Abstract
Objective:
Duchenne muscular dystrophy (DMD) is the most common single-gene lethal disorder. Substantial patient–patient variability in disease onset and progression and response to glucocorticoids is seen, suggesting genetic or environmental modifiers.
Methods:
Two DMD cohorts were used as test and validation groups to define genetic modifiers: a Padova longitudinal cohort (n = 106) and the Cooperative International Neuromuscular Research Group (CINRG) cross-sectional natural history cohort (n = 156). Single nucleotide polymorphisms to be genotyped were selected from mRNA profiling in patients with severe vs mild DMD, and genome-wide association studies in metabolism and polymorphisms influencing muscle phenotypes in normal volunteers were studied.
Results:
Effects on both disease progression and response to glucocorticoids were observed with polymorphism rs28357094 in the gene promoter of SPP1 (osteopontin). The G allele (dominant model; 35% of subjects) was associated with more rapid progression (Padova cohort log rank p = 0.003), and 12%–19% less grip strength (CINRG cohort p = 0.0003).
Conclusions:
Osteopontin genotype is a genetic modifier of disease severity in Duchenne dystrophy. Inclusion of genotype data as a covariate or in inclusion criteria in DMD clinical trials would reduce intersubject variance, and increase sensitivity of the trials, particularly in older subjects.
Duchenne muscular dystrophy (DMD) is a lethal childhood muscular disease characterized by progressive muscle weakness leading to loss of ambulation and early death due to respiratory or cardiac insufficiency. DMD is due to dystrophin gene mutations resulting in loss of dystrophin protein at muscle plasma membrane.1 To date, the only proven palliative treatment is chronic glucocorticoids treatment.2,3 The mechanisms of action of steroids in DMD are largely unknown, and response is variable. The natural history of DMD is also heterogeneous, with interpatient variability in disease progression, motor, respiratory, and cardiac involvement.4 It is believed that genetic modifiers (multigenic polymorphisms remote from the dystrophin gene) or environmental factors influence variability in disease progression and response to steroids.
This study focused on 29 candidate loci identified through various genetic analyses that might modulate muscle function in DMD. We identified SPP1 (osteopontin) as a strong genetic modifier of disease severity in DMD.
METHODS
Patients.
Two DMD patient cohorts were studied: one from the University of Padova (Padova cohort) and one from the Cooperative International Neuromuscular Research Group (CINRG) international clinical trial network (CINRG cohort). DMD diagnosis was achieved by the absence of dystrophin in skeletal muscle or the presence of out-of-frame DMD mutations.
Padova cohort.
We included 106 patients with DMD (table e-1A on the Neurology® Web site at www.neurology.org). Muscle strength was evaluated with the Medical Research Council (MRC) scale. A composite MRC score for upper (deltoid and triceps) and lower (ileo-psoas and quadriceps) limbs were calculated. Age at the beginning of steroid therapy, steroid therapy duration, and age at loss of ambulation were recorded.
For mRNA profiling, muscle biopsies were from time at diagnosis prior to any steroid treatment. Disease severity was defined by age at loss of ambulation. Muscle from severe vs mild DMD was defined using an outlier approach (patients showing the latest time to wheelchair vs the earliest) and the archival muscle then accessioned from original diagnostic muscle biopsy 10–20 years previously (table e-1B).
CINRG cohort.
A total of 156 subjects from the CINRG provided genomic DNA (table e-2). A total of 104 of 156 were nonambulatory at the time of this study. Grip strength phenotypes at study entry (baseline), using a standardized quantitative muscle testing system with audiovisual feedback (CQMS), was used for the analyses.5 For power calculations, we utilized both baseline and year 1 visit values.
Single nucleotide polymorphism genotyping.
Single nucleotide polymorphisms (SNPs) were selected from 3 different sources: 1) mRNA profiling in patients with severe vs mild DMD; 2) polymorphisms influencing muscle phenotypes in normal volunteers; 3) genome-wide association studies (GWAS) in metabolism (table e-3). Sequence variations were genotyped by TaqMan allele discrimination assays, denaturing high-pressure liquid chromatography, amplification refractory mutation system, or restriction fragment length polymorphism analysis.
mRNA profiling.
Labeled RNA was used in competitive hybridization to oligonucleotide microarrays platforms (GEO database ID: GPL6647) printed in-house by the CRIBI Microarray service (http://microcribi.cribi.unipd.it) in 2 replicates. Fluorescence was read with the ScanArray LITE confocal laser scanner (PerkinElmer). Images were quantified with ScanArray Express (PerkinElmer) software using the fixed circle method. Data were normalized using the total, Lowess, and z score methods, implemented in the TIGR Microarray Data Analysis System (MIDAS) software.6 Genes presenting more than 35% of pixels above 1 SD of the local background were considered for analysis. The genes with expression values in at least 9 experiments were analyzed by TMEV software8 performing unsupervised supported tree hierarchical clustering (ST)7 and correspondence analysis (COA).8 We used Pearson correlation single linkage method and a bootstrap approach with 100 iterations to calculate the support for the node of the tree. Two class unpaired SAM analysis9 using 210 permutation was performed.
Quantitative real-time RT-PCR.
Total RNA was reverse transcribed using SuperScript™ III Reverse Transcriptase (Invitrogen). USMG5, SPP1, and LILRA2 (primer sequences available upon request) expression were quantitated by real-time PCR using a ABI PRISM™ 7500 Real time PCR system (Applied Biosystems) with the SYBR® Green PCR Master Mix system (Applied Biosystems). As internal controls, TATA box binding protein, β-tubulin, and glucuronidase-β expression were used.
Statistical analyses.
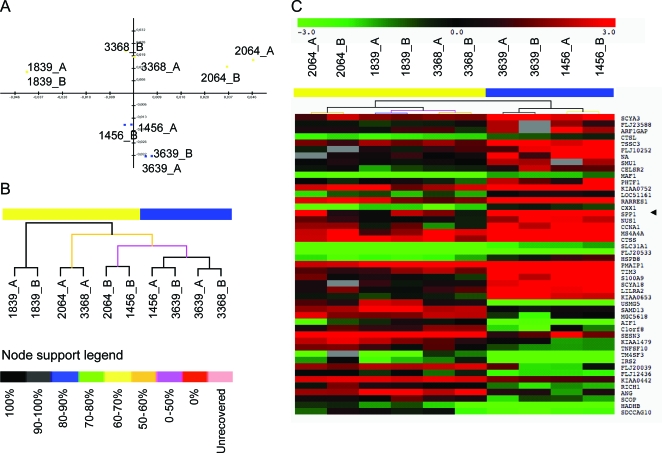
For the Padova cohort, 2 phenotypes, composite upper and lower limb MRC, and age at loss of ambulation were studied. Ordinal variables were analyzed using the Mann-Whitney U test, while for categorical variable the χ2 test was performed. Age at loss of ambulation, or age at last follow-up for ambulant patients, were analyzed using the Kaplan-Meier method; 95% confidence intervals (CIs) were calculated using the associated estimated SEs. In univariate analysis, the log-rank test was used to test the significance of each SNP. Multivariate analysis was performed using the Cox proportional hazards model, including the SNPs that showed a p < 0.1 in univariate analysis. For the CINRG cohort, grip CQMT strength was investigated. Both phenotypes were square root transformed to conform to normality. All analyses used the covariance models with age and current steroid use included as covariates. For those analysis of covariance models showing a significant p value, linear tests between each of the 3 genotype means were performed with the resulting p value adjusted for multiple comparisons using the Sidak method.
To determine the percent of variation in phenotype attributable to genotype, a likelihood ratio test compared the full model (phenotype, genotype, and all covariates) to the constrained model (phenotype and covariates only). A nominal p value of 0.05 was considered statistically significant; however, all p values were also assessed at a very conservative critical level of p = 0.0021 to account for multiple testing (24 tests; 3 cohorts, 2 genetic models, 2 phenotypes, 2 SNPs) (table 1).
Table 1.
Specific phenotype × genotype analyses
Abbreviations: CINRG = Cooperative International Neuromuscular Research Group; LL = lower limbs; MRC = Medical Research Council; QMT = quantitative muscle testing; UL = upper limbs.
Grip and total strength square root transformed.
All models adjusted for age and current steroid use.
The linear correlation between 2 variables was tested using the Pearson r or the Spearman rho.
Standard protocol approvals, registrations, and patient consents.
We received approval from ethical standards committees on human experimentation (institutional or regional) for any experiments using human subjects, and written informed consent was obtained from all patients (or guardians of patients) participating in the study (consent for research). The clinicaltrials.gov identifier number for the CINRG cohort is NCT00468832.
RESULTS
Selection of gene loci.
Twenty 9-gene loci were selected for testing as potential genetic modifiers. Two transcript units and associated polymorphisms were chosen from mRNA profiling comparing muscle biopsies (at diagnosis) from patients later experiencing severe vs mild disease progression; 8 loci were studied based upon previously established association with muscle or bone strength, size, or response to resistance training in normal populations (ACTN3 rs1815739; CHODL rs5842674; BC040297 rs2891837; DGKK rs7883609; GEMIN8 rs2168035 and rs7057480; KCDN1 rs2064034; TSPAN7 rs4826995)10–13 (Hoffman, unpublished data, in FAMuSS cohort of muscle strength, size, and response to training, and GWAS of bone volume); 19 loci were from GWAS studies of human metabolism (metabolic syndrome, circulating lipids, obesity). The rationale was that dystrophin-deficient muscle shows poor metabolic capacity,14–17 and that GWAS permitted sensitivity nonbiased scans of relevant loci (table e-3).
mRNA profiling in mild vs severe clinical course in DMD to identify candidate modifier loci.
The Padova cohort (106 patients) was studied longitudinally, and age at loss of ambulation was determined. The severe group included 2 patients (nos. 1,456, 3,639) showing loss of ambulation prior to age 10 years (mean 9.22 ± 0.6). The mild group included 3 patients (nos. 2,064, 1,839, 3,368) showing loss of ambulation >13 years (mean 16.7 ± 2.9) (table e-1B).
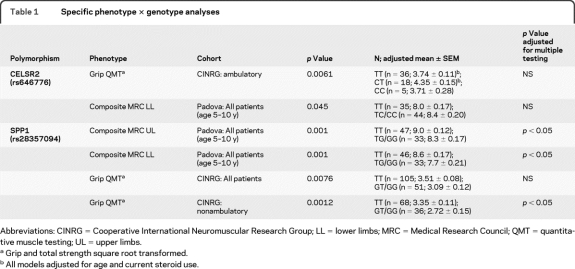
To identify differentially expressed mRNA transcripts, labeled RNA from muscle of patients with mild vs severe DMD was subjected to competitive hybridization with normal control pooled RNA on an oligonucleotide microarray platform. Correspondence analysis identified 2 groups by the microarray data corresponding to patients with DMD with late vs early loss of ambulation (figure 1A). ST analysis showed that patients with DMD with early loss of ambulation clustered together with 100% node support while patients with DMD with late loss of ambulation were clearly separated (with 0%–50% node support) (figure 1, B and C). Two-class SAM analysis showed 47 differentially expressed genes with 4.8 median false-positive genes (data available on request from authors).
Figure 1. Retrospective mRNA profiling of muscle biopsies from patients with Duchenne muscular dystrophy (DMD) showing mild vs severe clinical course.
(A) Component 1 (X-axis) and component 3 (Y-axis) plot of the correspondence analysis. Component 3 is seen to separate patients with DMD into 2 groups (yellow: 1,839, 3,368, 2,064; blue: 1,456, 3,639) corresponding to mild (yellow) vs severe (blue) clinical progression. (B) Bootstrap Pearson clustering analysis of the patients. Clustering of 1,456 and 3,639 patients is 100% bootstrap supported and is separated by the second cluster (3,668, 2,064, 1,839) linked to the first by a node presenting 0%–50% bootstrap supporting score. (C) Patients' bootstrap clusterization considering differentially expressed genes between the 2 patient groups. Gene expression values are expressed in log scale ratio relating to a common RNA control pool (log2 DMD patient/control pool). Right arrows highlight SSP1. All cluster tree presents the yellow/blue upper bar representing the subdivision of the patients with DMD in late loss of ambulation (yellow) and early loss of ambulation (red).
Real-time PCR confirmed the microarray mRNA expression of 3 genes tested: USMG5, SPP1, and LILRA2. SPP1 (secreted phosphoprotein 1, also called osteopontin) was selected based upon well-documented promoter polymorphisms involved in different human phenotypes (see Discussion) and in muscle remodeling.18 USMG5 (upregulated during skeletal muscle growth) was chosen due to its expression pattern. LILRA2 (leukocyte immunoglobulin-like receptor, subfamily A, member 2) has an inhibitory activity in monocytes, and thus may alter dystrophic muscle remodeling. Real-time PCR confirmed the decreased expression of SPP1 (−2.7-fold, p = 0.04) and LILRA2 (−3.2-fold, p = 0.023) and the increased expression of USMG5 (4.7-fold, p = 0.05) in patients with late vs early loss of ambulation.
Subsequent genotyping from mRNA-derived candidate genes was limited to 2 transcript units: SPP1 and USMG5. The SNP tested in SPP1 (rs28357094) has been previously shown to be associated with carotid intima thickness and type 1 diabetes.19,20 In USMG5, we studied a validated missense polymorphism (serine to proline) (rs11557060).
A test/validation cohort approach identifies SPP1/osteopontin (rs28357094) and CELSR2 (rs646776) as genetic modifiers of Duchenne dystrophy.
For the 29 candidate loci, we used a test cohort, then validation cohort approach. Each locus was initially tested for association with either the Padova longitudinal cohort (time to wheelchair [Kaplan-Meier] n = 106, and MRC score; n = 79) or the CINRG cross-sectional cohort (grip CQMS; n = 156). We established the threshold for significance in the test cohort as p = 0.01, and any locus meeting this threshold was subsequently tested in the second, validation cohort (data for all 29 loci in table e-4). Two polymorphisms were found associated with strength in the CINRG cohort as a test cohort: one muscle size/strength candidate, ACTN3 (rs1815739; p = 0.008), and one metabolism candidate, CELSR2 (rs646776; p = 0.006).21,22 One locus was found associated with disease severity in the Padova cohort as a test cohort: SPP1 (rs28357094; p = 0.001). These 3 loci were analyzed in the second cohort. When adjusting for multiple testing, only SPP1 remained significant in both cohorts (table 1).
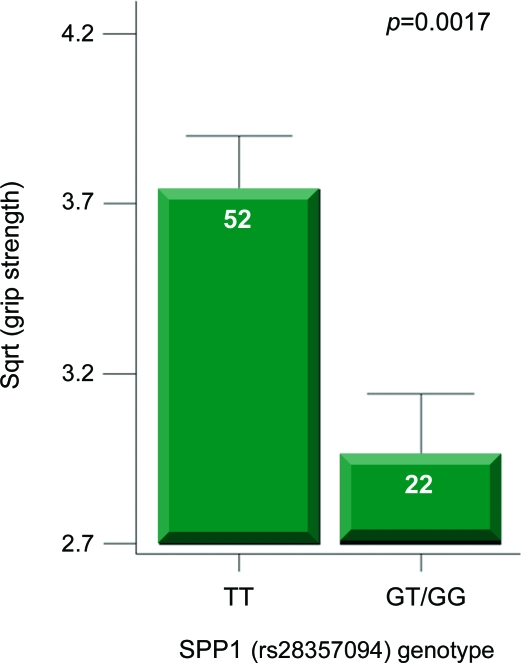
For SPP1, the promoter SNP studied (rs28357094) is immediately upstream of the transcriptional start site, and has been shown to alter transcriptional regulation of the gene through reporter assays.23 In both DMD cohorts, the rare G allele was found to be associated with greater weakness (table 1). In the Padova cohort, with assessment at age 5–10 years, both upper (p = 0.001) and lower (p = 0.001) limb MRC showed patients with the G allele to be significantly weaker. In the entire cross-sectional CINRG cohort, patients with the G allele showed weaker grip CQMS values using steroid use and age as covariates (p = 0.008). The highest effect size for genotype was seen in the grip strength values of nonambulatory steroid-treated boys with DMD (figure 2) (p = 0.0017).
Figure 2. SPP1 genotype is associated with decreased strength in steroid-treated patients with Duchenne muscular dystrophy.
Shown is the association of SPP1 genotype with muscle strength (grip quantitative muscle testing) in the Cooperative International Neuromuscular Research Group cross-sectional cohort. The strongest association with strength was seen in steroid-treated, nonambulatory patients.
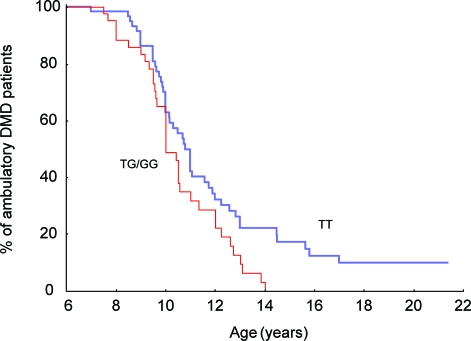
The muscle strength data showing association of the rare G allele with loss of strength suggested that the G allele carriers may show a more rapid disease progression. Kaplan-Meier plots of the longitudinal Padova cohort confirmed that patients with DMD with the G allele lost ambulation at younger age (p = 0.035; table 1; figure 3). The log rank confirmed also the relation (log rank, p = 0.0006) between median age at loss of ambulation and steroid therapy (treated patients with DMD, median age at loss of ambulation 11 years [95% CI 9.9–12 years]; untreated boys with DMD, 10 years [95% CI 9.8–10.2 years]). There was no significant difference in percentage of steroid-treated and untreated patients with respect to rs28357094 genotype (χ2 p = 0.34). In multivariate analysis (Cox proportional hazard method), both rs28357094 genotype (p < 0.05) and steroid therapy (p < 0.0005) were related to loss of ambulation. Spearman rho test showed that increasing age was associated with decreasing muscle strength (rho = −0.35, p < 0.002 for lower limbs and rho = −0.28, p < 0.02 for upper limbs).
Figure 3. SPP1 genotype is associated with greater severity of progression in Duchenne muscular dystrophy (DMD).
The proportion of patients with DMD in the Padova cohort remaining ambulatory at the specific age noted is shown (n = 106). The GT/GG genotype is associated with more rapid progression.
Effect of including genotype stratification on power calculations in clinical trials.
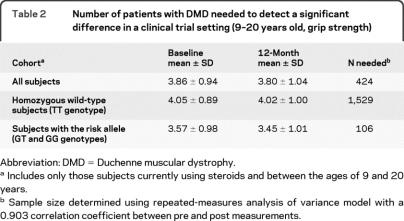
Data in both the Padova and CINRG cohorts showed that carriers of the SPP1 at-risk G genotype (35% of patients) had greater weakness and more rapid progression compared to the ancestral T genotype. Most clinical trials in DMD have studied the loss of strength as a function of time, where the goal of the therapeutic intervention was to slow the rate of strength loss. Table 2, columns 2 and 3, describes what would be expected to happen to a typical control arm in a future trial, showing little loss of strength over 12 months in the TT group, whereas G allele carriers showed greater strength loss. We calculated the number of subjects required to detect a 15% deflection in the loss of strength, based upon genotype (table 2, column 4). The TT patients required very large numbers of subjects (n = 1,529) to detect a 15% deflection of strength, whereas the GT/GG group required 1/15 the patient numbers (n = 106) to observe the same strength improvement over 1 year (table 2).
Table 2.
Number of patients with DMD needed to detect a significant difference in a clinical trial setting (9–20 years old, grip strength)
Abbreviation: DMD = Duchenne muscular dystrophy.
Includes only those subjects currently using steroids and between the ages of 9 and 20 years.
Sample size determined using repeated-measures analysis of variance model with a 0.903 correlation coefficient between pre and post measurements.
DISCUSSION
Two gene polymorphisms were identified as potential genetic modifiers of DMD in both the Padova and CINRG cohorts: SPP1/osteopontin and CELSR2. However, when adjusting for multiple testing, only SPP1/osteopontin remained statistically significant.
The effect of SPP1 genotype on progression was similar in magnitude to the pharmacologic use of steroids (about 1 year difference in the loss of ambulation). The Padova patients with the less common G allele were weaker using both upper and lower limbs MRC (p = 0.001) and also showed earlier loss of ambulation. At age 14 years, 20% of patients with DMD with TT genotype were ambulatory, whereas none with GT/GG genotype were ambulatory. In the CINRG cohort, patients with the G allele showed lower grip strength (p = 0.001), with the effect size largest within the nonambulatory, steroid-treated patients (figure 2). By grip strength, GT/GG patients showed a 12% decrease in strength relative to TT across the age range, and this increased to 19% difference in nonambulatory patients.
The rs28357094 polymorphism is 66 bp upstream of the transcriptional start site, and has been shown to alter SPP1 binding to the promoter. The rare G allele has been shown to decrease promoter strength, leading to lower SPP1 mRNA production from the gene, by in vitro cell transfection assays in nonmuscle cells.23 SPP1 has been intensively studied as a marker for tumor progression, and its role in inflammation and tissue remodeling in many pathologies and disorders.24–26
The goal of most clinical trials in DMD is to slow the progression of the disease. However, since the rate of progression is variable, large numbers are required for appropriate statistical powering. We used the CINRG cross-sectional data to carry out power calculations on genotype-stratified subcohorts. Assuming that a 15% deflection of strength loss is clinically significant, fewer patients with DMD with the G genotype are needed to detect a slowing of progression (n = 106), compared to those homozygous for the ancestral T genotype (n = 1,520 patients required).
The identification of genetic modifiers also provides insights into disease pathogenesis.27 Osteopontin is elevated in dystrophin-deficient muscle, both in humans14,28 and mdx mice.29 However, elevations of SPP1 are not specific to dystrophin deficiency, and are seen in multiple dystrophies, including Becker muscular dystrophy, calpain 3 (LGMD2A), and dysferlin deficiency (LGMD2B),18 and mouse models of muscular dystrophy.30
SPP1 is considered an inflammatory marker; however, inflammatory muscle diseases do not show striking OPN levels.18,31 Thus, SPP1 elevations may correspond with muscle remodeling rather than inflammation. In the mdx mouse, SPP1 was shown to be expressed by both muscle fibers and a subset of muscle inflammatory cells.18 Double knock-out (SPP1-null, dystrophin-null) mice showed less fibrosis and less functional deficits, suggesting that SPP1 expression is deleterious to dystrophic muscle.18 The mdx/SPP1 data and the DMD data presented here may seem contradictory; in mdx mice less SPP1 leads to improvement, whereas in patients with DMD less SPP1 leads to greater weakness. However, SPP1 is a multifunctional molecule that has positive roles in muscle regeneration32 and negative roles in muscle inflammation, and the mouse and human data may simply highlight these different roles for SPP1.
Taking into account all the findings presented here, the less common G allele of the rs28357094 SPP1/osteopontin promoter polymorphism is associated in DMD with greater weakness and younger age at loss of ambulation. Further research is needed to define the role of the rs28357094 promoter polymorphism on DMD pathophysiology. Regardless of the molecular action, it is important to note that stratification of patients with DMD by genotype has the potential to increase the statistical power and sensitivity of clinical trials. Given the data presented, the clinical trial planner would have 2 choices: either open the trial to all comers or restrict it to the G subgroup, in which case the savings (reduced numbers of patients needed for the trial) is roughly ¼. Conversely, the number of patients with the more severe G genotype is less common, necessitating pretrial screening of larger numbers of patients with DMD.
Supplementary Material
Editorial, page 208
Supplemental data at www.neurology.org
- CI
- confidence interval
- CINRG
- Cooperative International Neuromuscular Research Group
- COA
- correspondence analysis
- DMD
- Duchenne muscular dystrophy
- GWAS
- genome-wide association studies
- MRC
- Medical Research Council
- SNP
- single nucleotide polymorphism
- ST
- supported tree hierarchical clustering
AUTHOR CONTRIBUTIONS
Statistical analysis was conducted by Dr. Mario Ermani and Dr. Heather Gordish-Dressman.
COINVESTIGATORS
Data were collected by the Cooperative International Neuromuscular Research Group with clinical centers as follows: University of California, Davis—T. Abresch (Study Co-Chair), E. Henricson (Study Co-Chair), M. Cregan, M. Glicke, J. Han, L. Johnson, N. Joyce; Sundaram Medical Foundation—C. Chidambaranathan, S. Kumar; Bloorview MacMillan Medical Center—D. Biggar, L. Eliasoph, V. Harris, E. Hosaki; Alberta Children's Hospital—J. Mah, A. Chiu, E. Goia, L. Walker, C. Wright, M. Yousefi; Queen Silvia Children's Hospital—M. Tulinius, A.C. Ahlander, L. Berland, A.K. Kroksmark, U. Sterky; Children's National Medical Center—D. Escolar, R. Leshner, M. Birkmeier, S. Kaminski, K. Parker, C. Spurney, C. Tesi-Rocha; Royal Children's Hospital—A. Kornberg, K. Carroll, K. Devalle, R. Kennedy, M. Ryan, D. Villano; Hadassah Hebrew University Hospital—Y. Nevo, E. Wisband, D. Yaffe; Favaloro Foundation—A. Dubrovsky, J. Corderi, L. Levy, L. Mesa; Mayo Clinic—N. Kuntz, K. Coleman, S. Driscoll, A. Hoffman, W. Korn-Peterson, D. Selcen; IRCCS C Mondino Foundation Hospital—K. Gorni, L. Capone; University of Pittsburgh—P. Clemens, H. Abdel-Hamid, C. Bise, A. Craig, L. Hache, C. Nguyen; Washington University–St. Louis—A. Connolly, J. Florence, B. Malkus, A. Pestronk, R. Renna, J. Schierbecker, C. Siener, C. Wulf; Children's Hospital of Virginia—J. Teasley, S. Blair, B. Grillo, E. Monasterio; University of Tennessee–Memphis—T. Bertorini, J. Clifft, C. Feliciano, M. Igarashi; Children's Hospital at Westmead—K. North, T. Juarez, K. Rose, R. Webster, S. Wicks; University of Alberta—H. Kolski, L. Chen, C. Kennedy, G. Parks, J. Wohlers; University of Puerto Rico—J. Carlo, B. Deliz, S. Espada, P. Fuste, C. Luciano; Texas Children's Hospital—T. Lotze, H. Farber, A. Gupta, S. Habetz, J. Jeffries, R. McNeil; University of Minnesota—J. Day, A. Erickson, M. Margolis, G. Parry, D. Walk; and CINRG Coordinating Center—A. Cnaan, A. Arrieta, N. Bartley, P. Canelos, T. Duong, F. Hu, A. Zimmerman. The CINRG Web site is located at www.cinrgresearch.org.
DISCLOSURE
Dr. Pegoraro serves on a scientific advisory board for BioMarin Pharmaceutical Inc.; has received funding from MEDA Pharmaceuticals Inc.; and receives research support from Wellstone Grant 8568-01-01 and Italian Telethon. Dr. Hoffman, Dr. Piva, Dr. Gavassini, Dr. Cagnin, Dr. Ermani, Dr. Bello, Dr. Soraru, Dr. Pacchioni, Dr. Lanfranchi report no disclosures. Dr. Angelini serves on the editorial boards of Neurology, Neuromuscular Disorders, Journal of Neurological Sciences, Current Opinion of Neurology, and Therapeutic Advances in Neurological Disorders. Dr. Kesari, Dr. Lee, Dr. Devaney report no disclosures. Dr. Gordish-Dressman receives research support from the NIH (NICHD and NINDS). Dr. McDonald serves on scientific advisory boards for PTC Therapeutics, Inc., GlaxoSmithKline, BioMarin Pharmaceutical Inc., and Gilead Sciences, Inc.; and receives research support from PTC Therapeutics, Inc., the Muscular Dystrophy Association, and Clinical Research Network in Duchenne Muscular Dystrophy.
REFERENCES
- 1. Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987; 51: 919–928 [DOI] [PubMed] [Google Scholar]
- 2. Bushby K, Muntoni F, Urtizberea A, et al. Report on the 124th ENMC International Workshop: treatment of Duchenne muscular dystrophy: defining the gold standards of management in the use of corticosteroids. Neuromuscul Disord 2004; 14: 526–534 [DOI] [PubMed] [Google Scholar]
- 3. Moxley RT, Ashwal S, Pandya S, et al. Practice parameter: corticosteroid treatment of Duchenne dystrophy. Neurology 2005; 64: 13–20 [DOI] [PubMed] [Google Scholar]
- 4. Desguerre I, Christov C, Mayer M, et al. Clinical heterogeneity of Duchenne muscular dystrophy (DMD): definition of sub-phenotypes and predictive criteria by long-term follow-up. PLOS One 2009; 4: e4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayhew JE, Florence JM, Mayhew TP, et al. Reliable surrogate outcome measures in multicenter clinical trials of Duchenne muscular dystrophy. Muscle Nerve 2007; 35: 36–42 [DOI] [PubMed] [Google Scholar]
- 6. Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34: 374–378 [DOI] [PubMed] [Google Scholar]
- 7. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998; 95: 14863–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fellenberg K, Hauser NC, Brors B, et al. Correspondence analysis applied to microarray data. Proc Natl Acad Sci USA 2001; 98: 10781–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarkson PM, Hoffman EP, Zambraski E, et al. ACTN3 and MLCK genotype associations with exertional muscle damage. J Appl Physiol 2005; 99: 564–569 [DOI] [PubMed] [Google Scholar]
- 11. Thompson PD, Moyna N, Seip R, et al. Functional polymorphisms associated with human muscle size and strength. Med Sci Sports Exerc 2004; 36: 1132–1139 [DOI] [PubMed] [Google Scholar]
- 12. Yang N, MacArthur DG, Gulbin JP, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet 2003; 73: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walsh S, Liu D, Metter EJ, et al. ACTN3 genotype is associated with muscle phenotypes in women across the adult age span. J Appl Physiol 2008; 105: 1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen YW, Nagaraju K, Bakay M, et al. Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology 2005; 65: 826–834 [DOI] [PubMed] [Google Scholar]
- 15. Wehling-Henricks M, Oltmann M, Rinaldi C, et al. Loss of positive allosteric interactions between neuronal nitric oxide synthase and phosphofructokinase contribute to defects in glycolysis and increased fatigability in muscular dystrophy. Hum Mol Genet 2009; 18: 3439–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Onopiuk M, Brutkowski W, Wierzbicka K, et al. Mutation in dystrophin-encoding gene affects energy metabolism in mouse myoblasts. Biochem Biophys Res Commun 2009; 386: 463–466 [DOI] [PubMed] [Google Scholar]
- 17. Hsieh TJ, Jaw TS, Chuang HY, et al. Muscle metabolism in Duchenne muscular dystrophy assessed by in vivo proton magnetic resonance spectroscopy. J Comput Assist Tomogr 2009; 33: 150–154 [DOI] [PubMed] [Google Scholar]
- 18. Vetrone SA, Montecino-Rodriguez E, Kudryashova E, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-β. J Clin Invest 2009; 119: 1583–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de las Fuentes L, Gu CC, Mathews SJ, et al. Osteopontin promoter polymorphism is associated with increased carotid intima-media thickness. J Am Soc Echocardiogr 2008; 21: 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marciano R, D'Annunzio G, Minuto N, et al. Association of alleles at polymorphic sites in the Osteopontin encoding gene in young type 1 diabetic patients. Clin Immunol 2009; 131: 84–91 [DOI] [PubMed] [Google Scholar]
- 21. Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans [erratum 2008;40:1384]. Nat Genet 2008; 40: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sandhu MS, Waterworth DM, Debenham SL, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet 2008; 371: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giacopelli F, Marciano R, Pistorio A, et al. Polymorphisms in the osteopontin promoter affect its transcriptional activity. Physiol Genomics 2004; 20: 87–96 [DOI] [PubMed] [Google Scholar]
- 24. Fedarko NS, Jain A, Karadag A, et al. Elevated serum bone sialoprotein and osteopontin in colon, breast and prostate, and lung cancer. Clin Cancer Res 2001; 40: 4060–4066 [PubMed] [Google Scholar]
- 25. Scatena M, Liaw L, Giachelli CM. Osteopontin: a multifunctional molecule regulating chronic inflammation and vascular disease. Arterioscl Thromb Vasc Biol 2007; 27: 2302–2309 [DOI] [PubMed] [Google Scholar]
- 26. Golledge J, Muller J, Shephard N, et al. Association between osteopontin and human abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol 2007; 27: 655–660 [DOI] [PubMed] [Google Scholar]
- 27. Collaco JM, Cutting GR. Update on gene modifiers in cystic fibrosis. Curr Opin Pulm Med 2008; 14: 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell Biol 2000; 151: 1321–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Porter JD, Khanna S, Kaminski HJ, et al. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet 2002; 11: 263–272 [DOI] [PubMed] [Google Scholar]
- 30. Turk R, Sterrenburg E, van der Wees CG, et al. Common pathological mechanisms in mouse models for muscular dystrophies. FASEB J 2006; 20: 127–129 [DOI] [PubMed] [Google Scholar]
- 31. Marhaug G, Shah V, Shroff R, et al. Age-dependent inhibition of ectopic calcification: a possible role for fetuin-A and osteopontin in patients with juvenile dermatomyositis with calcinosis. Rheumatology 2008; 47: 1031–1037 [DOI] [PubMed] [Google Scholar]
- 32. Uaesoontrachoon K, Yoo HJ, Tudor EM, et al. Osteopontin and skeletal muscle myoblasts: Association with muscle regeneration and regulation of myoblast function in vitro. Int J Biochem Cell Biol 2008; 40: 2303–2314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.