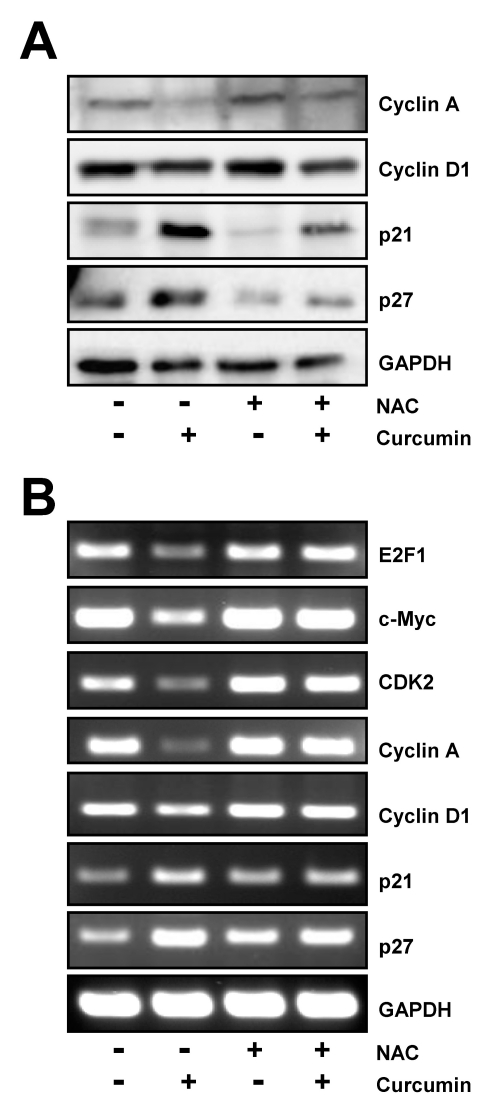
Recently, Garneau et. al reported that E2F4 knockdown led to the decreased expression of several genes such as cyclin A, p21 and p27 which regulate cell cycle progression and slowed proliferation rate in HCT116 cells [11]. Based on their observation, we examined the effect of curcumin and NAC on these genes which is regulated in E2F4-dependent manner.
Western blot analysis showed that curcumin decreased the level of cyclin A, but increased the expression of p21, and p27, cyclin dependent kinase inhibitors. Reduction of cyclin A and induction of p21 and p27 expression by curcumin was blocked by pre-incubation of cells with NAC (Fig. 3A), although there was no significant modulation of cyclin D1 expression which was a bit suppressed by curcumin in the presence of NAC.
Fig. 3.
Regulation of E2F4 target genes by curcumin is restored by NAC pretreatment. HCT116 cells were treated with 10 mM NAC for 2 h and then exposed to 20 µM curcumin for 20 h. (A) Western blot analysis was conducted to assess the expression of several E2F4 target genes such as cyclin A, cyclin D1, p21, and p27. (B) In parallel, the level of several E2F4 target genes including E2F1, c-Myc, CDK2, cyclin A, cyclin D1, p21, and p27 was examined by RT-PCR using each gene specific primers. GAPDH was used as an internal control. Results are from three independent experiments.
We also examined that the effect of NAC on curcumin-mediated modulation of other genes which has known to be affected by E2F4 level at the transcriptional level. In accordance with Western blot results, RT-PCR result showed that curcumin caused considerable decrease of cyclin A and cyclin D1 mRNA expression as well as increase of p21, and p27 mRNA expression. Amplification of E2F1, c-myc and CDK2 mRNAs was also found to be downregulated by curcumin, indicating that they might be the other E2F4-regulated genes. Removal of curcumin-generated ROS with NAC almost completely restored inhibition or induction of these E2F4 target gene expression (Fig. 3B).
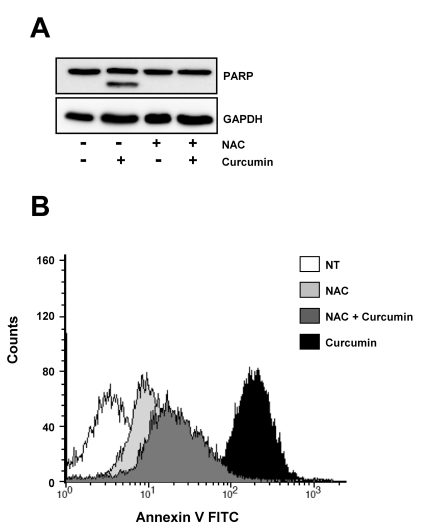
Pretreatment of antioxidant prevents curcumin-derived PARP cleavage and the following apoptotic cell death
Given the effect curcumin and NAC on the expression of E2F4 and its target proteins controlling cell cycle progression and cell death, we examined curcumin-induced PARP cleavage and the following apoptotic cell death in the absence or presence of NAC. As shown in Fig. 4A, curcumin induced PARP cleavage which is a marker of caspase-dependent apoptosis, but NAC pretreatment completely abolished this PARP cleavage of curcumin.
Fig. 4.
Curcumin-derived PARP cleavage and the following apoptotic cell death are blocked by pretreatment of cells with NAC. HCT116 cells were treated as in Fig. 3 and subjected to immunoblotting and flow cytometry analysis. (A) Cleavage of PARP was determined by Western blot analysis. GAPDH was used as a loading control. (B) Cells were stained with Annexin V-FITC and analyzed by flow cytometry. Results are from three independent experiments.
Consistent with the PARP cleavage by curcumin, the percentage of Annexin V positive cells which correspond to apoptotic cells were increased after curcumin treatment, but fairly reduced in the presence of NAC (Fig. 4B).
E2F4 overexpression diminishes the curcumin-induced cytotoxicity
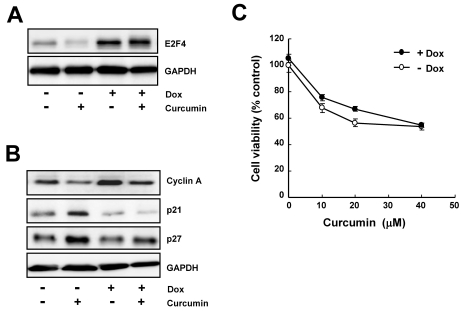
To confirm the role of E2F4 in the curcumin-induced cytotoxicity, we established doxycycline-inducible system which expresses E2F4 protein by the addition of doxycycline. Western blot results showed that E2F4 expression was tightly regulated by doxycycline (Fig. 5A) and the decreased (cyclin A) and increased (p21 and p27) expression by curcumin was significantly restored in E2F4-overexprssing cells (Fig. 5B). By cell viability assay, E2F4 overexpression was found to ameliorate curcumin-induced growth inhibition. These results seem to be a supportive evidence that E2F4 can be an important determinant of curcumin-induced cytotoxicity via regulation of E2F4-dependent expression of several target genes.
Fig. 5.
The overexpression of E2F4 protein is restored the regulation of E2F4 target genes and inhibition of cell proliferation in response to curcumin. HCT116 cells were treated with 1 µg/ml doxycyclin for 18 h and then stimulated with 20 µM curcumin for 20 h. (A) Induction of E2F4 protein was determined by immunoblotting. (B) The level of E2F4 target genes such as cyclin A, cyclin D1, p21, and p27 was assessed by Immunoblotting to determine the effect of E2F4 overexpression. GAPDH was used as a loading control. (C) Cells were treated with the indicated concentration of curcumin in the presence or absence of doxycyclin. Cell proliferation was measured by WST-8 assay. Results are from three independent experiments.
DISCUSSION
Curcumin has been known to have antioxidant and anti-inflammatory properties, which is mainly due to its ability to remove free-radicals such as ROS which mediates peroxidation of various cell components including membrane lipids, DNA, proteins [23]. However, the effect of curcumin on cancer cells is opposite to its activity of free-radical scarvenger. Because it was demonstrated that curcumin rapidly induced ROS, resulting in the release of cytochrome C and apoptosis inducing factor from the mitochondria which are involved in caspase 3-dependent and -independent apoptosis, respectively [17,24]. Our findings also support that anticancer activity of curcumin is mediated by ROS generation, since curcumin-induced inhibition of HCT116 cell growth were suppressed by pretreatment of antioxidant, NAC. Both PARP cleavage and an increase of Annexin V positive cells after curcumin treatment were also blocked by NAC pretreatment.
E2F, a family of transcription factor that plays an important role in controlling development, proliferation, differentiation, and survival, have been divided into either activator or repressor, based on their ability to induce E2F-responsive gene expression which causes the quiescent cells to progress into S-phase [5]. However, many studies manifested that this simple classification is not good enough to explain unique and diverse biological activities of each E2F family members. For example, in tissue culture cells and transgenic mice, overexpression of typical activator E2Fs, E2F1 - 3, has been demonstrated to induce entry into S phase as well as apoptotic cell death, which is accounted a separable and independent function of these E2Fs [25-27].
E2F4, one of the repressor E2F, is another example of E2F with opposing regulatory roles. In normal intestinal epithelial crypt cells and colorectal cancer cells, it was shown that E2F4 was a determinant in cell cycle progression, in contrast to its cell growth arrest potential in other settings [11]. In addition, cyclin-dependent kinase inhibitors and DNA damaging agents has been reported to reduce E2F4 level [28], suggesting that E2F4 deficiency facilitates drug-induced apoptosis. In accordance with these previous reports, we found the involvement of E2F4 in the curcumin-induced cytotoxicity. In curcumin-treated HCT116 colon cancer cells, expression of E2F4 itself and several E2F4 downstream genes, E2F1, c-myc, CDK2, cyclin A, cyclin D1, p21, and p27 which are involved in cell cycle control and related to the apoptotic cell death was reduced. This reduction was restored by pretreatment of NAC and doxycyclin-induced E2F4 expression. Moreover, E2F4 overexpression was found to partially restore curcumin-induced growth inhibition, confirming the role of E2F4 in the cell proliferation. It is noteworthy that partial compensation of curcumin-induced cell death by E2F4 overexpression implies that E2F4 is not the only determinant of curcumin to exert its anti-proliferative activity.
In summary, we demonstrated that curcumin caused ROS generation, which resulted in the downregulation of E2F4 expression and subsequent modulation of E2F4 downstream, which in turn inhibited proliferation of HCT116 colon cancer cells through apoptotic cell death mechanism. Our data propose E2F4 as a novel mediator to facilitate anti-proliferative activity of curcumin.
ACKNOWLEDGEMENTS
This work was supported by Future-based Technology Development Program (Nano Fields, Grant No. 2010-0020603) and the Aging-Associated Vascular Disease Research Center at Yeungnam University (Grant No. 2010-0007352) through the National Research Foundation of Korea (NRF) funded by the Ministry of Korean Government Education, Science and Technology.
ABBREVIATIONS
- ROS
reactive oxygen species
- NAC
N-acetylcystein
- PARP
poly (ADP-ribose) polymerase
- pRb
retinoblastoma protein
- PA
plasminogen activator
References
- 1.Bandara LR, La Thangue NB. Adenovirus E1a prevents the retinoblastoma gene product from complexing with a cellular transcription factor. Nature. 1991;351:494–497. doi: 10.1038/351494a0. [DOI] [PubMed] [Google Scholar]
- 2.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 3.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y, Armanious MK, Thomas MJ, Cress WD. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene. 2000;19:3422–3433. doi: 10.1038/sj.onc.1203682. [DOI] [PubMed] [Google Scholar]
- 5.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 7.Morkel M, Wenkel J, Bannister AJ, Kouzarides T, Hagemeier C. An E2F-like repressor of transcription. Nature. 1997;390:567–568. doi: 10.1038/37507. [DOI] [PubMed] [Google Scholar]
- 8.Cartwright P, Muller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 9.Trimarchi JM, Fairchild B, Verona R, Moberg K, Andon N, Lees JA. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koziczak M, Krek W, Nagamine Y. Pocket protein-independent repression of urokinase-type plasminogen activator and plasminogen activator inhibitor 1 gene expression by E2F1. Mol Cell Biol. 2000;20:2014–2022. doi: 10.1128/mcb.20.6.2014-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau H, Paquin MC, Carrier JC, Rivard N. E2F4 expression is required for cell cycle progression of normal intestinal crypt cells and colorectal cancer cells. J Cell Physiol. 2009;221:350–358. doi: 10.1002/jcp.21859. [DOI] [PubMed] [Google Scholar]
- 12.Abe Y, Hashimoto S, Horie T. Curcumin inhibition of inflammatory cytokine production by human peripheral blood monocytes and alveolar macrophages. Pharmacol Res. 1999;39:41–47. doi: 10.1006/phrs.1998.0404. [DOI] [PubMed] [Google Scholar]
- 13.Oyama Y, Masuda T, Nakata M, Chikahisa L, Yamazaki Y, Miura K, Okagawa M. Protective actions of 5'-n-alkylated curcumins on living cells suffering from oxidative stress. Eur J Pharmacol. 1998;360:65–71. doi: 10.1016/s0014-2999(98)00635-9. [DOI] [PubMed] [Google Scholar]
- 14.Nagano T, Oyama Y, Kajita N, Chikahisa L, Nakata M, Okazaki E, Masuda T. New curcuminoids isolated from Zingiber cassumunar protect cells suffering from oxidative stress: a flow-cytometric study using rat thymocytes and H2O2. Jpn J Pharmacol. 1997;75:363–370. doi: 10.1254/jjp.75.363. [DOI] [PubMed] [Google Scholar]
- 15.Cho Jy, Kang PJ, Chun W, Moon YO, Park YT, Lim SY, Kim SS. Curcumin attenuates glial cell activation but cannot suppress hippocampal CA3 neuronal cell death in i.c.v. Kanic Acid Injection Model. Korean J Physiol Pharmacol. 2003;7:307–310. [Google Scholar]
- 16.Kong PJ, Kwon OY, Han YH, Kim SY, Lee SN, Son HJ, Kim SS. Comparison of Inhibitory Potency of Various Antioxidants on the Activation of BV2 Microglial Cell Lines Induced by LPS. Korean J Physiol Pharmacol. 2007;11:9–13. [Google Scholar]
- 17.Cheng AL, Hsu CH, Lin JK, Hsu MM, Ho YF, Shen TS, Ko JY, Lin JT, Lin BR, Ming-Shiang W, Yu HS, Jee SH, Chen GS, Chen TM, Chen CA, Lai MK, Pu YS, Pan MH, Wang YJ, Tsai CC, Hsieh CY. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or premalignant lesions. Anticancer Res. 2001;21:2895–2900. [PubMed] [Google Scholar]
- 18.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, Marczylo TH, Morgan B, Hemingway D, Plummer SM, Pirmohamed M, Gescher AJ, Steward WP. Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clin Cancer Res. 2004;10:6847–6854. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 19.Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, Abbruzzese JL, Ng CS, Badmaev V, Kurzrock R. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 20.Watson JL, Hill R, Yaffe PB, Greenshields A, Walsh M, Lee PW, Giacomantonio CA, Hoskin DW. Curcumin causes superE2F4 Downregulation by Curcumin 397 oxide anion production and p53-independent apoptosis in human colon cancer cells. Cancer Lett. 2010;297:1–8. doi: 10.1016/j.canlet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Moragoda L, Jaszewski R, Majumdar AP. Curcumin induced modulation of cell cycle and apoptosis in gastric and colon cancer cells. Anticancer Res. 2001;21:873–878. [PubMed] [Google Scholar]
- 22.Lee DH, Choi HC, Lee KY, Kang YJ. Aprotinin Inhibits Vascular Smooth Muscle Cell Inflammation and Proliferation via Induction of HO-1. Korean J Physiol Pharmacol. 2009;13:123–129. doi: 10.4196/kjpp.2009.13.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menon VP, Sudheer AR. Antioxidant and anti-inflammatory properties of curcumin. Adv Exp Med Biol. 2007;595:105–125. doi: 10.1007/978-0-387-46401-5_3. [DOI] [PubMed] [Google Scholar]
- 24.Woo JH, Kim YH, Choi YJ, Kim DG, Lee KS, Bae JH, Min DS, Chang JS, Jeong YJ, Lee YH, Park JW, Kwon TK. Molecular mechanisms of curcumin-induced cytotoxicity: induction of apoptosis through generation of reactive oxygen species, down-regulation of Bcl-XL and IAP, the release of cytochrome c and inhibition of Akt. Carcinogenesis. 2003;24:1199–1208. doi: 10.1093/carcin/bgg082. [DOI] [PubMed] [Google Scholar]
- 25.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young AP, Nagarajan R, Longmore GD. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene. 2003;22:7209–7217. doi: 10.1038/sj.onc.1206804. [DOI] [PubMed] [Google Scholar]
- 27.Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 28.DuPree EL, Mazumder S, Almasan A. Genotoxic stress induces expression of E2F4, leading to its association with p130 in prostate carcinoma cells. Cancer Res. 2004;64:4390–4393. doi: 10.1158/0008-5472.CAN-03-3695. [DOI] [PubMed] [Google Scholar]