Abstract
The surface glycoprotein (SU) of most gammaretroviruses contains a conserved histidine at its amino terminus. In ecotropic murine leukemia virus SU, replacement of histidine 8 with arginine (H8R) or deletion of H8 (H8del) abolishes infection and cell-cell fusion but has no effect on binding to the cellular receptor. We report here that an aromatic ring side chain is essential to the function of residue 8. The size of the aromatic ring appears to be important, as does its ability to form a hydrogen bond. In addition, infection by all of the nonaromatic amino acid substitutions could be partially rescued by the addition of two suppressor mutations (glutamine 227 to arginine [Q227R] and aspartate 243 to tyrosine [D243Y]) or by exposure to chlorpromazine, an agent that induces fusion pores in hemifusion intermediates to complete fusion, suggesting that, like the previously described H8R mutant, the mutants reported here also arrest membrane fusion at the hemifusion state. We propose that H8 is a key switch-point residue in the conformation changes that lead to membrane fusion and present a possible mechanism for how its substitution arrests fusion at the hemifusion state.
Enveloped viruses, including retroviruses, enter a host cell by fusing their membrane with that of the cell through interactions between the viral envelope protein and its cellular receptor. Retroviral envelope proteins (Env) consist of two subunits derived from a single protein precursor: SU, which directly interacts with the host receptor, and a transmembrane protein (TM), which promotes viral and cellular membrane fusion. It is thought that newly synthesized Env proteins initially fold into a metastable, fusion-inactive conformation and then assemble in this state onto virions. During the entry process, the metastable conformation is induced to change to a stable, fusion-active one. In the case of gammaretroviruses, binding of SU to receptors initiates conformational changes in both SU and TM (23). These changes are thought to release the energy that drives membrane fusion (5, 6). However, details of how these changes in Env activate fusion are still unknown.
The SU of gammaretroviruses contain three functional domains: the amino-terminal receptor-binding domain (RBD) (4, 8), a proline-rich region (PRR), and a carboxy-terminal domain. Although the RBD is not thought to be involved in the actual fusion per se, the histidine residue in a conserved SPHQV motif near its amino terminus has been shown to be essential for postbinding events. Deletion or substitution of this critical histidine residue in either ecotropic (H8del, H8A, H8K, and H8R) or amphotropic (H5del) murine leukemia virus (MLV) Env results in a fusion defect but has no effect on receptor recognition or virus binding (3, 13, 26). This defect in fusion can be partially overcome either by adding two suppressor mutations located in and near PRR or by providing in trans soluble RBD fragment with a histidine at residue 8 (4, 13, 26). In addition, we recently showed that the fusion defect of H8R mutant also can be partially rescued by chlorpromazine, a membrane-curving reagent that promotes fusion once the hemifusion stage is reached (27).
We investigated further the molecular role of this essential histidine residue in membrane fusion. We report here that there are at least three key biochemical characteristics for the side chain of residue 8 in ecotropic MLV SU: (i) an aromatic ring; (ii) the size of aromatic ring, with small being better; and (iii) the ability to hydrogen bond. Substitution of tyrosine for H8 (H8Y) resulted in 2-fold less infection than wild-type (WT) virus, whereas replacement with phenylalanine (H8F) and tryptophan (H8W) resulted in 20- and 200-fold less infection, respectively. In contrast, substitution of isoleucine (H8I), leucine (H8L), methionine (H8 M), alanine (H8A), or aspartate (H8D) abolished infection of mouse NIH 3T3 cells. In addition, like the H8R mutant, infection by the newly identified down mutations can also be partially rescued by chlorpromazine, suggesting that they also arrest membrane fusion at the hemifusion stage.
An aromatic ring is essential at position 8.
We replaced H8 with three aromatic residues (tyrosine [Y], phenylalanine [F], or tryptophan [W]), one negatively charged residue (aspartate [D]), and four aliphatic residues (alanine [A], leucine [L], isoleucine [I], or methionine [M]) at this site. Specific mutations were introduced into the env gene by using oligonucleotide-directed mutagenesis (QuikChange kit; Stratagene); each mutant env gene was introduced into the plasmid pcDNA-MoMLV, providing WT gag and pol genes and transiently transfected into H1-BAG cells that stably express a Moloney MLV-derived BAG genome encoding β-galactosidase (26). Virus particles carrying mutant Envs were tested for their infectivity on NIH 3T3 cells by endpoint dilution titration. Incorporation and processing of mutant Env was analyzed by Western blotting of virus pellets and lysates of transfected cells as previously described (26).
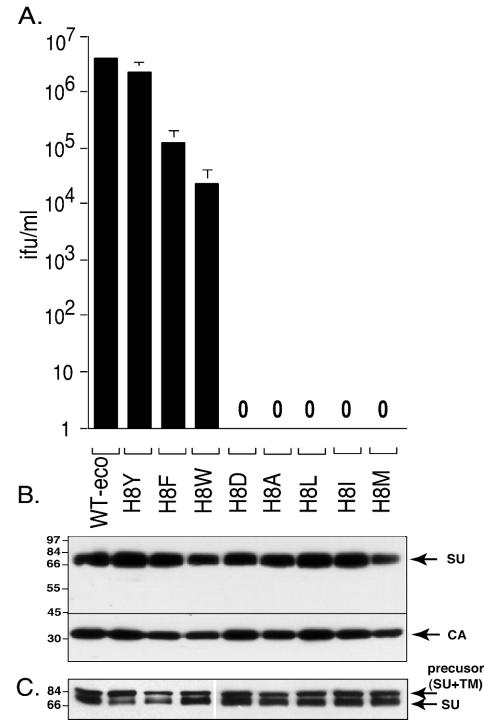
The aromatic residue substitutions (H8Y, H8F, and H8W) resulted in infection but to different extents (Fig. 1A). H8Y viruses were ∼2-fold less infectious than WT, whereas H8F and H8W substitutions resulted in 20-fold and 200-fold less infection than WT Env. In contrast, substitution of an aliphatic residue (H8A, H8L, H8I, and H8 M) or a negatively charged residue (H8D) completely abolished infection, indicating that an aromatic ring is essential to the function of the eighth residue of SU. The reduction in infectivity was not due to poor expression or to poor incorporation into virions since equal volumes of each virus stock contained comparable amounts of SU and virions (i.e., capsid protein) (Fig. 1B), and equal masses of producer cell lysates contained comparable amounts of precursor and mature envelope proteins (Fig. 1C). Our results for H8A are in agreement with the previous report of Bae et al. (3). The information regarding H8Y, H8F, H8W, H8D, H8L, H8I, and H8M is new data.
FIG. 1.
An aromatic ring is essential for histidine 8 function. (A) Infection of NIH 3T3 cells. Cells were exposed to 10-fold serial dilution of virus stocks, and the virus titer was calculated from the end point dilution (n = 4). Mean results of three independent experiments are shown ± the standard errors. The standard errors for the WT are too small to be seen in the scale of the figure. (B) Incorporation of mutant Envs into virions. Equal volumes of viral pellets were separated on a 8% sodium dodecyl sulfate-polyacrylamide gel and transferred to nitrocellulose membranes. Blots were cut into two parts at 45 kDa. The top part was reacted with anti-SU antisera; the lower part was incubated with anti-CA antisera. (C) Western blot analysis of virus producer cell lysates. The blot was reacted with anti-SU antisera. The numbers at the left of panels B and C indicate the molecular mass in kilodaltons.
The membrane fusion capability correlates with infection of the pseudotype viruses.
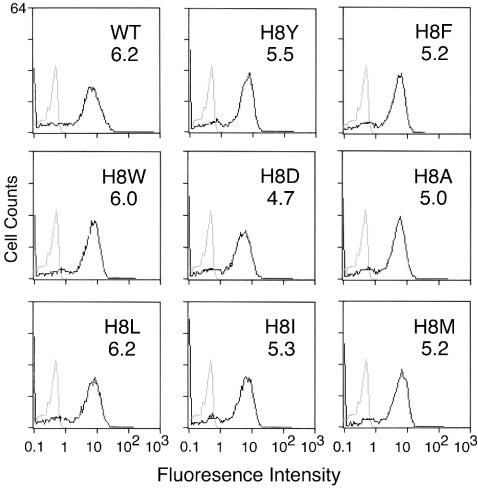
Bae et al. first showed that virus binding was not affected by deletion of H8 but that cell-cell fusion was blocked (3). Each of the new substitutions was characterized for receptor binding as well. All of the viruses pseudotyped with mutant Env bound to receptor and binding was comparable to WT Env pseudotyped virus (Fig. 2). These results support the previous conclusion that residue 8 is not required for virus attachment (3, 26).
FIG. 2.
Virus binding. Virus supernatant concentrated to 15- to 20-fold by using Centricon Plus-80 (100-kDa cutoff) to remove shedded SU was incubated at 4°C with human 293 cells stably expressing the exogenous ecotropic receptor. Cells were then stained with goat anti-SU antisera (anti-Rauscher gp70; Quality Biotech, Inc.) and mouse anti-goat antisera conjugated to fluorescein isothiocyanate. The level of binding was analyzed by flow cytometry. Gray lines represent the basal level of fluorescence determined by incubation of WT virus with parent human 293 cells lacking ecotropic receptor. For comparison these basal values are shown in each panel. Black lines represent the fluorescence intensity of binding of WT or mutant virus to human 293 cells expressing ecotropic receptor. The value of the mean fluorescence intensity is shown in upper right corner of each panel. Values shown are from a representative of at least two independent binding experiments.
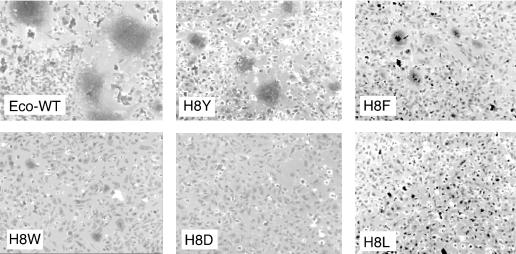
The ability of the H8 mutant Env to mediate membrane fusion was evaluated by using an XC rat sarcoma cell-cell fusion assay. Since the R-peptide has been cleaved from the majority of Env molecules on virions (10) and cleavage of R-peptide is required for virus-cell membrane fusion and greatly enhances Env-induced cell-cell fusion (20), the R-less form of each Env was used in this assay. Human 293 cells transiently transfected with plasmids encoding the R-less versions of WT and mutant Envs were cocultivated with XC cells, which display the ecotropic MLV receptor on their surface. After 12 h, cells were fixed and stained with basic fuchsin. Large syncytia containing 10 to 20 nuclei in the same cytoplasm were readily visible throughout cultures expressing WT Env (Fig. 3). Mutant H8Y Env also induced syncytia at a frequency comparable to that of WT Env, but the number of nuclei per syncytium was decreased severalfold. The H8F change gave limited syncytium formation; both the number of syncytia and the number of nuclei per syncytium were reduced, whereas the H8W change gave even less and was just slightly above background. None of the other H8 mutant Envs induced syncytia above the background level observed in the parent 293-XC cell cocultures (Fig. 3 and data not shown).
FIG. 3.
Cell-cell fusion. Cell-cell fusion assay was performed as previously described (26). XC cells were cocultivated with human 293 cells expressing the indicated Env/R-less. The panels shown are representative of two independent experiments. No syncytia were observed in cocultures of parental 293 and XC cells. The photomicrographs of the cells were taken by using an Axiophot microscope (Zeiss). Magnification, ×100.
Infection by all defective H8 mutants except H8D can be partially suppressed by two additional changes at positions 227 and 243.
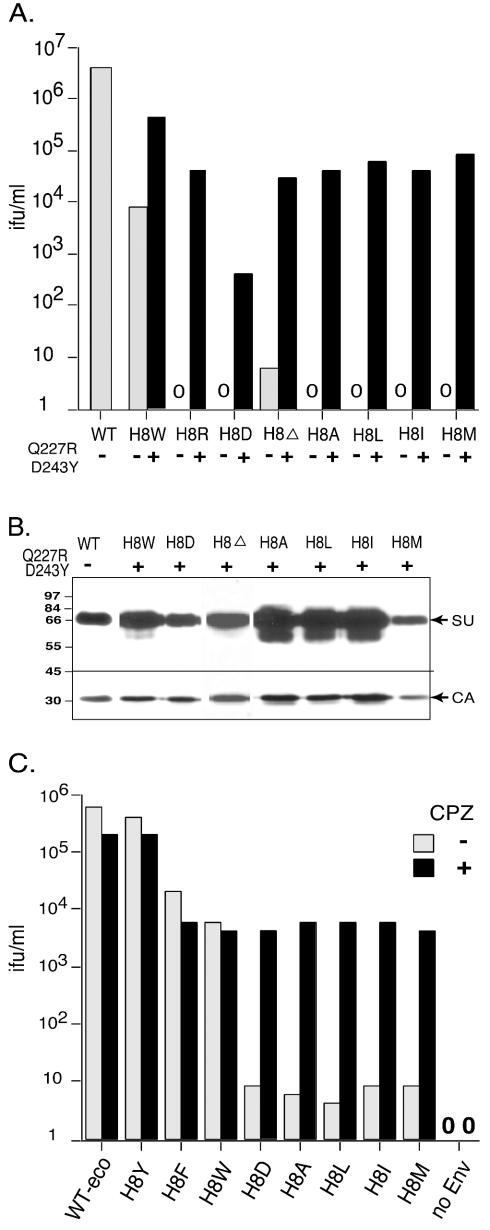
We previously reported that the fusion defect of an H8R mutant can be suppressed by two additional changes at positions 227 and 243, although alone the two changes do not affect infection (26). We investigated whether these two changes can suppress the newly identified down mutations. A representative of two independent experiments is shown in Fig. 4. Interestingly, we observed that the addition of Q227R and D243Y to H8W increased infection ∼50-fold over infection by H8W alone (Fig. 4A). This value was 10-fold greater than those for other defective mutants combined with Q227R and D243Y. The addition of Q227R and D243Y to the H8 mutants increased infection, but the increase for H8D was consistently less than with all of the other defective H8 mutants. All of the triple mutant Envs were incorporated into virions as efficiently as WT Env (Fig. 4B).
FIG. 4.
All of the H8 down mutations can be partially suppressed by two additional mutations, Q227R and D243Y, and rescued by chlorpromazine. (A) Titers of H8 down mutant viruses with (▪) or without () the suppressor mutations Q227R and D243Y. Mouse NIH 3T3 fibroblast cells were exposed to 10-fold serial dilutions of virus stocks.Titers were calculated based on an endpoint dilution (n = 4). (B) Western blot analysis of virion containing mutant envelope proteins. Proteins were separated by sodium dodecyl sulfate-8% polyacrylamide gel. Membranes were cut into two parts at 45 kDa. The top part was probed with anti-SU antisera, and the bottom portion was reacted with anti-CA antisera. The experiment was performed twice; a representative is shown. (C) XC cells were incubated on ice for 30 min with 10-fold serial dilutions of virus stocks in BES (N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid)-buffered medium (pH 7.4) containing Polybrene (20 μg/ml; Sigma) and then shifted to 37°C for another 30 min. Cells were rinsed twice to remove unbound virus and then incubated with (▪) or without () 0.4 mM chlorpromazine (pH 7.4) for 1 min. Chlorpromazine was immediately removed, and the cells were washed twice again. After 40 h, cells were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for transduction of β-galactosidase activity, and the infectious titers were calculated from the endpoint dilution (n = 4). The experiments were performed at least twice. A representative is shown. No infection of XC cells by particles lacking Env was observed in the presence of chlorpromazine.
Defective H8 mutants can be partially rescued by chlorpromazine.
Hemifusion is an intermediate stage in the fusion process that is characterized by merging activity in the outer monolayers without the opening of fusion pores (11, 14). Chlorpromazine is a chemical membrane-curving agent that preferentially partitions to and destabilizes the inner membrane monolayers of lipid bilayers (14). Application of chlorpromazine to cell membranes arrested at hemifusion transforms the intermediate into complete fusion (14). We recently showed that the H8R mutant arrests virus-cell fusion at the hemifusion state and demonstrated that chlorpromazine resolves hemifusion-arrested virus-cell membrane fusion to give infection (27). Thus, if the defective H8 mutants give the same defect in infection, then application of chlorpromazine to virus-cell complexes should relieve the block and rescue infection. Brief exposure to chlorpromazine resulted in comparable infection of WT, H8Y, H8F, and H8W viruses, whereas infection of the other defective mutants was increased 500- to 1,000-fold (Fig. 4C), a finding consistent with each arresting fusion at the hemifusion state.
A pi-electron interaction is “key.”
We propose that the high degree of conservation of residue 8 within the highly mutable Env results from the selection of histidine as the best functioning amino acid. Our reasons are as follows. Since aromatic but not aliphatic amino acids can replace its function, the histidine at position 8 appears to function primarily through interaction of pi-electron in the unprotonated imidazole form, a form that has delocalized pi-electrons and aromatic character (21). The differences between H8Y, H8F, and H8W were observed in every titration, suggesting that the small rings of histidine, tyrosine, and phenylalanine side chains function better than the bulky indole ring of tryptophan. In addition, the greater infection by H8Y than H8F virus suggests that the hydroxyl group on tyrosine may be of some importance, possibly because it can participate in hydrogen bonds. The slightly greater infection of WT over H8Y virus may result from a more favorable geometry of histidine versus tyrosine (21). The increase in H8W virus infection upon addition of the second site suppressors is also consistent with size and hydrogen bonding capability being important, that is, infection by H8W virus increased when tyrosine replaced aspartic acid at position 243.
What might H8 interact with?
Considering that a pi-electron is essential, the most likely possibilities are other residues known to interact principally through pi-electrons and also by hydrogen bonding, that is, other histidine, tyrosine, or tryptophan residues. We favor the proposal of Lavillette et al. (13) and Barnett et al (4) that the carboxy-terminal domain of SU is the best candidate for the following reasons. The sequences of the carboxy-terminal domain of gammaretrovirus SU are highly conserved and include 17 aromatic ones as good candidates that we speculate are the interacting partners for H8. Although PRR sequences influence the fusigenicity and infectivity of Env (12, 24, 25), we do not favor this segment because few of its residues are conserved between ecotropic, amphotropic, and xenotropic MLVs, and none of these are aromatic residues. In addition, the possibilities that H8 interacts with lipids such as glycosphingolipids as has been shown for the fusion of human immunodeficiency virus type 1 (HIV-1) (18) or even with an unidentified coreceptor common to gammaretroviruses cannot be ruled out.
Q227R and D243Y suppression of an H8D substitution was much less than for the aliphatic changes and H8del.
We previously proposed a molecular model in which the presence of an arginine at position 227 in the RBD extends β-strand 9, bringing downstream residue 243 close enough to displace residue 8 (26) on the flexible amino terminus (it was disordered in the crystal structure). The level of infection of the triple H8D Q227R D243Y virus would be expected if the negative charge on the H8D side chain influenced the flexibility of the amino terminus. For example, a strong interaction between this aspartic acid with the nearby arginine 232 might reduce the ability of the D243Y substitution to replace residue 8.
Defective H8 mutants appear to arrest entry at the hemifusion stage.
Studies of the hemagglutinin (HA) protein of influenza virus have established that a hemifusion state is an intermediate on the pathway to complete fusion (11). The transmembrane domain and fusion peptide are involved in resolving the hemifusion intermediate into complete fusion (2, 7, 11, 14, 16, 17, 19). Deletion or substitution(s) of either segment of HA protein arrests cell-cell fusion at the hemifusion stage, but this block can be resolved by the membrane-curving agent chlorpromazine to give complete fusion (7, 14). We recently showed that replacement of histidine 8 by arginine (H8R) in Moloney MLV surface protein blocks infection by arresting membrane fusion at the hemifusion state. In addition, we showed that brief exposure to chlorpromazine not only relieves this block to cell-cell fusion but also rescues H8R virus infection of XC cells, NIH 3T3 cells, and human 293 cells expressing the Moloney MLV receptor cDNA (27). By applying chlorpromazine to virus-cell complexes of the H8 mutants, we observed a significant increase in infectivity (Fig. 4C), suggesting that each mutant arrested the fusion process at the hemifusion state, similar to our findings for the H8R mutant.
Speculation on the molecular mechanism of H8 function.
What is the essential role of H8 in virus-cell membrane fusion? One clue is that its function is conserved in the amphotropic MLV Env (13), suggesting that it is essential to a fundamental step of gammaretrovirus membrane fusion. Another clue is that the essential function can be provided in trans. Moreover, purified ecotropic RBD rescues infection by virus containing ecotropic, amphotropic, and xenotropic Env from which most of the RBD has been deleted (4, 13). In addition, T-cell-tropic feline leukemia virus has a proline instead of a histidine in its amino-terminal segment (22), and its host range is restricted to cells that secrete a protein, called FeLIX, that is nearly identical to the RBD of feline leukemia virus subgroup B (1). Thus, T-cell-tropic feline leukemia virus appears to be a naturally occurring histidine mutant whose spread is possible partly because it is rescued in trans by the presence of FeLIX.
We propose that an interaction of H8 is a key switchpoint in the conformation changes that occur after fusion peptide exposure and just prior to six-helix bundle formation. There is precedence for proposing a role of this type: hisitidine is often found as a controllable element in conformation changes (21). Studies of HIV-1 have suggested that the energy from transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, is sufficient to disrupt the hemifusion state and promote complete fusion (9, 15). Thus, one possibility for how the H8 down mutations arrest infection at the hemifusion state is that they prevent or slow down the conformation change just prior to six-helix bundle formation. For example, it may be necessary to separate SU from TM before the bundles can form and H8 may be a key residue in that dissociation. If the dissociation does not occur, is delayed, or occurs asynchronously in the absence of H8, then the bundles might not form or they might be induced at the wrong time so that the energy from their formation dissipates instead of being focused to resolve the hemifusion state.
Acknowledgments
We thank Bin Fang and Alpa Khatri for technical assistance with the virus-binding assays and XC cell-cell fusion assays and Alan Rein for the pRR88-p2E− plasmid encoding a truncation of Env at the normal place of R-peptide cleavage.
This study was supported by grant AI33410 from the NIAID, NIH (L.M.A.).
REFERENCES
- 1.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 7.Chernomordik, L. V., V. A. Frolov, E. Leikina, P. Bronk, and J. Zimmerberg. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol. 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doms, R., and J. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, L. E., R. Sowder, T. D. Copeland, G. Smythers, and S. Oroszlan. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 52:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 12.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F. L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 72:9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavillette, D., A. Ruggieri, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melikyan, G. B., R. M. Markosyan, M. G. Roth, and F. S. Cohen. 2000. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol. Biol. Cell 11:3765-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puri, A., J. Winick, R. J. Lowy, D. Covell, O. Eidelman, A. Walter, and R. Blumenthal. 1988. Activation of vesicular stomatitis virus fusion with cells by pretreatment at low pH. J. Biol. Chem. 263:4749-4753. [PubMed] [Google Scholar]
- 19.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson, J. S., and D. C. Richardson. 1989. Principles and patterns of protein conformation, p. 72-73. In G. D. Fasman (ed.), Prediction of protein structure and the principles of protein conformation. Plenum Press, Inc., New York, N.Y.
- 22.Rohn, J. L., M. S. Moser, S. R. Gwynn, D. N. Baldwin, and J. Overbaugh. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J. Virol. 72:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valsesia-Wittmann, S. 2001. Role of chimeric murine leukemia virus env β-turn polyproline spacers in receptor cooperation. J. Virol. 75:8478-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, B. W., P. M. Cannon, E. M. Gordon, F. L. Hall, and W. F. Anderson. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J. Virol. 72:5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zavorotinskaya, T., and L. M. Albritton. 1999. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J. Virol. 73:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zavorotinskaya, T., Z. Qian, J. Franks, and L. M. Albritton. 2004. A point mutation in the surface protein of murine leukemis virus arrests membrane fusion at the hemifusion state. J. Virol. 78:437-481. [DOI] [PMC free article] [PubMed]