Abstract
The Mucin 1 (MUC1) oncoprotein is aberrantly overexpressed in diverse human malignancies including breast and lung cancer. Although MUC1 modulates the activity of several transcription factors, there is no information regarding the effects of MUC1 on global gene expression patterns and the potential role of MUC1-induced genes in predicting outcome for cancer patients. We have developed an experimental model of MUC1-induced transformation that has identified the activation of gene families involved in oncogenesis, angiogenesis, and extracellular matrix remodeling. A set of experimentally derived MUC1-induced genes associated with tumorigenesis was applied to the analysis of breast and lung adenocarcinoma cancer databases. A 35-gene MUC1-induced tumorigenesis signature predicts significant decreases in both disease-free and overall survival in patients with breast (n = 295) and lung (n = 442) cancers. The data show that the MUC1 oncoprotein contributes to the regulation of genes that are highly predictive of clinical outcome in breast and lung cancer patients.
Introduction
Mucin 1 (MUC1) is a member of a family of extensively O-glycosylated proteins that are predominantly expressed by epithelial cells. MUC1 and other mucins constitute a physical barrier that protects epithelia from damage induced by exposure to the external environment (1). MUC1 consists of two subunits that form a stable heterodimer after translation as a single polypeptide and autocleavage in the endoplasmic reticulum (2, 3). The MUC1 NH2-terminal subunit (MUC1-N) is the mucin component of the dimer containing variable numbers of 20 amino acid tandem repeats that are subject to O-glycosylation (4). At the apical border of the normal epithelial cell, MUC1-N extends beyond the glycocalyx through a noncovalent complex with the transmembrane MUC1 COOH-terminal subunit (MUC1-C; ref. 5). In concert with transformation and loss of polarity, MUC1 is aberrantly overexpressed over the entire surface of breast carcinoma cells (6). Importantly in this regard, overexpression of MUC1 is sufficient for the induction of anchorage-independent growth and tumorigenicity (7). Specifically, other studies have shown that the MUC1-C cytoplasmic domain (MUC1-CD) is responsible for the induction of transformation and that the MUC1-N subunit is dispensable for this response (8). Overexpression of MUC1 also confers resistance to stress-induced cell death, including that conferred by exposure to genotoxic anticancer agents (9–11). Targeting of the MUC1-C subunit to the nucleus attenuates p53-mediated apoptosis in the response to DNA damage (12). However, it is not known to what extent MUC1-C can alter transcriptional patterns of cells overexpressing this gene and how these patterns may relate to MUC1-C associated tumorigenesis. As well, it is not known whether MUC1-C–induced genes are associated with poor prognosis in breast cancer and potentially other types of cancer.
Materials and Methods
Cell culture and tumor models
Rat 3Y1 embryonic fibroblasts were transfected by an empty vector (3Y1/Vector) or one expressing the cytoplasmic domain of MUC1 (3Y1/MUC1-CD) and maintained as described (8). 3Y1/Vector and 3Y1/MUC1-CD cells were injected s.c. into the hind limbs of female athymic mice (FCRI-Taconic) in increasing concentrations from 102 to 107 cells in 100 µL of PBS per mouse. Ten mice were injected at each cell concentration. Tumor volume was determined by direct measurement with calipers and calculated using the formula (length × width × depth/2). When the tumor volume was >2,000 mm3, mice were euthanized using CO2 followed by cervical dislocation. Tumors were excised, snap frozen in liquid nitrogen, and stored at −80°C until RNA extraction. All animal experiments were conducted in accordance with institutional guidelines at The University of Chicago.
RNA extraction and purification
RNA was collected and purified from confluent cell cultures using TRIzol reagent (Invitrogen Life Sciences) according to the manufacturer’s instructions. Frozen xenografts were sectioned into pieces ~5 mm3 in size and soaked overnight in RNAlater-ICE solution (Applied Biosystems-Ambion). Samples were spun, washed in RLT buffer (QIAGEN), and homogenized on ice using a mechanical glass-Teflon homogenizer set at 3,000 rpm. Further purification was performed using a combination of RNeasy spin columns and TRIzol reagent, as previously described (13). The quality of samples was assessed using gel electrophoresis in 1.8% agarose and spectrophotometry, and samples of high quality were transferred to the Functional Genomics Facility of The University of Chicago for labeling and hybridization with GeneChip Rat Genome 230 2.0 Arrays (Affymetrix).
Statistical analysis of DNA microarray data
The selection and analysis of genes differentially expressed in 3Y1/Vector and 3Y1/MUC1-CD cells in vitro and 3Y1/MUC1-CD xenografts was based on previously detailed approaches (14–17). Briefly, each array was hybridized with a pooled sample normalized to total RNA and consisting of RNA obtained from three independent xenografts or cell lines. After data retrieval and scaling using MAS 5.0 suit (Affymetrix), data were rescaled using “global median normalization” across the entire data set (17) and filtered using a multistep filtration method, which involves the application of Receiver Operating Characteristic analysis for the estimation of cutoff signal intensity values (14). Subsequent analysis was based on pair-wise comparisons (3Y1/Vector in vitro versus 3Y1/MUC1-CD in vitro and 3Y1/MUC1-CD in vitro versus 3Y1/MUC1-CD in vivo) of duplicated arrays using Significance Analysis of Microarrays (18) version 3.0. Differentially expressed probe set IDs were selected using a 2.0-fold induction cutoff level with a False Discovery Ratio of 0. Selected probe set IDs were gene annotated and functionally designated using Ingenuity Pathways Analysis (Ingenuity Systems, Inc.).
Statistical analysis of clinical cancer databases
We analyzed two publicly available cancer databases containing expressional data from breast cancer (19) and adenocarcinoma of the lung (20) to determine if the genes representing MUC1-CD–induced tumorigenesis have predictive value in determining the outcome for each patient sample. Statistical analyses were performed using samples for which survival data were available, which included 295 breast cancer and 442 lung adenocarcinoma cases. All statistical analyses were performed using JMP 7.1 (SAS Institute, Inc.). The raw signal intensity for each probe set ID of interest for each patient was normalized to the median value of the probe set ID across the entire database and subsequently log2 transformed. Multiple probe set IDs for a given gene were averaged for each patient sample to obtain a representative expression value for each gene. Expression data were clustered using hierarchical clustering via Ward’s method to visualize gene expression patterns across each database. Genes having uniform expression across the patient samples were eliminated and not used for further analyses. K-means clustering was performed to partition the patient samples into two clusters, and principal component analysis was used to visualize the clusters. To identify the genes that were differentially expressed between the two patient clusters, quantile-quantile plots were used to confirm that the gene expression distribution of each gene was approximately normally distributed across the database. Subsequently, F tests were used to test the null hypothesis of equal variance for each gene between the two patient clusters. The result of the F test (equal or unequal variance) was input into an unpaired two-tailed Student’s t test to test the null hypothesis of equal magnitude of expression of each gene between the two patient clusters. The α level for each t test was 0.05 but corrected for multiple comparisons using a Bonferroni correction that adjusted the α level to 0.05 divided by the number of comparisons of interest (i.e., the number of genes in the analysis). Kaplan-Meier survival analysis was performed to determine whether clustering based on the differential expression of the MUC1-CD–induced genes could identify patients with decreased survival. Survival analysis was performed on clusters defined by k-means clustering, and log-rank tests were used to test the null hypothesis of equal survival functions between the two patient clusters. To derive the 35-gene MUC1-CD–induced Tumorigenesis Signature (MTS), the approaches outlined above were used to select the genes that were differentially expressed in each database but similarly regulated (either overexpressed or underexpressed) in both databases.
Results
To assess the effects of MUC1 on gene expression patterns, 3Y1 fibroblasts stably transfected with an empty vector (3Y1/Vector) or MUC1-CD (3Y1/MUC1-CD) were injected into the hind limbs of athymic mice. Tumor formation was not detectable when 107 3Y1/Vector cells were injected (Fig. 1A). By contrast, tumors were formed after injection of 104 3Y1/MUC1-CD cells (Fig. 1A). Tumor formation was also detectable when injecting 103, but not 102, 3Y1/MUC1-CD cells (Fig. 1A). To investigate the genes associated with MUC1-CD–induced tumorigenesis, we performed expression profiling of 3Y1/Vector and 3Y1/MUC1-CD cells growing in vitro, and of 3Y1/MUC1-CD cells growing as tumor xenografts. Using Significance Analysis of Microarrays, we found 299 probe set IDs corresponding to 290 genes differentially expressed in 3Y1/MUC1-CD compared with 3Y1/Vector cells grown in vitro. Moreover, 322 probe set IDs corresponding to 282 unique genes were differentially expressed in 3Y1/MUC1-CD cells growing as tumor xenografts compared with that in vitro (Supplementary Table S1). Importantly, the transition from growth of 3Y1/MUC1-CD cells in vitro to in vivo was associated with significant increases (10- to 1,000-fold) in the expression of genes encoding proteins involved in cytoskeletal organization and extracellular matrix formation (Fig. 1B). In addition, there were substantial increases in genes encoding cytokines and growth factors (Fig. 1B). These findings indicated that MUC1-CD–induced tumorigenicity is associated with distinct changes in gene expression.
Figure 1.
MUC1-CD induces tumorigenesis that is accompanied by distinct changes in gene expression. A, tumor formation of 3Y1/Vector and 3Y1/MUC1-CD cells. Points, mean; bars, SE. B, significant increases in gene expression in transition of 3Y1/MUC1-CD cells grown in vitro to in vivo.
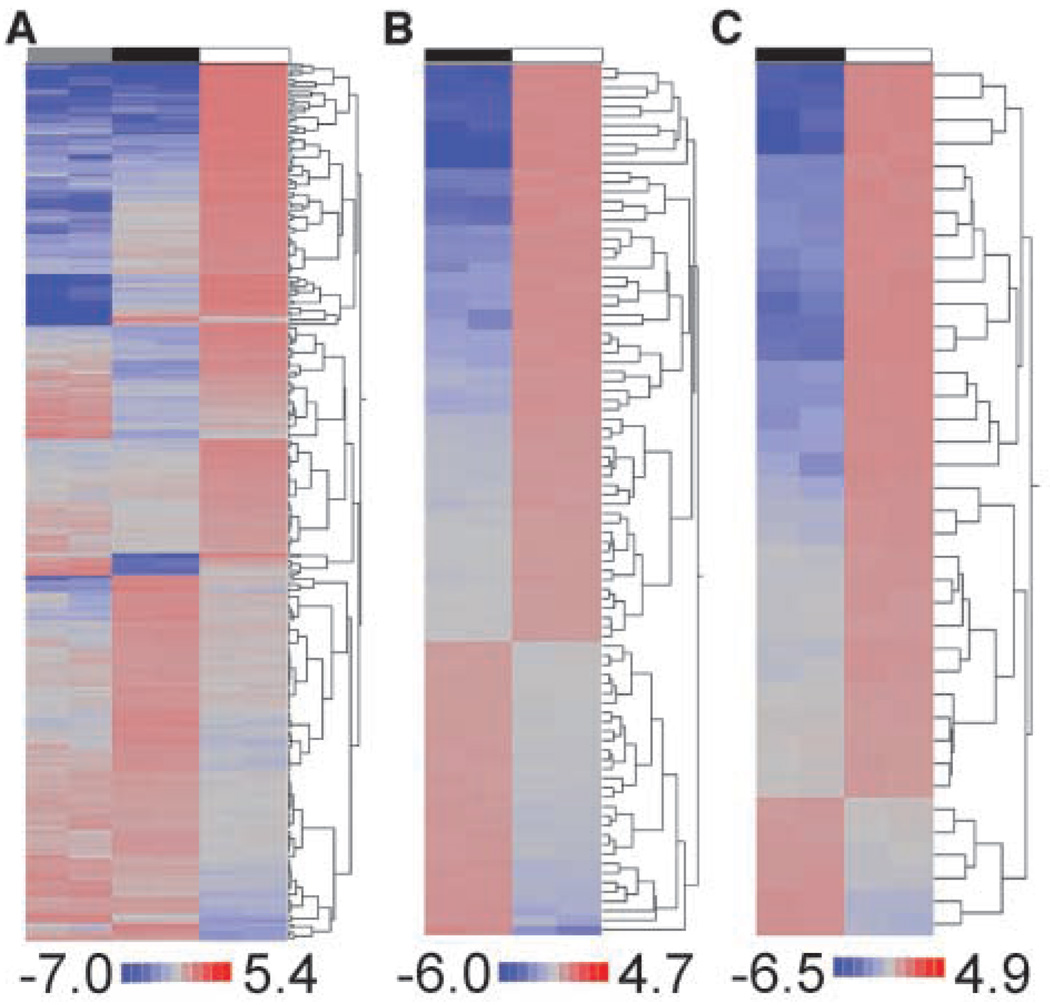
Expressional clustering of Significance Analysis of Microarrays–selected genes identified unique patterns for the 3Y1/Vector and 3Y1/MUC1-CD cells grown in vitro and for the 3Y1/MUC1-CD cells grown as tumors (Fig. 2A). The combination of expressional and functional gene clustering identified two broad subgroups of genes functionally representing tumorigenesis (subgroup 1) and cell motility, metastasis, and angiogenesis (subgroup 2). Subgroup 1 is composed of 92 tumorigenesis-related genes with 62 up-regulated and 30 down-regulated in vivo (Fig. 2B). Up-regulated genes included those linked to cell cycle regulation and proliferation (Fig. 2B). Up-regulated genes in subgroup 1 are also those involved in stress responses (Egr1, Gadd45), the Wnt signaling pathway (Axin2), cell-cell and cell–extracellular matrix interactions, and inflammation (Fig. 2B). Down-regulated genes include those involved in DNA repair (Xrcc5), checkpoint control (Bub1b), cell adhesion (Cdh2), and steroid metabolism (12 genes; Fig. 2B). Subgroup 2 included 38 genes that were predominantly up-regulated in vivo and involved in promoting cell motility, induction of angiogenesis, and activation of cell-cell and cell–extracellular matrix interactions (Fig. 2C). Ingenuity Pathway Analysis was further used to assign known functions to 175 of the genes that were up-regulated in vivo. Based on this analysis, 100 of these genes are functionally linked to cancer. For example, 44 genes are associated with cell proliferation and another 39 genes encode proteins that control cell death (Supplementary Table S2). Moreover, 36 genes have been functionally linked to the development of metastases (Supplementary Table S2). Eight genes related to tumor angiogenesis (Angptl4, Col18a1, Cst3, Ctss, Itga1, Mmp2, Mmp14, and Serpine1) were also up-regulated in the 3Y1/MUC1-CD tumor xenografts compared with that obtained when these cells were grown in vitro. These findings thus collectively indicate that MUC1-CD–induced tumorigenesis is associated with the activation of genes that functionally contribute to the growth of tumors in vivo.
Figure 2.
MUC1-CD–induced tumorigenesis is associated with the activation of genes that functionally contribute to tumor growth. A, expressional clustering of all differentially expressed genes. B, tumorigenesis subset. C, metastasis, cell motility, and angiogenesis subset. Gray, 3Y1/Vector in vitro; black, 3Y1/MUC1-CD in vitro; white, 3Y1/MUC1-CD in vivo. Red, overexpressed. Values are mean-normalized log2-transformed signals.
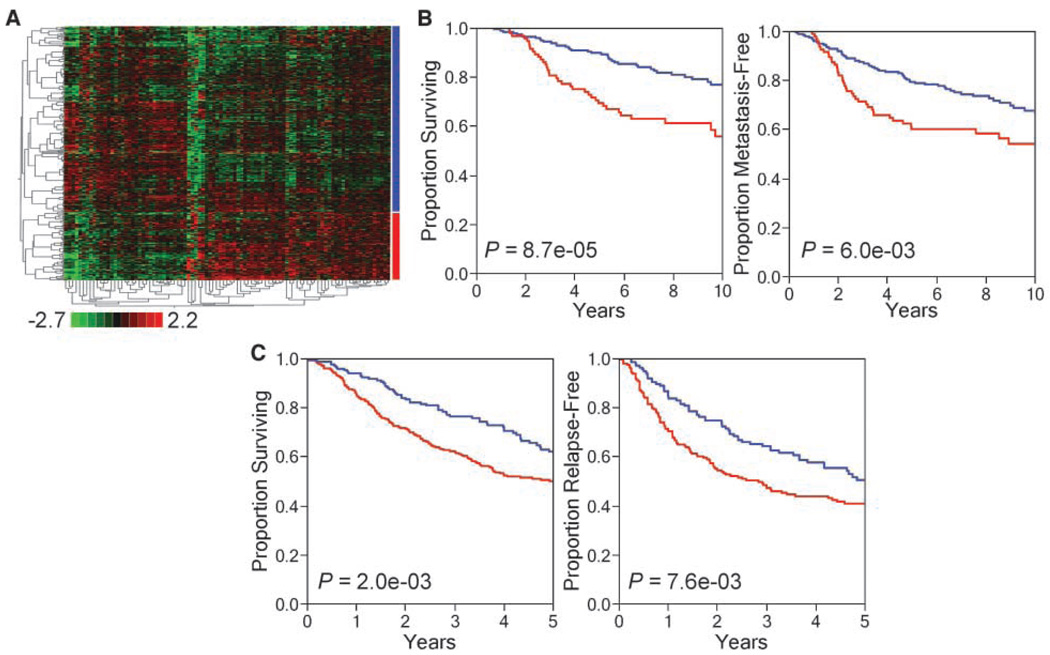
We hypothesized that the genes representing MUC1-CD–induced tumorigenesis predict a worse prognosis in cancer patients. To test this hypothesis, we used two independent expressional databases representing 295 cases of breast cancer and 442 cases of lung adenocarcinoma (see Materials and Methods). First, we used the 282 genes differentially expressed in 3Y1/MUC1-CD cells growing as tumor xenografts compared with genes expressed in vitro and representing MUC1-CD–induced tumorigenesis (see Supplementary Table S1) for analysis of the breast cancer database. We derived a set of 93 MUC1-CD–induced genes (marked by * in Supplementary Table S1) using gene expression clustering and the statistical selection of differentially expressed genes (see Materials and Methods). Figure 3A shows stratification of the breast cancer samples into two distinct subgroups differing in expression of the 93-gene set. Using Kaplan-Meier statistics, we found that tumors from breast cancer patients expressing this 93-gene set had a significantly worse prognosis than those from patients not expressing these genes (overall survival, P = 8.7e-05; metastasis-free survival, P = 6.0e-03; Fig. 3B). We further analyzed these 93 genes in a composite database of patients with adenocarcinoma of the lung. These 93 genes were also prognostic in this database (overall survival, P = 2.0e-03; relapse-free survival, P = 7.6e-03; Fig. 3C). Using the statistical approaches outlined above, we narrowed the 93 genes based on similarities in gene expression between the breast and lung cancer databases (see Materials and Methods). Table 1 lists the 35 genes that remained after this analysis. This subset of genes was named the MUC1 tumorigenesis signature (MTS). The results show k-means clustering of the breast (Fig. 4A) and lung cancer (Fig. 4B) databases into two distinct groups based on expression of the MTS, which we defined as MTS+ (expressor) and MTS− (nonexpressor). The MTS+ group represents 37% and 42% of breast and lung cancer patients, respectively. Importantly, in both types of cancers, the MTS+ group had a statistically significant decrease in overall survival (breast, P = 8.0e-04; lung, P = 4.0e-04; Fig. 4C and D) as well as in disease-free survival (breast, P = 0.028; lung, P = 0.011; Fig. 4E and F).
Figure 3.
A 93-gene subset of MUC1-CD–induced tumorigenesis genes predicts poor outcome in breast and lung cancers. A, expressional clustering of the 93-gene subset in breast cancer (n = 295) partitions patient samples into 2 subgroups (red and blue bars). Vertical axis, patients; colored bars to the right of the cluster diagram correspond to the survival curves in B. For lung cancer, patient subgroups were defined based on differential expression of the 93-gene subset and determined using k-means clustering. Kaplan-Meier survival curves of disease-free and overall survival for breast (B; n = 295) and lung (C; n = 442) cancers show patient subgroups defined by differential expression of the 93-gene subset have significant differences in outcome.
Table 1.
Expression of a 35-gene MTS in breast and lung cancers
| Gene symbol | MTS+/MTS− (breast) | MTS+/MTS− (lung) |
|---|---|---|
| ADA | 1.37 | 1.30 |
| ASPM | 1.46 | 4.51 |
| BUB1B | 1.27 | 3.36 |
| CDC20 | 1.66 | 5.93 |
| CENPE | 1.32 | 2.11 |
| CST3 | 0.81 | 0.74 |
| CTSC | 2.23 | 1.19 |
| DHCR7 | 1.55 | 1.33 |
| ECT2 | 1.37 | 2.45 |
| FADS1 | 1.33 | 1.45 |
| FAM64A | 2.07 | 1.63 |
| FDPS | 1.25 | 1.20 |
| GBP2 | 1.45 | 1.44 |
| IDI1 | 1.47 | 1.19 |
| IFI44L | 2.97 | 1.58 |
| IMPA1 | 1.26 | 1.24 |
| ISG15 | 3.34 | 1.80 |
| KIF20A | 2.24 | 3.70 |
| MKI67 | 1.20 | 1.71 |
| MTHFD1 | 1.24 | 1.49 |
| NET1 | 1.22 | 1.26 |
| NSDHL | 1.25 | 1.14 |
| PGD | 1.21 | 1.30 |
| PSAT1 | 1.34 | 1.18 |
| RNASE4 | 0.58 | 0.66 |
| RRM2 | 1.45 | 3.22 |
| SIDT2 | 0.87 | 0.85 |
| SLIT2 | 0.75 | 0.70 |
| SOAT1 | 1.38 | 1.34 |
| SQLE | 1.56 | 1.37 |
| STAT1 | 2.46 | 1.67 |
| TFRC | 1.47 | 1.55 |
| UBD | 2.90 | 1.78 |
| UBL3 | 0.74 | 0.81 |
| VCAM1 | 2.09 | 1.61 |
Figure 4.
A 35-gene MTS is prognostic in breast and lung cancers. K-means clustering of breast (A) and lung (B) cancers by expression of MTS. Kaplan-Meier curves for overall survival for breast (C) and lung (D) cancers. Kaplan-Meier curves for disease-free survival for breast (E) and lung (F) cancers. For graphs (A–F): red, MTS+; blue, MTS−. See Table 1 for further details.
Discussion
The present studies show the significant tumorigenic potential of MUC1-CD. Injection of 104 MUC1-CD–transfected 3Y1 cells was sufficient to establish tumors in athymic mice, whereas for most tumor cell lines, it is necessary to inject at least 106 to 107 cells (21). To define the genes involved in MUC1-CD–induced tumorigenesis, we used large-scale expressional profiling and compared the cell lines transfected by MUC1-CD in vitro with tumors established from these cell lines in hind limb xenografts. The genes differentially expressed in MUC1-CD–transfected xenografts were significantly enriched by functional groups associated with tumorigenesis. Among them were gene families that contribute to transformation and induction of the tumor microenvironment. Importantly, we also identified gene families associated with cell motility, angiogenesis, and metastasis—functions previously associated with overexpression of the MUC1 protein. These results suggest that some of the tumor-related functions of MUC1 are associated with induction of specific transcriptional cascades by the MUC1 cytoplasmic domain.
To further investigate the potential relevance of MUC1-dependent genes identified in our experiments, we applied them to analyses of clinical expressional databases. Overexpression of the MUC1 protein has been associated with a poor prognosis in several human adenocarcinomas including breast and lung cancer (22, 23); however, these studies did not identify mechanisms by which MUC1 confers poor prognosis or whether MUC1 is associated with the regulation of gene subsets linked to cancer. Here, we report that the MUC1-CD induces broad changes in the pattern of gene expression in a rodent experimental model that remarkably predicts a poor prognosis in patients with adenocarcinoma of the breast or lung, suggesting that the gene expression patterns induced by MUC1-CD, unlike many empirically derived gene “signatures,” may be important for tumorigenesis in diverse types of human cancers. The importance of MUC1-CD in tumorigenesis and cancer therapy has recently been reviewed (24, 25). These reports emphasized the physical interaction of MUC1-CD with proteins important in cell transformation and survival after cytotoxic stress and the posttranslational modifications of MUC1-CD. Our findings emphasize the downstream transcriptional programs associated with MUC1-CD–induced transformation and are consistent with the association of MUC1 with key transcription factors such as estrogen receptor-α and p53 (12, 26). In summary, our data provide strong support for the oncogenic potential of the transcriptional programs activated by MUC1-CD and a new biologically derived molecular classifier for the stratification of patients with breast and lung cancer.
Supplementary Material
Acknowledgments
Grant support: Lung Cancer Foundation and the NIH grants RO1 CA1114231 and RO1 CA97098.
Footnotes
Reprints and Subscriptions To order reprints of this article or to subscribe to the journal, contact the AACR Publications Department at pubs@aacr.org.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
D.W. Kufe has interest in and is a consultant to Genus Oncology. The other authors disclosed no potential conflicts of interest.
References
- 1.Duraisamy S, Kufe T, Ramasamy S, Kufe D. Evolution of the human MUC1 oncoprotein. Int J Oncol. 2007;31:671–677. [PubMed] [Google Scholar]
- 2.Ligtenberg MJ, Kruijshaar L, Buijs F, van Meijer M, Litvinov SV, Hilkens J. Cell-associated episialin is a complex containing two proteins derived from a common precursor. J Biol Chem. 1992;267:6171–6177. [PubMed] [Google Scholar]
- 3.Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui J, Abe M, Hayes D, Shani E, Yunis E, Kufe D. Isolation and sequencing of a cDNA coding for the human DF3 breast carcinoma-associated antigen. Proc Natl Acad Sci U S A. 1988;85:2320–2323. doi: 10.1073/pnas.85.7.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merlo GR, Siddiqui J, Cropp CS, et al. Frequent alteration of the DF3 tumor-associated antigen gene in primary human breast carcinomas. Cancer Res. 1989;49:6966–6971. [PubMed] [Google Scholar]
- 6.Kufe D, Inghirami G, Abe M, Hayes D, Justi-Wheeler H, Schlom J. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 9.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–20612. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 11.Yin L, Li Y, Ren J, Kuwahara H, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278:35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 12.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Khodarev NN, Yu J, Nodzenski E, et al. Method of RNA purification from endothelial cells for DNA array experiments. Biotechniques. 2002;32:316. doi: 10.2144/02322mt02. 8, 20. [DOI] [PubMed] [Google Scholar]
- 14.Khodarev NN, Park J, Kataoka Y, et al. Receiver operating characteristic analysis: a general tool for DNA array data filtration and performance estimation. Genomics. 2003;81:202–209. doi: 10.1016/s0888-7543(02)00042-3. [DOI] [PubMed] [Google Scholar]
- 15.Khodarev NN, Kataoka Y, Murley JS, Weichselbaum RR, Grdina DJ. Interaction of amifostine and ionizing radiation on transcriptional patterns of apoptotic genes expressed in human microvascular endothelial cells (HMEC) Int J Radiat Oncol Biol Phys. 2004;60:553–563. doi: 10.1016/j.ijrobp.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 16.Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci U S A. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimchi ET, Posner MC, Park JO, et al. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauceri HJ, Hanna NN, Beckett MA, et al. Combined effects of angiostatin and ionizing radiation in anti-tumour therapy. Nature. 1998;394:287–291. doi: 10.1038/28412. [DOI] [PubMed] [Google Scholar]
- 22.Inata J, Hattori N, Yokoyama A, et al. Circulating KL-6/MUC1 mucin carrying sialyl Lewisa oligosaccharide is an independent prognostic factor in patients with lung adenocarcinoma. Int J Cancer. 2007;120:2643–2649. doi: 10.1002/ijc.22613. [DOI] [PubMed] [Google Scholar]
- 23.Al-azawi D, Kelly G, Myers E, et al. CA 15-3 is predictive of response and disease recurrence following treatment in locally advanced breast cancer. BMC Cancer. 2006;6:220. doi: 10.1186/1471-2407-6-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carson DD. The cytoplasmic tail of MUC1: a very busy place. Sci Signal. 2008;1:pe35. doi: 10.1126/scisignal.127pe35. [DOI] [PubMed] [Google Scholar]
- 25.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor α. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.