Abstract
The identification of animals in an inbred miniature swine herd that consistently fail to produce replication- competent humantropic porcine endogenous retrovirus (PERV) has prompted studies on the biology of PERV in transmitter and nontransmitter animals. We analyzed PERV RNA transcript profiles in a family of inbred miniature swine (SLAd/d haplotype) in which individual members differed in their capacity to generate humantropic and ecotropic (i.e., pigtropic) virus. We identified unique HaeIII and HpaII gag restriction fragment length polymorphism (RFLP) profiles resulting from single nucleotide polymorphisms in blood cells; these were found only in animals that produced humantropic PERV. These HaeIII and HpaII gag RFLP profiles proved to be components of humantropic PERV as they were transmitted to 293 human target cells in vitro. The humantropic HaeIII and HpaII gag RFLP genotypes in the family of study were not present in other miniature swine in the herd that produced humantropic PERV, indicating that these RFLP profiles relate specifically to this family's lineage.
Porcine endogenous retrovirus (PERV) is a type C retrovirus found in the genomic DNA of all pigs (5, 9-11, 20, 21). Although PERV can infect human cell lines in vitro (16, 21, 26), there is no evidence that PERV is transmitted to human cells in cases where patients have come into contact with living porcine tissues or organs (4, 8, 18, 19, 22). Studies on cross-species infection are hampered by the fact that there is no predictive in vivo model for humantropic PERV transmission. Therefore, the risk of clinical PERV transmission cannot be estimated. Some of the concerns regarding cross-species infection are derived from the knowledge of related retroviruses like feline leukemia virus (FeLV), and murine leukemia virus (13, 25). In addition, there are precedents where retroviruses are thought to have crossed species barriers, for example gibbon ape leukemia virus to koala bears (14) and simian immunodeficiency virus to humans (6). While endogenous retroviruses (ERV) are typically transmitted as DNA provirus sequences from parent to offspring (12), studies have shown that some ERV can exist as exogenous agents and thereby also mediate horizontal transmission. Examples are mouse mammary tumor virus (15) and jaagsiekte sheep retrovirus (27). Indeed, it is assumed that the evolutionary origin of some retroviruses is via the “endogenization” of exogenous retrovirus strains (12).
Genomic mapping studies have shown that there are between approximately 10 and 100 proviral PERV loci in the genome of various pigs (1, 3, 7, 9, 10, 20; Linda Scobie, unpublished data). However, most of these loci possess gross deletions or frameshift or point mutations rendering them replication defective. In our studies on miniature swine a number of full-length proviruses were found which upon transfection into human (293) and porcine (ST-IOWA) cells were unable to replicate (9). Furthermore, reporter gene analysis of the long terminal repeat (LTR) sequences from these proviruses revealed that their transcriptional activity was very low compared with tissue-culture adapted LTR sequences (24; Linda Scobie, personal communication). These studies are currently being pursued with a more detailed PERV LTR analysis, based on the demonstration for FeLV that flanking genomic DNA can profoundly influence LTR transcriptional activity (2). Collectively, the presently available data indicate that pig genome analysis of PERV provirus gives little information on the profile of potential PERV expression. Therefore, proviral mapping studies need to be integrated with PERV gene expression analysis in specific cellular and tissue compartments before a meaningful assessment of the risk of PERV transmission can be made.
We have previously identified animals in a miniature swine herd inbred for the major histocompatibility complex, which do not transmit PERV in vitro to human cells (16, 23). This prompted us to study characteristics of animals that produce humantropic PERV by looking at patterns of PERV expression. This was performed in a selected family in which the parents produced five offspring, which displayed various phenotypes with respect to production of humantropic PERV. Using a gene expression profiling strategy to analyze PERV, we identified transmission-specific fingerprints using restriction fragment length polymorphism (RFLP) analysis of single nucleotide polymorphisms (SNPs) in the gag region. This strategy was based on the PERV gag gene because (i) the number of gag-positive proviral loci present in Large White (9) and miniature swine (Linda Scobie, personal communication) genomes is less than that of env-positive loci; (ii) the gag sequence is highly conserved in PERV; and (iii) gag expression is essential for virus particle production from a cell.
MATERIALS AND METHODS
Derivation of inbred miniature swine.
The derivation of the herd of inbred miniature swine starting with two founder animals in 1975 has been described previously (23). The ongoing miniature swine inbreeding program focuses on increasing the coefficient of inbreeding and maintaining distinct swine leukocyte antigen (SLA) haplotypes within the herd. Human decay accelerating factor (hDAF) transgenic Large White pigs were obtained from Imutran/Novartis Pharma (facility at Harlan Sprague Dawley Inc., Madison, Wis.). All animal procedures were undertaken in compliance with national and institutional regulations.
In vitro transmission assays.
Peripheral blood mononuclear cells (PBMC) were the only pig tissue used for the studies described in this manuscript due to their (i) relative accessibility and (ii) high responsiveness to induction of PERV expression upon treatment with mitogens. Cocultures between porcine PBMC and human or porcine target cells with subsequent analysis of virus transmission to the target cells were performed as described previously (16). Briefly, before coculture with human kidney epithelial 293 and porcine testis fibroblast ST-IOWA target cells, PBMC were stimulated with mitogens (phytohemagglutinin P [2.5 μg/ml] and phorbol 12-myristate-13-acetate [1 ng/ml]). Infection of target cells was measured by the presence of reverse transcriptase (RT) activity in the culture supernatants (HS-kit Mn2+ RT; Cavidi Tech AB, Uppsala, Sweden); for transmitting animals this typically became detectable after 10 to 14 days in ST-IOWA cells and after 20 to 30 days in 293 cells. In the case where RT activity was not detectable, 293 cocultures were maintained for a maximum of about 60 days before being classified as negative for PERV transmission.
PCR and sequence analysis.
RNA was extracted from cell lines and PBMC using Trizol (Invitrogen Life Technologies, Baltimore, Md.) according to the manufacturer's instructions. For analysis of 293 cells, RNA was extracted from cells 2 weeks after initial detection of RT activity in the cell supernatant by enzyme-linked immunosorbent assay. Single step RT-PCRs were performed on 0.3 to 0.7 μg of total RNA using the OneStep RT-PCR kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. The PCR cycling conditions for gag sequences consisted of 30 min at 50°C followed by 35 cycles of 96°C for 2 s, 55°C for 30 s, and 72°C for 30 s. Conditions for porcine glyceraldehyde 3-phosphate dehydrogenase (pGAPDH) PCR were the same except that 20 amplification cycles were performed. All PCRs were performed using a 2400 thermocycler (Perkin-Elmer BioSciences, Atlanta, Ga.). For sequencing, gag PCR products were cloned into pCRII-TOPO (Invitrogen Life Technologies) according to the manufacturer's guidelines and sequenced using the following four sequencing primers: gag sense 2, AAGATTTGGCAGAGGATCCT; gag antisense 2, AGAGCTAGGATCCTTGGTCC; gag sense; and gag antisense. DNA sequencing was performed using a CEQ 2000 XL (Beckman Instruments, Palatine, Fla.) and associated Dye Terminator Cycle Sequencing with Quick Start kit (Beckman Instruments) according to the manufacturer's instructions. Degenerate gag PCR primers were designed based on alignments of multiple GenBank PERV gag sequences and are therefore capable of annealing to gag sequences from a wide variety of PERV of all subtypes (PERV-A, -B, and -C). The PCR primers used were as follows: gag sense, TAATATGGGACAGACWGTGACWACCCC; gag antisense GTTACCCGTCCCACTWCCAAGTCA; pGAPDH sense, CGTCAAGCTCATTTCCTGGTACG; and pGAPDH antisense, GGGGTCTGGGATGGAAACTGGAAG.
RFLP analysis.
Gag PCR products were resolved on a 1% agarose gel in 1× TAE buffer and gel purified using the QIAquick gel extraction kit (Qiagen). The gel-purified gag PCR product was digested for 1 h at 37°C with HpaII and HaeIII restriction enzymes (New England BioLabs, Beverly, Mass.), either separately or in combination, using reaction buffers recommended by the manufacturer. Following digestion, Hi-Density TBE Sample Buffer (Invitrogen Life Technologies) was added to the reaction and the sample was immediately resolved on a prerun precast 10% polyacrylamide TBE Gel (Invitrogen Life Technologies) in 0.5 × TBE running buffer (Invitrogen Life Technologies). Gels were run for 90 min at 130 V and dried overnight using the Dry Ease gel-drying system (Invitrogen Life Technologies). Following drying, the 35S-labeled gel was exposed for 3 to 6 days at room temperature to BioMax MS film (Perkin-Elmer BioSciences).
Bioinformatics.
DNA sequence analyses were carried out using the Vector NTI Suite 7.0 software (Informax, Inc, Bethesda, Md.). Multiple alignments were performed using AlignX.
RESULTS
PERV transmission characteristics of a family of miniature swine.
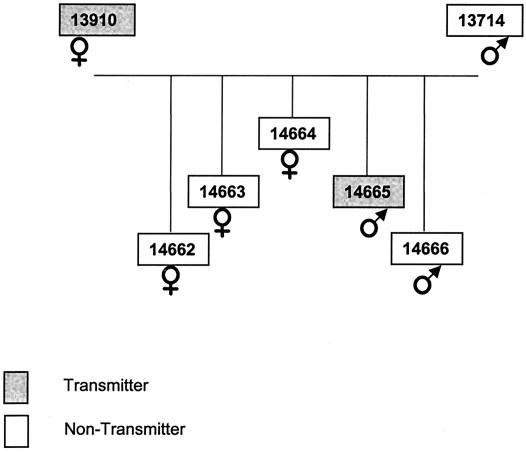
The initial analysis focused on one family in which parents and offspring (n = 5) differed with respect to production of humantropic and ecotropic virus (Fig. 1; Table 1.) Multiple transmission assays were performed using PBMC from these animals, which confirmed that the PERV transmission phenotypes (as detected with human 293 or porcine ST-IOWA target cells) were stable over time (Table 1). One parent (animal 13910) produced humantropic virus, and one parent (animal 13714) failed to produce humantropic virus in vitro. In the offspring, one animal (animal 14665) produced human- and ecotropic virus, and four others (animals 14662-4 and 14666) failed to produce human- or ecotropic virus.
FIG. 1.
Family tree showing the relationships between the miniature swine family members analyzed in the study. A transmitter animal causes PERV transmission to human 293 cells by PBMC during in vitro coculture; the nontransmitters do not.
TABLE 1.
RFLP and transmission analysis of inbred miniature swine and Large White pigsa
| Pig no. | PERV transmission to 293 cells | Origin |
|---|---|---|
| 13714 | 0/11 | Family |
| 13910 | 6/6 | Family |
| 14662 | 0/2 | Family |
| 14663 | 0/1 | Family |
| 14664 | 0/1 | Family |
| 14665 | 3/4 | Family |
| 14666 | 0/5 | Family |
| 10715 | 0/4 | Unrelated |
| 13519 | 4/4 | Unrelated |
| 12550 | 0/4 | Unrelated |
| 15101 | 0/4 | Unrelated |
| 11619 | 2/2 | Unrelated |
| 238d | 0/1 | Large White |
| 791c | 0/1 | Large White |
All animals with the exception of animals 791c, 14661-4, and 14666 infected porcine (ST-IOWA) cells in vitro. Values are the ratio of independent occasions that animals transmitted, or failed to transmit, humantropic PERV to 293 cells in vitro to total number of attempts to transmit PERV. The origin of the miniature swine is defined as being from either within the family study or from more distantly related animals.
PERV expression and RFLP analysis in a family of miniature swine.
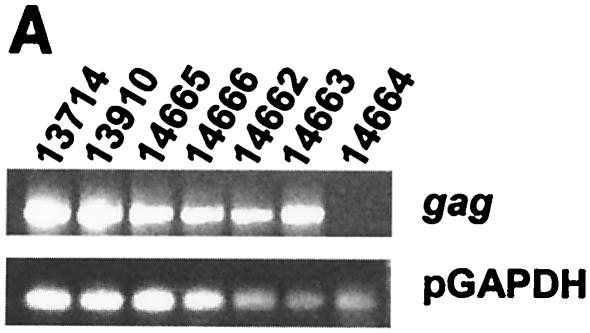
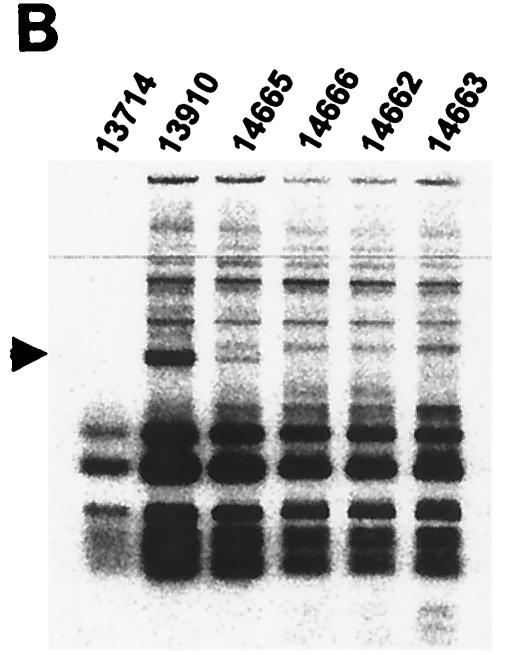
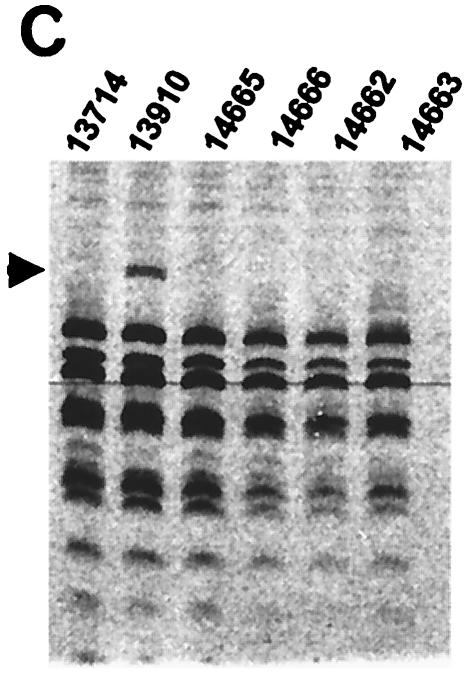
In addition to four offspring animals (animals 14662-4 and 14666) not producing ecotropic (data not shown) or humantropic PERV (Table 1), one of these animals (animal 14664) failed to express gag mRNA (Fig. 2A). Thus, it is likely that within the members of the inbred herd, gag expression by other family members is likely to originate from a small number of transcriptionally active genomic loci. RT-PCR was performed on RNA extracted from mitogen-stimulated PBMC (Fig. 2A). RT-PCR products were digested with the restriction enzymes HaeIII alone or in combination with HpaII and unique RFLPs were resolved by polyacrylamide gel electrophoresis and autoradiographical exposure of 35S-labeled fragments. In the parent animal 13910, a unique RFLP band was observed for HaeIII (Fig. 2B) and also for the HaeIII/HpaII combination (Fig. 2C). The offspring animal 14665 also expressed the HaeIII RFLP band (Fig. 2B) but not the HaeIII/HpaII RFLP band (Fig. 2C). These two pigs were the only animals in the family producing humantropic PERV in transmission assays (Table 1). None of the other family members expressed either one of these two RFLP bands. Therefore, the unique RFLP bands correlated with the in vitro transmitter phenotype. In a separate analysis, it was demonstrated that the HaeIII/HpaII RFLP band was also present in the restriction enzyme digest of HpaII alone (data not shown).
FIG. 2.
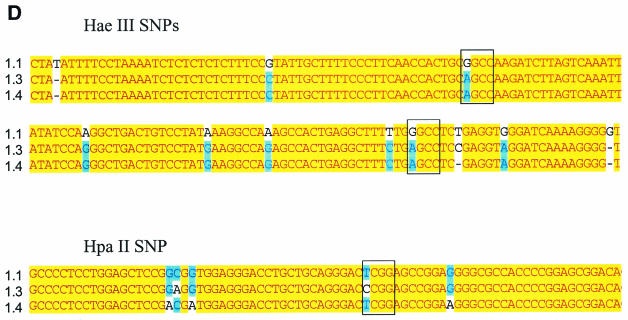
Genotyping of a miniature swine family by RFLP analysis of gag mRNA. (A) RT-PCR of PERV gag gene from PBMC of family members. (B) HaeIII RFLP analysis of 35S-labeled PERV gag RT-PCR products. (C) HaeIII/HpaII RFLP analysis of gag RT-PCR products. (D) Nucleotide basis for HpaII and HaeIII RFLPs; clone 1.1, nonhumantropic PERV; clone 1.3, humantropic PERV with HaeIII and HpaII RFLP; clone 1.4, humantropic PERV with HaeIII RFLP only. Restriction sites are indicated by the open box. HaeIII site, GGCC; HpaII site, CCGG.
Nucleotide basis for HaeIII and HpaII RFLPs.
To define the sequence basis for the deduced HaeIII and HpaII polymorphisms, individual gag PCR products were cloned that were amplified from the PBMC of animal 13910. This animal expressed a mixture of PERV species including humantropic PERV containing the HaeIII and HpaII RFLPs. RFLP analysis was first performed on individual clones to identify one clone containing the HaeIII RFLP alone, another with both the HaeIII and HpaII RFLPs, and a third with neither RFLP (data not shown). Secondly, each of these three PERV gag clones was sequenced and aligned to identify the locations and nature of the nucleotide polymorphisms responsible for determining the observed unique RFLP bands. The HaeIII RFLP is derived from one out of two possible candidate locations in the gag PCR fragment; both locations had a purine (G/A) SNP in a HaeIII site, approximately 1 and 1.2 kb, respectively, from the start of the gag PCR product (Fig. 2D). Here, in both locations, the SNP abolishes the HaeIII site in the putative humantropic PERV. The unique HpaII RFLP band results from a pyrimidine (T/C) SNP associated with a new HpaII restriction site approximately 550 nucleotides from the start of the gag PCR product (Fig. 2D). This is the only polymorphism found for a HpaII site within the entire fragment. One of the putative humantropic clones contained both the HpaII SNP and the HaeIII SNPs while the other humantropic clone contained the HaeIII SNPs but not the HpaII SNP. These data provide the sequence basis for the unique RFLP banding pattern and demonstrate that the HaeIII RFLP can exist alone or along with the HpaII RFLP in a single PERV clone.
Analysis of PERV expression in distantly related miniature swine and Large White pigs.
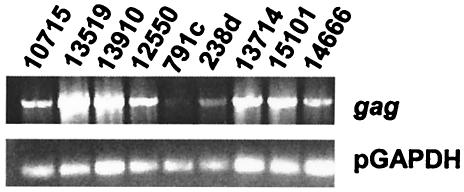
In order to determine the distribution of the gag HaeIII and HaeIII/HpaII RFLPs, we extended our RFLP analysis to other miniature swine in the herd that were distantly related to the SLAd/d family described above, as well as to hDAF-transgenic Large White pigs. In transmission assays Large White pigs did not produce ecotropic or humantropic PERV (Table 1). Interestingly, PERV was not detectable in one of the hDAF animals (Fig. 3a, animal 791c). None of the family-unrelated animals displayed the RFLP bands present in animal 13910 (Table 2), despite the capacity of producing humantropic PERV in one of these animals (animal 13519). These data indicate that the HaeIII and the HaeIII/HpaII RFLPs correlate with the transmitter phenotype within our family only but are not generally found among the miniature swine herd or the Large White pigs.
FIG. 3.
RT-PCR of PERV in Large White pigs (animals 791c and 238d) and unrelated miniature swine (all other animals).
TABLE 2.
RFLP analysis of unrelated immature swine and Large White pigs
| Animal | HaeIII | HpaII |
|---|---|---|
| Transmitters | ||
| 13519 | − | − |
| 13910 | + | + |
| Nontransmitters | ||
| 10715 | − | − |
| 12550 | − | − |
| 791c | − | − |
| 238d | − | − |
| 13714 | − | − |
| 15101 | − | − |
| 14666 | − | − |
Effect of PBMC stimulation on expression of PERV RFLP bands.
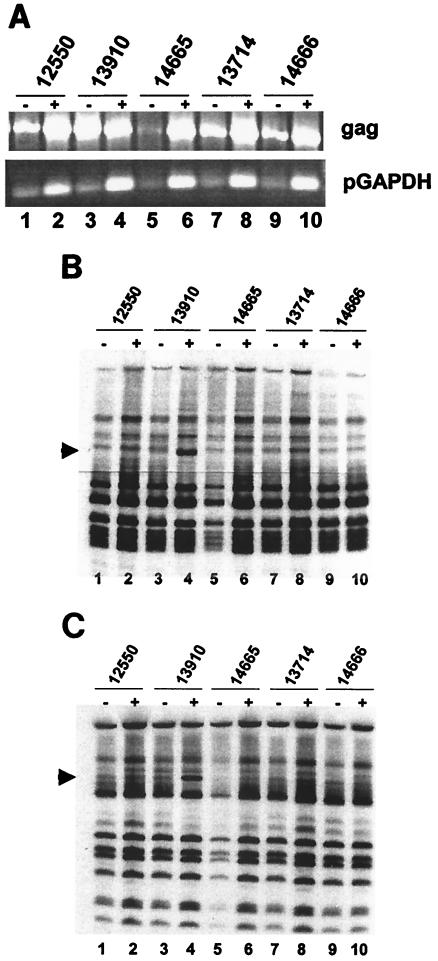
Transmission of PERV from pig to human cells in vitro is enhanced by stimulation of PBMC with mitogens (26), via nonspecific transcriptional activation of some proviral LTRs. We investigated the effect of mitogenic stimulation on expression of the RFLP bands described above, using HaeIII and HpaII separately in restriction enzyme digestion. Despite high PERV gag gene expression (Fig. 4A, lane 3), RFLP bands were not detectable in unstimulated PBMC (Fig. 4B, lane 3; 4C, lane 3) for the parent animal 13910. However, RFLPs were detectable after stimulation of PBMC with mitogens (Fig. 4B and C). These results indicate that the generation of the HaeIII and HpaII RFLP bands requires transcriptional activation of a proviral locus.
FIG. 4.
Effect of mitogenic stimulation on RFLP expression patterns in miniature swine. Animals were analyzed before (−) and after (+) stimulation with PHA and PMA. (A) RT-PCR of PERV gag gene from PBMC. (B) HaeIII RFLP analysis of PERV gag RT-PCR products. (C) HpaII RFLP analysis of PERV gag RT-PCR products.
Link between RFLP profile and production of humantropic PERV.
To determine whether the unique RFLP banding is a constituent of humantropic PERV, we examined the RFLPs from infected 293 cells. We evaluated 293 cells following in vitro coculture with PBMC from two members in the family (animals 13910 and 14665) and from two unrelated transmitting animals (animals 13519 and 11619). The unique HaeIII and HpaII RFLP bands were identified in infected 293 cells after coculture with PBMC from animals 13910 and 14665 (Table 3), but not in 293 cells after coculture with PBMC from the other two unrelated animals. For pig 14665, the HaeIII RFLP band detected in PBMC (Fig. 2B) was similarly present in 293 cells (Table 3): the HpaII RFLP band was not detectable in either PBMC or the infected 293 cells (Table 3). These observations were confirmed in repeat analyses on separately performed cocultures (Table 1) and indicate that the HaeIII RFLP is present in humantropic PERV isolated from these animals. For pig 13910, which expresses the HpaII and HaeIII RFLPs in PBMC (Fig. 2B and C), both RFLP bands were detected in the infected 293 cells (Table 3). However, during one other coculture analysis the HpaII RFLP was detected in 293 cells in the absence of the HaeIII RFLP band. Thus, both the HaeIII and HpaII RFLP bands represent components of humantropic PERV and can be transmitted as part of humantropic PERV to 293 cells. It seems likely that HaeIII and HpaII RFLPs are located in different PERVs representing different replication-competent PERV proviruses.
TABLE 3.
RFLP analysis of human 293 cells infected with humantropic PERV
| Pig no. | No. of positive RFLP analysis results/total no. of RFLP analysesa
|
|
|---|---|---|
| HaeIII | HpaII | |
| 13910 | 2/3 | 3/3 |
| 14665 | 3/3 | 0/3 |
| 13519 | 0/1 | 0/1 |
| 11619 | 0/1 | 0/1 |
Data are presented from animals from one family (animals 13910 and 14665) and two unrelated animals (animals 13519 and 11619).
DISCUSSION
As PERV can infect human cells in vitro (21, 26), there has been concern about the risk of PERV transmission to human xenograft recipients. We have previously identified pigs that consistently do not transmit PERV to human cells in vitro (16). It is relevant to know whether there is a genomic basis for a transmission phenotype and whether there is a molecular basis for variation between individual animals in production of humantropic PERV. We addressed this issue by identification and tracking humantropic PERV based on unique RFLP profiles in the gag gene. We demonstrate that such a genotyping approach is a feasible strategy for monitoring humantropic PERV in at least one family of miniature swine and could potentially be used to monitor humantropic PERV distribution per se if additional restriction enzymes were used.
Given that there are a high number of proviral loci in pig cells, it is of interest to estimate how many transcriptionally active PERV loci are in the pig genome. There is preexisting evidence that the number of active PERV loci is likely to be low, although the role of tissue-specific and genetic factors controlling locus expression remains unknown. For example, by Northern blot the ST-IOWA cell line does not express PERV gag mRNA (data not shown). In addition, env proteins are expressed at sufficiently low levels that allow these cells to be used in cocultures to assess the ability of pigs to produce ecotropic PERV (data not shown). In this study, we describe further evidence that the number of heritable transcriptionally active PERV proviruses is likely to be low based on the distribution pattern of transcriptionally active PERV, as measured by gag RT-PCR, in a miniature swine family (Fig. 2A).
Currently, the in vitro coculture of porcine PBMC and human 293 cells is the most sensitive assay for assessing pig-to-human PERV transmission. However, during in vitro culture humantropic PERV transmission viruses can acquire adaptive mutations. The present results appear not to be confounded by new viruses that might be created during propagation in tissue culture. This is illustrated by the observation that the same RFLP profile is present in porcine PBMC before coculture and in 293 cells after coculture, and the absence of new bands in infected 293 cells. Our results are consistent with previous findings that demonstrate that the RFLP profile of replication-competent virus can remain stable during prolonged culture periods. For the example of FeLV, RFLPs develop when a molecular FeLV clone is cultured in feline embryo fibroblasts cells as a result of recombination with endogenous retroviral sequences (17). In a scenario more akin to the PERV-293 cell situation, unique RFLPs did not emerge in parallel cultures of the FeLV clone in a permissive dog cell line (D17), which does not contain FeLV-related sequences, despite extended periods in culture following FeLV infection. Our data demonstrate that animals that produce humantropic PERV can express unique PERV RFLP profiles that are different from that of the pigs unable to produce humantropic PERV. Moreover, the unique RFLP profile is maintained during transmission of PERV to 293 cells and hence can be used in tracking humantropic replication-competent PERV in members of distinct families of the herd.
The two unique RFLP profiles observed using the HaeIII and HpaII restriction enzymes most likely represent two different replication-competent proviruses. This is evident from the in vitro transmission data using cells from the parent animal 13910, showing that the HpaII RFLP band is present in infected 293 cells irrespective of the HaeIII RFLP band (Table 3), and data from animal 14665 showing the reverse situation, namely the presence of the HaeIII RFLP band in PBMC 293 cells upon coculture with PBMC in the absence of the HpaII RFLP band (Fig. 2; Table 3). In addition, the different susceptibility of the PERV associated with the unique RFLP bands from animals 13910 and 14665 to mitogenic activation suggests that the associated provirus has different genomic integration sites in each animal (Fig. 4). The relationship between animals 13910 and 14665 in the miniature swine family investigated is noteworthy in this respect. The offspring animal 14665 only expressed the HaeIII RFLP profile, while both the HaeIII and HpaII RFLP bands were present in the parent animal 13910; thus, we conclude that only the humantropic PERV with the HaeIII RFLP profile was acquired by this offspring. However, these two animals differed in expression signal and responsiveness to stimulation of the HaeIII proviral loci between these animals. This suggests that the provirus containing the HaeIII RFLP band is present at a different locus in animals 13910 and 14665. Thus, it can be questioned whether the humantropic PERV in these animals was transmitted through the germ line or as an exogenous agent. If PERV were acquired as an exogenous agent, mosaicism with respect to regulation of gene expression due to different genomic integration sites is to be expected.
In conclusion, the present study demonstrates that humantropic PERV from miniature swine that can be transmitted from PBMC to human 293 cells in vitro are identifiable by the presence of unique PERV gag RFLP profiles. The RFLP profiles described represent intrinsic components of replication-competent proviruses and are specific to particular families. Such links warrant further study addressing whether the acquisition of humantropic PERV is due to Mendelian inheritance or due to horizontal transmission of PERV between family members. In addition care must be taken when extrapolating expression patterns of PERV to other tissues until potential tissue-specific patterns of expression have been investigated. The data contribute to a better understanding of the mechanisms by which humantropic PERV present themselves in miniature swine and the pattern with which PERV may be transmitted. The development of methodologies such as RFLP analysis will enable the tracking of distinct replication-competent humantropic PERV, which serves as a relevant tool in the specific breeding of animals for xenotransplantation that are free from replication-competent humantropic PERV.
Acknowledgments
We thank Linda Scobie and David Onions (University of Glasgow) for their contributions to this study.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry, B. T., A. K. Ghosh, D. V. Kumar, D. A. Spodick, and P. Roy-Burman. 1988. Structure and function of endogenous feline leukemia virus long terminal repeats and adjoining regions. J. Virol. 62:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinsmore, J. H., C. Manhart, R. Raineri, D. B. Jacoby, and A. Moore. 2000. No evidence for infection of human cells with porcine endogenous retrovirus (PERV) after exposure to porcine fetal neuronal cells. Transplantation 70:1382-1389. [DOI] [PubMed] [Google Scholar]
- 5.Ericsson, T., B. Oldmixon, J. Blomberg, M. Rosa, C. Patience, and G. Andersson. 2001. Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J. Virol. 75:2765-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 7.Gunzburg, W. H., and B. Salmons. 2000. Xenotransplantation: is the risk of viral infection as great as we thought? Mol. Med. Today 6:199-208. [DOI] [PubMed] [Google Scholar]
- 8.Heneine, W., A. Tibell, W. M. Switzer, P. Sandstrom, G. V. Rosales, A. Mathews, O. Korsgren, L. E. Chapman, T. M. Folks, and C. G. Groth. 1998. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet 352:695-699. [DOI] [PubMed] [Google Scholar]
- 9.Herring, C., G. Quinn, R. Bower, N. Parsons, N. A. Logan, A. Brawley, K. Elsome, A. Whittam, X. M. Fernandez-Suarez, D. Cunningham, D. Onions, G. Langford, and L. Scobie. 2001. Mapping full-length porcine endogenous retroviruses in a Large White pig. J. Virol. 75:12252-12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, J. H., G. C. Webb, R. D. Allen, and C. Moran. 2002. Characterizing and mapping porcine endogenous retroviruses in Westran pigs. J. Virol. 76:5548-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 12.Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackey, L. 1975. Feline leukaemia virus and its clinical effects in cats. Vet. Rec. 96:5-11. [DOI] [PubMed] [Google Scholar]
- 14.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, R., M. Dixon, R. Smith, G. Peters, and C. Dickson. 1987. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J. Virol. 61:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 76:3045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overbaugh, J., N. Riedel, E. A. Hoover, and J. I. Mullins. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731-734. [DOI] [PubMed] [Google Scholar]
- 18.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, E. Otto, et al. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 19.Patience, C., G. S. Patton, Y. Takeuchi, R. A. Weiss, M. O. McClure, L. Rydberg, and M. E. Breimer. 1998. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet 352:699-701. [DOI] [PubMed] [Google Scholar]
- 20.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 22.Pitkin, Z., and C. Mullon. 1999. Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif. Organs 23:829-833. [DOI] [PubMed] [Google Scholar]
- 23.Sachs, D. H., G. Leight, J. Cone, S. Schwarz, L. Stuart, and S. Rosenberg. 1976. Transplantation in miniature swine. I. Fixation of the major histocompatibility complex. Transplantation 22:559-567. [DOI] [PubMed] [Google Scholar]
- 24.Scheef, G., N. Fischer, U. Krach, and R. R. Tonjes. 2001. The number of a U3 repeat box acting as an enhancer in long terminal repeats of polytropic replication-competent porcine endogenous retroviruses dynamically fluctuates during serial virus passages in human cells. J. Virol. 75:6933-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teich, N., J. Wyke, T. Mak, A. Bernstein, and W. Hardy. 1984. Pathogenesis of retrovirus-induced disease, p. 901-923. In R. A. Weiss, N. Teich, H. E. Varmus, and J. M. Coffin (ed.), RNA tumor viruses, 2nd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 26.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.York, D. F., R. Vigne, D. W. Verwoerd, and G. Querat. 1992. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J. Virol. 66:4930-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]