Abstract
The Bacillus anthracis genome reflects its close genetic ties to B. cereus and B. thuringiensis but has been shaped by its own unique biology and evolutionary forces. The genome is comprised of a chromosome and two large virulence plasmids, pXO1 and pXO2. The chromosome is mostly co-linear among B. anthracis strains and even with the closest near neighbor strains. An exception to this pattern has been observed in a large inversion in an attenuated strain suggesting that chromosome co-linearity is important to the natural biology of this pathogen. In general, there are few polymorphic nucleotides among B. anthracis strains reflecting the short evolutionary time since its derivation from a B. cereus-like ancestor. The exceptions to this lack of diversity are the variable number tandem repeat (VNTR) loci that exist in genic and non genic regions of the chromosome and both plasmids. Their variation is associated with high mutability that is driven by rapid insertion and deletion of the repeats within an array. A notable example is found in the vrrC locus which is homologous to known DNA translocase genes from other bacteria.
Keywords: Variable Number Tandem Repeats, chromosomal inversion, phylogeny
1. The evolutionary context of the B. anthracis Genome
Bacillus anthracis is a clonal species that is nested inside of the B. cereus and B. thuringiensis group of strains. It has been suggested that this group be reclassified as a single species (B. cereus) and that subspecies designation could then be assigned to various specialized groups within the species (Helgason et al., 2000). Due to its unique pathogenic biology, B. anthracis would probably be simply a subspecies of B. cereus. The traditional systematic nomenclature has been driven by extreme phenotypes in the group including the effects from insect-toxin genes found in B. thuringiensis strains and, of course, the catastrophic pathology from anthrax. These phenotypes are clinically and biologically important but make poor taxonomic guides due to their non-monophyletic distribution amongst strains. This is doubtlessly due to the intense adaptive fitness advantage offered by these traits and their control by mobile genetic elements. These critical and distinct phenotypes are frequently controlled by genes on unique extra chromosomal plasmids. For example, B. thuringiensis strains kill insect using cry and other toxins, while B. anthracis must produce both the anthrax protein toxins and synthesize the rare poly-gamma-D-glutamic acid capsule. In the end, the evolutionary tree of these bacilli can be more accurately predicted from chromosomal nucleotide sequences (Hegalson et al., Priest et al (2004), rather that the complex, diverse and horizontally transferred family of large plasmids.
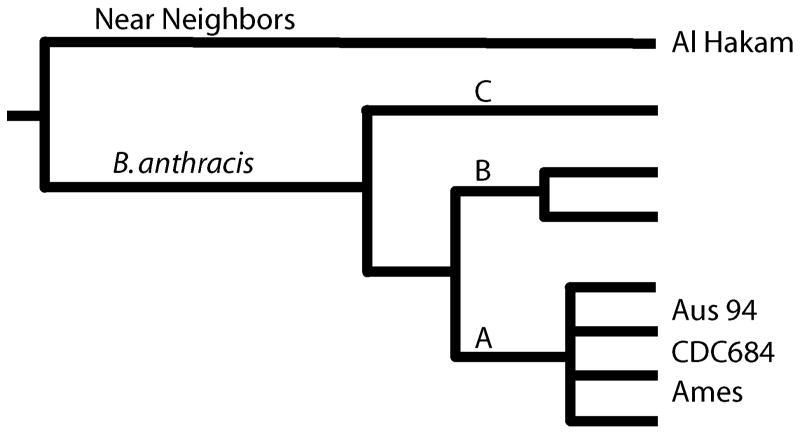
The availability of whole genome sequences has made the reconstruction of the B. anthracis phylogeny extremely accurate. In a clonal organism, phylogenetic analysis methods (e.g., maximum parsimony) are the best approach for estimating population structure. Figure 1 is a cartoon reproduction of the detailed work of Pearson (Pearson et al., 2004; Pearson et al., 2009), which may represent the most accurate phylogenetic reconstruction for any species. This is possible in B. anthracis primarily due to its low genetic diversity and the absence of measurable lateral transfer of genetic material since its derivation as a species. The lack of diversity is presumably due to a short evolutionary history that has precluded mutational saturation in the single nucleotide polymorphism (SNP) characters that generally have been discovered by whole genome sequencing in B. anthracis. A short evolutionary time frame is not necessarily a short chronological timeframe. The important soil spore reservoir stage of the B. anthracis lifecycle could have greatly influenced the evolutionary rate of this organism, making long temporal periods appear considerably shorter. Most genomic mutations occur during DNA replication and a spore may lay dormant for years. Calibrating the evolutionary rate to the chronological rate is always problematic, particularly in a spore forming bacterium. Hence, the lack of diversity in B. anthracis is due in part to this pathogen’s lifestyle and in part to its recent emergence from a non pathogenic relative.
Figure 1.
Phylogenetic structure of B. anthracis.
1.1 B. anthracis Population Structure
The B. anthracis substructure is divided into three major lineages (A, B, C) with the A clade being the most important, globally dispersed causative form of anthrax (>90% of all cases, see Van Ert et al, 2007). The A sub-linage radiates into multiple closely related and widely dispersed subgroups (Keim et al., 1997; Keim et al., 2000; Van Ert et al., 2007). While there is topological complexity in this group, it is only resolved by using whole genome sequences to discover a small number of SNPs (a few hundred). However, once found, these SNPs are highly dependable characters and with high consistency in phylogenetic reconstructions (Pearson et al., 2004; Pearson et al., 2009). The two B lineages are important in certain geographic regions but do not have the wide global distribution as the A lineage. There are two important subclades within the B group. The B1 subclade has been previously found in Southern Africa (Gierczynski et al., 2004; Keim et al., 2000; Smith et al., 2000; Van Ert et al., 2007) where it co-exists in space and time with strains from the A clade. The B2 clade has been reported more widely in southern and eastern Europe (Gierczynski et al., 2004; Keim et al., 2000; Van Ert et al., 2007) and in a single report from California in the USA (Van Ert et al., 2007). The basally derived C lineage is an enigma due to its source and rarity. Two independent isolates have been found in the United States (Van Ert et al., 2007), perhaps indicating a New World source population for the other two major clades of B. anthracis. However, the relatively recent introduction of the A and B clades from the Old World (Kenefic et al., 2008; Simonson et al., 2009), is inconsistent with this conclusion and multiple transports of the ancestral population across the continents lacks parsimony. North America is the most heavily sampled continent for B. anthracis evolutionary studies and this may bring bias into our view of the C lineage. More sampling in different global locales will help resolve this quandary.
1.2 B. anthracis Near Neighbors
The near-neighbors to B. anthracis are a mixture of both B. thuringiensis and B. cereus strains (Hill et al., 2004) illustrating the problematic nature of species designations in this bacterial group. These two species are not monophyletic and their differentiation is based upon phenotypes that poorly represent their evolutionary history.
The Al Hakam B. thuringiensis strain was isolated from an Iraqi spore production facility and represents one of the very closest relatives to B. anthracis (Challacombe et al., 2007). Even so, distinguishing B. anthracis from its nearest relatives can be readily accomplished using phylogenetically identified characters on the branch leading to the B. anthracis clade. These characters have included SNPs (Bode et al., 2004) and a particularly important mutation in the plcR gene (Easterday et al., 2005; Gohar et al., 2008). This frame-shift mutation may have a pleiotrophic effect, due to its regulatory function, that may biologically differentiate B. anthracis from its relatives (Gohar et al., 2008). It is now clear that the presence of toxin and capsule genes are not reliable B. anthracis identifier as these are being observed in other Bacilli strains (Hoffmaster et al., 2004; Leendertz et al., 2004; Pannucci et al., 2002a; Pannucci et al., 2002b). These relatives of B. anthracis may be pathogenic but it is not clear if they are causing anthrax. Detailed studies in animal models are needed to resolve this issue of “Anthrax, but not Bacillus anthracis?” (Okinaka et al., 2006).
2. A Chromosome and Two Plasmids
The B. anthracis genome is tripartite and comprised of a single circular chromosome and two circular virulence plasmids. The genome nucleotide composition is highly biased towards adenine and thymine, with only ~35% of the bases from guanine and cytosine. The prevalence of A+T means that this DNA has a higher buoyant density and lower melting temperatures than many others. The plasmids are relatively large and code for many different genes, but importantly they carry the toxin and capsule determinants. The pXO1 plasmid (Okinaka et al., 1999b) has pagA (protective antigen and a intra membrane toxin transporter), lef (lethal factor, which is a Zn2+-dependent endoprotease) and cyaA (edema factor, which is a calmodulin-sensitive adenylate cyclase). The pXO2 plasmid (Okinaka et al., 1999a) carries the capsule biosynthesis genes found in a cluster and is essential for full anthrax disease. The Sterne strain that is commonly used as a live veterinarian vaccine is missing pXO2 but still has the pXO1 plasmid with its toxins. As denoted by its name, the protective antigen protein elicits an immune response that is very effective in preventing disease. Its role as the toxin transporter can be disrupted by specific antibodies. While virulence involvement of specific chromosomal genes is not as definitive, there are many candidates. The chromosomal background of the plasmid may be the difference between an opportunistic pathogen and one that causes catastrophic disease, with a global distribution.
2.1 The Ames Genome
While the quality of a particular genomic sequence is often hard to assess, it is important to recognize that most genome sequences have multiple errors that can range from just a few nucleotide mistakes to large-scale assembly problems. Draft sequences in particular have not had the same level of annotation nor curaton, and verification as complete closed genomes. The best publicly available bacterial genome sequence is from the Ames ancestor strain (Ravel et al., 2009). The intense focus on the Ames strain during the anthrax letter attack investigation lead to a highly accurate sequence determination that may be completely free of errors. The DNA was generated from the frozen stock used for strain distribution to many different laboratories, worldwide. Hence, it is an important reference genome for research purposes. This also represents a stock that is relatively close to the original 1981bovine isolate and should be representative of wild type strains due to the limited number of passages separating it from a natural event.
Overall, the ancestral Ames genome is 5,503,926 nucleotides in size with 5,775 protein coding genes identified (Ravel et al., 2009; Read et al., 2003). In addition, there are 33 ribosomal RNA genes (23S, 16S and 5S) arranged in 11 operons and, along with 95 tRNA genes, found exclusively on the chromosome. The chromosome itself represents about 95% of the genome with the two large plasmids containing the remaining coding capacity (pXO1=181,677 bp; pXO2=94,830 bp).
2.2 Chromosome Structure
The chromosome structure of B. anthracis is relatively conserved with few large rearrangements. In certain other pathogens, relatively recent, large and numerous chromosomal rearrangements occur after the pathogens emerge and “niche shifts” to become obligate pathogens. Yersinia pestis (Achtman et al., 2004; Parkhill et al., 2001) and Francisella tularensis (Beckstrom-Sternberg et al., 2007; Champion et al., 2009; Larsson et al., 2009) are two notable examples where IS-element numbers increased dramatically and chromosomal rearrangements mediated by these repeated sequences occur in populations, which otherwise have very few polymorphisms. The interpretation of the increased frequency of such changes is that much of the genome is released from selective pressure due to the “niche shift”. In the relatively narrow new niche, much of the genome is no longer needed and is open for random mutation.
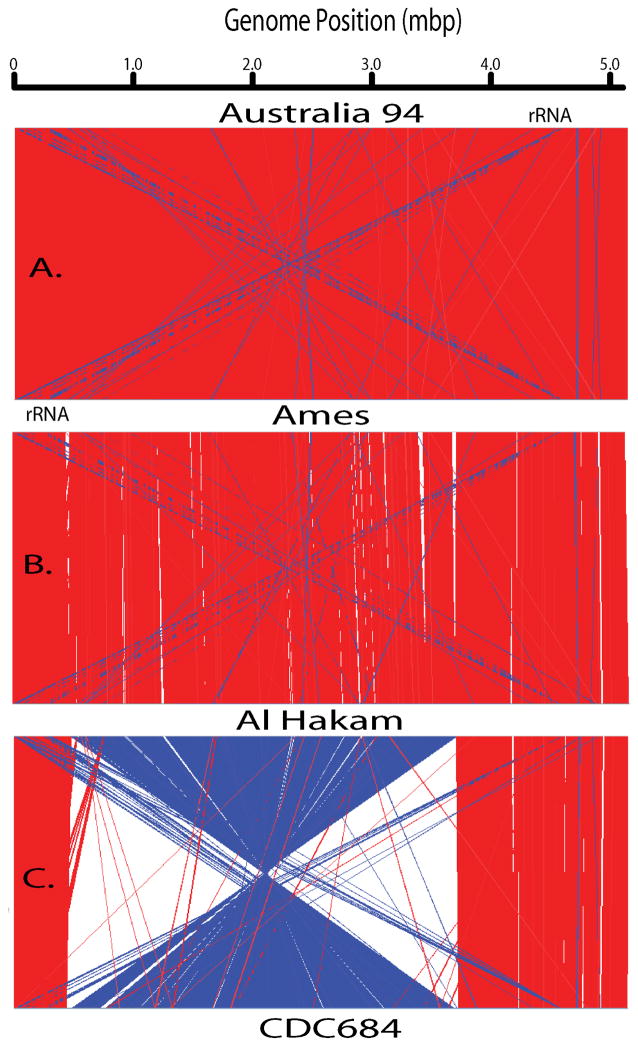
Figure 2 illustrates the relatively high conservation of gene order by aligning the several genomes and then graphically connecting regions of great similarity. Panel A is a comparison of two genomes (Australia 94 and Ames) from the A-clade that represent a relatively close relationship. This comparison has the fewest differences with a dominance of red lines indicative of high co-linearity between the genomes. Under these analytical conditions, there are essentially no indels (white sectors) observed and only a few inversions (blue). Examination of the blue-line regions annotation indicates the presence of repeated sequence elements that are oriented in opposite directions. These are not true chromosomal inversions, but rather paralogous repeated elements oriented in an inverted fashion. Most notable are the rRNA gene operons (5S, 16S and 23S), where one rRNA operon near 4.5 mbp is inverted relative to multiple other rRNA operons between 0 and 1.0 Mb. This creates two blue diagonal “spotlight” patterns evident in all three panels due to the conserved positions of these genes. The other inversions in panel A include protein genes such as a sodium/alanine symporter family protein, a UDP-N-acetulglucosamine 2-epimerase, and hypothetical proteins. The repeated and inverted nature of the rRNA operons is to be expected due to their great sequence conservation and their distribution on both sides of the replication origin. But, the inversion of these other protein genes appears to be more happenstance and may be due to stochastic events and adjacent DNA sequence structure.
Figure 2.
Alignments of four whole chromosomes.
The Artemis software (Rutherford et al., 2000) was used to analyze four closed genomes. This figure is a series of lines connecting pairwise regions of each genome. The red lines indicate sequence similarity between directly repeated regions while the blue lines are indicative of sequence similarity in a reverse orientation. White regions are indels, indicating the presence/absence of a particular region between the genome pairs.
A more distant relationship is shown in Figure 2B where the near neighbor B. thuringiensis Al Hakam and the B. anthracis Ames genomes are compared. Based upon the phylogenetic tree (Figure 1), this is a relatively distant comparison, but the chromosomal co-linearity is still very apparent by the massive red regions across the entire chromosome. In addition to the previously mentioned inversions, there are additional indels apparent on both sides of this comparison. The largest and most evident insertions in Ames relative to Al Hakam represent prophage differences. Other insertions involve metabolic genes and perhaps adaptive genomic islands. But relative to other pathogens, these are very small differences and argue for the importance of subtle changes in the chromosome that defines B. anthracis.
The final chromosomal structure comparison is to the laboratory strain CDC684, which has both virulence plasmids (Ezzell et al., 1990) and yet is avirulent (Figure 1C). In addition to the small repeated and inverted sequences observed in the other two panels, CDC684 has a massive inversion involving more than half of the genome. While the inversion is represented as if it occurred across the replication terminus (ca. position 2.5 mbp), it just as well could have occurred across the origin of replication (position 0). The strand orientation of the genome position is set by origin and, hence, it appears to have occurred across the terminus in this figure.
Could such an inversion affect virulence? It is known that gene orientation in the chromosome is correlated to their expression level and all the inverted genes would now be altered in orientation. While this change is clearly not a lethal alteration, it could have disrupted gene expression sufficiently to make this strain avirulent in the animal models.
Given the otherwise conservation of gene arrangement, is seems likely that the gene order is important for the biology of the pathogen. Major alterations are possibly not tolerated by B. anthracis when under natural ecological pressures. The CDC684 chromosomal inversion demonstrates that such changes are possible, though there may be biological consequences.
3. Variable Number Tandem Repeats
Despite the paucity of SNPs (Pearson et al., 2004) and rarity of larger genome rearrangements, there are regions in the genome that vary greatly from strain to strain. Variable number of tandem repeated sequences (VNTRs) are found in many different chromosomal and plasmid locations and may exhibit a few or even dozens of different allelic states when global strain populations are examined. The basis for this diversity is a greatly elevated mutational rate that is based primarily upon slip strand mismatch repair. The rules for these replication errors have been extensively studied in other bacteria (Vogler et al., 2006; Vogler et al., 2007) and they appear similar in B. anthracis. As the DNA polymerase replicates across short tandem repeated sequences, it may “slip backwards” and replicate a particular repeat twice; or alternatively “hop forward” and not replicate a particular repeat. The errors most commonly occur for single repeat units and less commonly for two or more repeat units during a single mutational event (Vogler et al., 2006; Vogler et al., 2007). The repeated regions behave as small populations (Ohno, 1970) where substitution mutations may occur within a repeat unit and then multiply by spreading to adjacent individuals or, alternatively, go extinct as a particular repeat is deleted. These arrays expand and contract in a cyclic fashion that will homogenize the different repeats. A cis-dependent homogenization of sequences has been documented for tandem arrays in many different species. The homogenization process is in competition with substitution mutations that would diversify and slowly degrade the tandem arrays. Highly homogenous tandem arrays are evidence that the expansion and contraction cycle is rapid, dynamic and purifying (fixing or removing) the rare substitution mutation. This rapid evolution results in greatly differing numbers of repeated units at any particular locus, across the global or even local populations of B. anthracis (Keim et al., 2000; Van Ert et al., 2007).
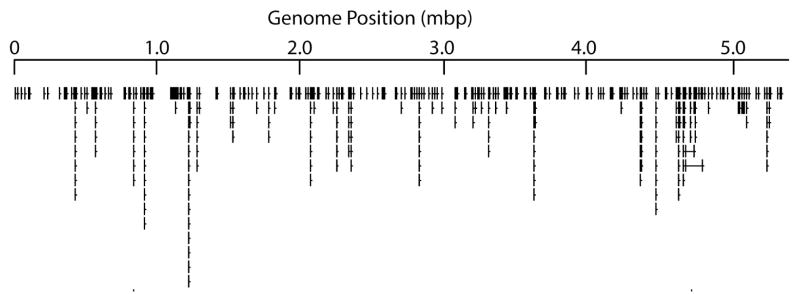
Bacillus anthracis VNTRs were first discovered in the hypothetical protein gene called vrrA (variable repeat region A) by Andersen (Andersen et al., 1996) and later characterized across multiple strains by Jackson and Keim (Jackson et al., 1997). Turnbull’s group was doubtlessly observing variation in this same genomic region with their use of arbitrarily primed PCR (Henderson et al., 1994), which lead to Andersen’s discovery of vrrA. Additional variable number tandem repeats were discovered at the “wet bench” after genomic variation was detected with AFLP markers (Keim et al., 1997; Schupp et al., 2000). This was laborious “wet bench” work involving marker analysis of multiple DNAs followed by marker fragment isolation and sequencing. All of this changed dramatically once whole genome sequences were available and candidate VNTRs could discovered in silico. The first complete sequences were from the plasmids pXO1 and pXO2 (Okinaka et al., 1999a; Okinaka et al., 1999b), that quickly lead to the identification of the “AAT” and “AT” repeats in each, respectively (Keim et al., 1999; Keim et al., 2000). The Porton Downs “Ames” chromosome sequence came later (Read et al., 2003), but its availability lead to the identification of numerous repeated loci (Lista et al., 2006). These regions can be graphically represented by an “icicle plot” where the tandem arrays are projected perpendicular from the genome sequence and where the repeat number of each tandem array is shown by the length of the “icicles.” In figure 3, only the longest tandem arrays are shown, but regardless there are more than two dozen or more apparent.
Figure 3.
“Icicle” plot of tandemly repeated sequences.
This is a graphical representation of the tandemly repeated loci in the Ames genome. The longer “icicles” are indicative of longer tandem arrays at those loci.
3.1 VNTRs as subtyping tools
VNTRs were critical to differentiating among strains of B. anthracis, when all other methods were inadequate. The use of Multiple Locus VNTR Analysis (MLVA) was pivotal to the identification of different subpopulations and the precise identification of particular strains (Hoffmaster et al., 2002; Keim et al., 1999; Keim et al., 2000). The MLVA8 sub-typing system characterized strains and allowed for the judicious selection of strains for whole genome analysis (Pearson et al., 2004). In search of additional discriminatory power and to provide better phylogenetic estimation, additional VNTR loci have been added by different research groups (Lista et al., 2006; Van Ert et al., 2007). This can be problematic, as rapidly evolving loci can lead to homoplasy due to convergent evolution. Keim et al. (Keim et al., 2004) have suggested that a combination of VNTRs and SNPs represents a better approach, termed PHRANA. Select highly stable and phylogenetically informative SNPs (canonical SNPs) can be used to categorize an unknown isolate into a defined clade, while subsequent use of MLVA discriminates among closely related isolates. Hierarchical use of these subtyping systems maximizes speed, phylogenetic accuracy and resolution, while minimizing cost.
3.2 Single Nucleotide Repeat VNTRs
Perhaps, the most mutable and variable VNTRs are a special category known as single nucleotide repeat sequences (SNRs). The repeat length in these VNTRs is only a single nucleotide, which in most cases is an A/T single nucleotide. This is due to the high A+T content of the B. anthracis genome. In the Ames genome there are more than 50 SNRs known that are greater than 9 nucleotides in length. One locus on the pXO2 plasmid, HM1, is notable for its large number of alleles and large repeat number. HM-1 has some alleles that exceed 50 nucleotides and many different allele size between 9 and 60 has been observed. Kenefic (Kenefic et al., 2008) was able to differentiate even very closely related isolates from a North American outbreak due to the rapid evolution of these sequences. These SNR loci are almost always in non protein coding regions and their biological effect is still obscure.
3.4 VNTRs in Genes
However, many VNTRs can be found in protein coding regions of the genome. When triplet, or multiples of three (3X), repeat sizes are observed, it is almost always indicative of a protein gene location. As the VNTR expands or contracts, a repeat size of 3X will not change the translational reading frame, though the amino acid composition of the protein will still be altered. The collagen-like protein encoded by the bclA gene is one of the better characterized examples of VNTR protein gene in B. anthracis. This glycoprotein is a structural constituent of the exosporium filaments on the spore surface (Sylvestre et al., 2002). As the tandem arrays within the gene become longer, the filaments also increase in length (Sylvestre et al., 2003). While VNTRs have been implicated in many biological phenomena including phase variation (Weiser et al., 1989), this is the only current B. anthracis example of a phenotypic effect controlled by changes in a VNTR. Even here the variation is only detected by special staining of electron micrographs, so we clearly have much to learn about the biological importance of VNTRs in this pathogen.
4. vrrC Evolution
The evolution of VNTRs within genes can be illustrated by taking a detailed look the vrrC locus in B. anthracis. This gene has a notable tripartite VNTR structure (Figure 4) that greatly alters the predicted protein, a DNA translocase. The VNTR attribute of this gene is not observed in the B. subtilis homologs, though the VNTR does occupy a coding region that has lower evolutionary conservation when compared across species. In other words, the vrrC VNTR is not required by other bacteria and its presence in the B. anthracis gene may not be biologically important but simply tolerated in a flexible region or the protein.
Figure 4.
A dotplot homology analysis of the vrrC locus.
The VNTR region of the vrrC gene was analyzed using a dot plot algorithm, which compares a particular sequence against itself. The diagonal lines are indicative of directly repeated DNA sequences in this region of the genome. The strong diagonal line in the middle is the identity line and each half of the box contains the same information, in a mirrored fashion. Note that there are three separate tandemly repeated subregions, termed vrrC1, vrrC2, and vrrC3. The strong diagonal lines near the identity line, show the level of identity between direct repeats and the spacing also is indicative or the repeat length. Each subregion is compared to the others in a pair-wise fashion as indicated: a) vrrC1 × vrrC2; b) vrrC1 × vrrC3; and c) vrrC2 × vrrC3.
4.1 vrrC is a DNA translocase
The predicted vrrC protein shows homology to members of the FtsK-SpoIIIE DNA translocase protein family. The large vrrC ORF spans 3,651 bp of the genome, and encodes a putative 1,217 amino acid protein (Ames genome). The putative vrrC polypeptide is rich in glutamic acid and valine, common in the VNTR repeat motifs. SpoIIIE is a DNA-dependent ATPase (Wu and Errington, 1994) required for post-septational translocation of the forespore chromosome during sporulation. In addition, SpoIIIE has been shown to aid chromosome separation when normal vegetative cell division is defective (Britton and Grossman, 1999; Sharpe and Errington, 1995). A role for SpoIIIE has been implicated in the final stages of spore engulfment (Sharpe and Errington, 1995). The vrrC protein contains the conserved ATP binding P-loop motif found in the FtsK/SpoIIIE DNA translocase proteins. This nucleotide-binding domain confers functionally important ATPase activity to B. subtilis SpoIIIE (Bath et al., 2000). The conserved nucleotide-binding domain is found close to the carboxyl end of vrrC, away from the variable region which is in central portion. The greatest similarity is between vrrC and the Ytpt protein, a B. subtilis SpoIIIE homolog of unknown function. A putative B. anthracis SpoIIIE sequence, distinct from vrrC, was also identified in the Ames genome. Hence, the exact cellular role for the vrrC protein has yet to be identified.
4.2 vrrC has a tripartite VNTR
The full vrrC VNTR is spread across three separated repeat arrays (Figure 4). The repeat motif length is 36 bp in vrrC1, 18 bp in vrrC2, and 42 bp in vrrC3. A sequence alignment dot plot illustrates the homology within and among discrete repeat arrays (Figure 1). The dominant motif 5′ to 3′ in vrrC1 is: GAA(G/A) (A/T)(A/G)(T/C)(T/C) AGAAGAAGT (G/A)GAAGTA(A/G)(T/C) TGC(A/G)GAA (A/G)C(A/G). There is a nine bp insertion (GAAAAATTA) between two repeats to the 3′ end of the array. There are three dominant vrrC2 motifs, which appear to alternate at random. In order of descending frequency within the array the motif sequences 5′ to 3′ are: G(T/C)AGAAG(A/G)ACAA(C/T)CAGTT, G(C/T)AGA(G/A)GAA(G/A)CA(C/T)C(G/A)TC, and GTAGAAGAA(A/G)CACCGATT. In vrrC3 the dominant 5′ to 3′ motif is: CAAGTAG(T/A)(A/G) G(T/A)(G/A)(G/C)A(A/G) CC(A/G)CAAGTGGAAGA(A/G)AA(A/G) C(C/A)A(A/G)TGCA(G/A). There is a three bp deletion in one of the repeats in the sequence array.
Because of high levels of polymorphism observed at two of the vrrC tandem repeat sequences, vrrC1 and vrrC2, these loci were employed in a large MLVA study of 426 global B. anthracis isolates (Keim et al., 2000). Polymorphism in vrrC may be measured as the number of haplotypes (alleles) at each array locus among diverse isolates of B. anthracis. As previously described, 11 vrrC1 haplotypes and four vrrC2 haplotypes were identified (Keim et al., 2000). Three haplotypes have been observed for vrrC3 in a set of 94 diverse strains. Locus polymorphism in vrrC1 and vrrC2 determined by sequence analysis shows insertion/deletion events ranging from 18 to 72, and 3 to 36 nucleotides, respectively. The Nei’s Diversity Index, calculated as 1 − Σ (allele frequency)2, is 0.58, 0.50, and 0.12, for vrrC1, vrrC2, and vrrC3, respectively. By random chance, any two strains will have different alleles at these loci with the probability of 0.58, 0.50 and 0.12. These are very high values for a species where strains were impossible to differentiate by more traditional methods.
Regardless of the biological role, we can use vrrC to understand how VNTR loci are created and continue to evolve. In all three subloci, the repeat structure is a multiple of three and mutational changes do not disrupt the reading frame. The 36, 19 and 42 nucleotide repeat structures, however, is more complex as degenerate sub-repeat structure is obvious in all three subloci. That these repeat structures are always in multiple of three suggests that each sub-region went through an evolutionary progression. A single codon was repeated to generated an initial tandem array, but eventually substitution mutation converted this into a longer-repeat (multiple codon) array. The original single-codon array degenerated but is still evident in the subrepeat structure.
While the three adjacent VNTRs are now independently evolving, they have similar sequences arguing for a common ancestral VNTR. In figure 4, there are pairwise comparisons among the subloci labeled a, b and c. The relationship between vrrC2 and vrrC3 seems the closest (c), with vrrC2 and vrrC3 next (a), and the vrrC1 and vrrC3 (b) the most distant. A simple interpretation would be suggest that vrrC1 and vrrC2 evolved from a common ancestral VNTR first, followed later by the divergence of vrrC2 and vrrC3. Homogenization is a cis-dependent process in the chromosome and it is possible that once a tandem array surpasses some critical size, substitution mutations and within-array processes promote the development of two independent adjacent VNTRs. In the case of vrrC, this happened twice, sequentially, to create a tripartite sequence structure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen GL, Simchock JM, Wilson KH. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J Bacteriol. 1996;178:377–384. doi: 10.1128/jb.178.2.377-384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Beckstrom-Sternberg SM, Auerbach RK, Godbole S, Pearson JV, Beckstrom-Sternberg JS, Deng Z, Munk C, Kubota K, Zhou Y, Bruce D, et al. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PLoS One. 2007;2:e947. doi: 10.1371/journal.pone.0000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode E, Hurtle W, Norwood D. Real-time PCR assay for a unique chromosomal sequence of Bacillus anthracis. J Clin Microbiol. 2004;42:5825–5831. doi: 10.1128/JCM.42.12.5825-5831.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Grossman AD. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J Bacteriol. 1999;181:5860–5864. doi: 10.1128/jb.181.18.5860-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challacombe JF, Altherr MR, Xie G, Bhotika SS, Brown N, Bruce D, Campbell CS, Campbell ML, Chen J, Chertkov O, et al. The complete genome sequence of Bacillus thuringiensis Al Hakam. J Bacteriol. 2007;189:3680–3681. doi: 10.1128/JB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, Borowsky M, Young S, Koehrsen M, Engels R, et al. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterday WR, Van Ert MN, Simonson TS, Wagner DM, Kenefic LJ, Allender CJ, Keim P. Use of single nucleotide polymorphisms in the plcR gene for specific identification of Bacillus anthracis. J Clin Microbiol. 2005;43:1995–1997. doi: 10.1128/JCM.43.4.1995-1997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzell JW, Jr, Abshire TG, Little SF, Lidgerding BC, Brown C. Identification of Bacillus anthracis by using monoclonal antibody to cell wall galactose-N-acetylglucosamine polysaccharide. J Clin Microbiol. 1990;28:223–231. doi: 10.1128/jcm.28.2.223-231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierczynski R, Kaluzewski S, Rakin A, Jagielski M, Zasada A, Jakubczak A, Borkowska-Opacka B, Rastawicki W. Intriguing diversity of Bacillus anthracis in eastern Poland--the molecular echoes of the past outbreaks. FEMS Microbiol Lett. 2004;239:235–240. doi: 10.1016/j.femsle.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Gohar M, Faegri K, Perchat S, Ravnum S, Okstad OA, Gominet M, Kolsto AB, Lereclus D. The PlcR virulence regulon of Bacillus cereus. PLoS One. 2008;3:e2793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto AB. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis--one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–2630. doi: 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I, Duggleby CJ, Turnbull PC. Differentiation of Bacillus anthracis from other Bacillus cereus group bacteria with the PCR. Int J Syst Bacteriol. 1994;44:99–105. doi: 10.1099/00207713-44-1-99. [DOI] [PubMed] [Google Scholar]
- Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, Bliss KA, Laker M, Pardington PE, Richardson AP, Tonks M, et al. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl Environ Microbiol. 2004;70:1068–1080. doi: 10.1128/AEM.70.2.1068-1080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmaster AR, Fitzgerald CC, Ribot E, Mayer LW, Popovic T. Molecular subtyping of Bacillus anthracis and the 2001 bioterrorism-associated anthrax outbreak, United States. Emerg Infect Dis. 2002;8:1111–1116. doi: 10.3201/eid0810.020394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PJ, Walthers EA, Kalif AS, Richmond KL, Adair DM, Hill KK, Kuske CR, Andersen GL, Wilson KH, Hugh-Jones M, et al. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl Environ Microbiol. 1997;63:1400–1405. doi: 10.1128/aem.63.4.1400-1405.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Kalif A, Schupp J, Hill K, Travis SE, Richmond K, Adair DM, Hugh-Jones M, Kuske CR, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Klevytska AM, Price LB, Schupp JM, Zinser G, Smith KL, Hugh-Jones ME, Okinaka R, Hill KK, Jackson PJ. Molecular diversity in Bacillus anthracis. J Appl Microbiol. 1999;87:215–217. doi: 10.1046/j.1365-2672.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, Okinaka R, Jackson PJ, Hugh-Jones ME. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Van Ert MN, Pearson T, Vogler AJ, Huynh LY, Wagner DM. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect Genet Evol. 2004;4:205–213. doi: 10.1016/j.meegid.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Kenefic LJ, Pearson T, Okinaka RT, Chung WK, Max T, Trim CP, Beaudry JA, Schupp JM, Van Ert MN, Marston CK, et al. Texas isolates closely related to Bacillus anthracis Ames. Emerg Infect Dis. 2008;14:1494–1496. doi: 10.3201/eid1409.080076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Elfsmark D, Svensson K, Wikstrom P, Forsman M, Brettin T, Keim P, Johansson A. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leendertz FH, Ellerbrok H, Boesch C, Couacy-Hymann E, Matz-Rensing K, Hakenbeck R, Bergmann C, Abaza P, Junglen S, Moebius Y, et al. Anthrax kills wild chimpanzees in a tropical rainforest. Nature. 2004;430:451–452. doi: 10.1038/nature02722. [DOI] [PubMed] [Google Scholar]
- Lista F, Faggioni G, Valjevac S, Ciammaruconi A, Vaissaire J, le Doujet C, Gorge O, De Santis R, Carattoli A, Ciervo A, et al. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 2006;6:33. doi: 10.1186/1471-2180-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin, New York: Springer-Verlag; 1970. [Google Scholar]
- Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, Keim P, Koehler T, Lamke G, Kumano S, Manter D, et al. Sequence, assembly and analysis of pX01 and pX02. J Appl Microbiol. 1999a;87:261–262. doi: 10.1046/j.1365-2672.1999.00883.x. [DOI] [PubMed] [Google Scholar]
- Okinaka R, Pearson T, Keim P. Anthrax, but not Bacillus anthracis? PLoS Pathog. 2006;2:e122. doi: 10.1371/journal.ppat.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, Koehler TM, Lamke G, Kumano S, Mahillon J, et al. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J Bacteriol. 1999b;181:6509–6515. doi: 10.1128/jb.181.20.6509-6515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci J, Okinaka RT, Sabin R, Kuske CR. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J Bacteriol. 2002a;184:134–141. doi: 10.1128/JB.184.1.134-141.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci J, Okinaka RT, Williams E, Sabin R, Ticknor LO, Kuske CR. DNA sequence conservation between the Bacillus anthracis pXO2 plasmid and genomic sequence from closely related bacteria. BMC Genomics. 2002b;3:34. doi: 10.1186/1471-2164-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Pearson T, Busch JD, Ravel J, Read TD, Rhoton SD, U’Ren JM, Simonson TS, Kachur SM, Leadem RR, Cardon ML, et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci U S A. 2004;101:13536–13541. doi: 10.1073/pnas.0403844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T, Okinaka RT, Foster JT, Keim P. Phylogenetic understanding of clonal populations in an era of whole genome sequencing. Infect Genet Evol. 2009 doi: 10.1016/j.meegid.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Ravel J, Jiang L, Stanley ST, Wilson MR, Decker RS, Read TD, Worsham P, Keim PS, Salzberg SL, Fraser-Liggett CM, et al. The complete genome sequence of Bacillus anthracis Ames “Ancestor”. J Bacteriol. 2009;191:445–446. doi: 10.1128/JB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Schupp JM, Klevytska AM, Zinser G, Price LB, Keim P. vrrB, a hypervariable open reading frame in Bacillus anthracis. J Bacteriol. 2000;182:3989–3997. doi: 10.1128/jb.182.14.3989-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe ME, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci U S A. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Okinaka RT, Wang B, Easterday WR, Huynh L, U’Ren JM, Dukerich M, Zanecki SR, Kenefic LJ, Beaudry J, et al. Bacillus anthracis in China and its relationship to worldwide lineages. BMC Microbiol. 2009;9:71. doi: 10.1186/1471-2180-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KL, DeVos V, Bryden H, Price LB, Hugh-Jones ME, Keim P. Bacillus anthracis diversity in Kruger National Park. J Clin Microbiol. 2000;38:3780–3784. doi: 10.1128/jcm.38.10.3780-3784.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol. 2002;45:169–178. doi: 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J Bacteriol. 2003;185:1555–1563. doi: 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J, Zanecki SR, Pearson T, Simonson TS, U’Ren JM, et al. Global genetic population structure of Bacillus anthracis. PLoS One. 2007;2:e461. doi: 10.1371/journal.pone.0000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AJ, Keys C, Nemoto Y, Colman RE, Jay Z, Keim P. Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7. J Bacteriol. 2006;188:4253–4263. doi: 10.1128/JB.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AJ, Keys CE, Allender C, Bailey I, Girard J, Pearson T, Smith KL, Wagner DM, Keim P. Mutations, mutation rates, and evolution at the hypervariable VNTR loci of Yersinia pestis. Mutat Res. 2007;616:145–158. doi: 10.1016/j.mrfmmm.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]