Abstract
The competence quorum-sensing system of Bacillus subtilis consists of two-component regulatory proteins, ComP (histidine kinase) and the response regulator, ComA, an extracellular pheromone (ComX), and a protein that is needed for the proteolytic cleavage and modification of pre-ComX (ComQ). ComQ and pre-ComX are both necessary and sufficient for the production of active pheromone, which is released as an isoprenylated peptide. Laboratory strain 168 and a number of natural isolates of bacilli differ in the primary sequences of their pheromones as well as in the masses of their isoprenyl adducts. We have shown that ComX, ComQ, and the membrane-localized sensor domain of ComP are highly polymorphic in natural isolates of bacilli all closely related to the laboratory strain of B. subtilis. In this study, we used two statistical tests (the ratio of synonymous and nonsynonymous substitution rates and the Tajima D test) to demonstrate that these polymorphic sequences evolved by diversifying selection rather than by neutral drift. We show that the choice of isoprenyl derivative is determined by the C-terminal (mature) sequence of pre-ComX rather than by the ComQ protein. The implications of these findings for the evolution of the quorum-sensing system and for the protein-protein interactions involved in determining specificity are discussed.
The ComP-ComA two-component system controls genetic competence in Bacillus subtilis 168 as well as the expression of a number of additional genes that appear to be unrelated to competence (18, 21, 23). ComP, a membrane-localized histidine kinase, functions as a cell density detector by responding to the extracellular ComX pheromone. ComX is released at a constant rate per cell during growth (3). In laboratory strains derived from B. subtilis 168, ComX is synthesized as a 55-residue precursor, which is then cleaved, while its C-terminal 10 amino acids are modified and exported from the cell (15). ComQ appears to be both necessary and sufficient for the cleavage and modification of pre-ComX (3, 15, 27, 31). In response to extracellular ComX, the ComP sensor protein autophosphorylates within its cytosolic transmitter domain and then donates its phosphate group to a conserved aspartate residue in the receiver domain of ComA. Phosphorylated ComA activates the transcription of comS (6, 7, 17, 18, 21, 23), which is required for accumulation of the key transcription factor ComK, needed for the development of competence. ComS, which is encoded by a gene in the srfA operon (6, 7), prevents the degradation of ComK by a proteosome-like complex composed of MecA, ClpC, and ClpP (29, 30). The adapter protein MecA binds to ComK, targeting it for degradation by the ClpCP protease. ComS binds to MecA, causing the release of ComK, thus protecting this protein from degradation. ComK then activates the transcription of the downstream competence genes, which specify binding and uptake proteins for transformation.
The genes encoding the quorum-sensing proteins are contiguous in B. subtilis 168, occurring in the order comQ, comX, comP, and comA. This locus, with the same organization, has been identified in other closely related Bacillus isolates, including those in the B. subtilis W23, B. subtilis 168, and B. mojavensis groups and also in the industrial strains B. natto NAF4 and B. licheniformis ATCC 14580 (2, 12, 16, 27, 28). Sequence analysis of quorum-sensing loci from 14 of these natural isolates revealed a striking variability compared to neighboring genes and the housekeeping genes gyrA and rpoB. This polymorphism, which is characteristic of ComQ, pre-ComX, and the sensor domain of ComP, was correlated with specificity in the quorum-sensing response (2, 27).
The strains analyzed could be placed in at least four pherotype groups, defined as clusters of isolates able to activate one another for the quorum-sensing response. The pheromones produced in the different pherotypes differed not only in the sequence of the mature peptides, but also in their modifications, which consist of isoprenylations (2). Since comQ and comX together are sufficient for the production of active pheromones in Escherichia coli (27), ComQ alone would seem to be responsible for the complete maturation of pre-ComX. In accordance with this conclusion, an isoprenoid binding site was recently identified in ComQ, and mutations in this site prevented the production of mature ComX (3).
Polymorphism is also exhibited by the pheromones of two other quorum-sensing systems in gram-positive bacteria; the Agr system of Staphylococcus spp. (10), which regulates virulence genes, and the competence pheromone system of Streptococcus spp. (9). It is remarkable that proteins involved in many eukaryotic sexual reproductive systems also exhibit marked diversity and that this variability extends to both ligand and receptor proteins in several cases (25). Thus, the Bacillus quorum-sensing proteins are among many involved in cell-cell communication that exhibit polymorphism. The variability in several of the eukaryotic reproductive genes has been shown to be due to positive Darwinian selection (25).
In this report, we used sequence analysis to show that the evolution of polymorphism at the quorum-sensing locus has been dominated by positive selective forces and consequently cannot be explained by neutral changes. We also explored the determinants of specificity during the modification of pre-ComX. We show that the nature of the isoprenoid modification is determined by the C-terminal domain of pre-ComX rather than by the putative isoprenyl transferase, ComQ. The implications of these findings for the evolution of the Bacillus quorum-sensing system are discussed.
MATERIALS AND METHODS
Strains and plasmid construction.
The natural Bacillus isolates and sequences used in this study, as well as the various pheromone producer and tester strains, are described elsewhere (2, 12, 22, 27). The Bacillus tester strains RO-H-1 (BD2962), RO-B-2 (BD2983), and RO-E-2 (BD3020) were grown either in liquid competence medium supplemented with glucose (0.5%) (1), l-histidine, l-leucine, and l-methionine (50 μg/ml) or on tryptose blood agar base (Difco) supplemented with chloramphenicol (5 μg/ml), erythromycin (5 μg/ml), kanamycin (5 μg/ml), spectinomycin (5 μg/ml), or tetracycline (20 μg/ml) as appropriate. Escherichia coli XL1 Blue (Stratagene) was used for cloning the comQ and comX genes into the pET22(b) vector, and transformants were selected on tryptose blood agar (Difco) plates containing ampicillin (100 μg/ml). E. coli producer strains were constructed by introducing these plasmids into E. coli BL21(DE3), permitting isopropyl-β-d-thiogalactopyranoside (IPTG) induction of transcription from the T7 promoter of the pET22(b) derivatives. DNA manipulation, cloning, and molecular biological procedures were carried out with standard protocols.
For the comX exchange experiments, plasmid pED409 (comQRO-B-2 comXRO-B-2)was amplified with the divergent primers Bam-5′X-B2 (5′-CGGGATCCAGGAGGATTTATATGCAGGAAATTGTTG) and Xba-lacO-rev (5′-TTTCTAGAGGGGAATTGTTATCCGC), yielding a product lacking comQ. The comQ gene from the RO-H-1 strain was then amplified with the primers Uni-comQ-Xba (27) and Bam-3′Q-H1 (5′-CGGGATCCTTATTTAATCATAACAGACAATAATTTTTCAGT) and cloned in the amplified vector cut with the same enzymes. The resulting plasmid, pED434 (comQRO-H-1 comXRO-B-2), contains a BamHI site between comQ and comX. The opposite procedure (cloning of comQRO-B-2 into pED368) was performed to obtain pED433 (comQRO-B-2 comXRO-H-1) with primers Uni-comQ-Xba, Xba-lacO-rev, Bam-5′X-H1 (5′-CGGGATCCGGAGGGATTTATTATGCAAGAAATGGTA), and Bam-3′Q-B2 (5′-CGGGATCCTTATTTTATTATTCCTGTTATTAATTCTTCTATCTT). Then pED434 (comQRO-H-1 comXRO-B-2) was used to exchange comXRO-B-2 with comXRO-C-2 or comXRO-E-2 with the BamHI site between comQ and comX.
comX genes from RO-C-2 and RO-B-2 were PCR amplified with primers containing a BamHI site at the 5′ end of the genes, Bam-5′XROC2 (5′-CGGGATCCAGGAGTGATGAAAATGATGCAAGACCTAA) and Bam-5′XROE2 (5′-CGGGATCCAGGAGTATACTTATGAAACAAGACATGATTG) and a SalI site at the 3′ end of the genes, Sal-3′XROC2 (5′-CGCGTCGACTTAACCATCCCATTCACGGGTTGTTG) and Sal-3′XROE2 (5′-CGCGTCGACTTATTGCTCCCAGAAAATTCCCTTGC). The amplified comX genes were then cut with BamHI and SalI and ligated into pED434 cut with the same enzymes.
The ComX chimera was constructed with a similar strategy except that the primer at the 3′ end of comX contained the entire sequence necessary for replacement of the 11 codons at the C-terminal end of comXRO-H-1 with the corresponding sequence (italic) from comXRO-E-2 fused to a few codons encoding the precursor part of comXRO-H-1 (ComXH1/E2, 5′-CGCAATTGTTATTGCTCCCAGAAAATTCCCTTGCATTTCTTTGAAATTTCTAAACCTTTAAAACCATTTATTATACATTC). This primer was used together with the Bam-5′X-H1 primer and with pED368 as the template, and after hydrolysis with BamHI and SalI the product was cloned into pED434 cut with the same enzymes to yield plasmid pED622.
All constructs were checked for fidelity of amplification by direct sequencing of the replaced genes.
Pheromone purification.
ComX pheromones were purified from culture supernatants of E. coli BL21(DE3) producer strains containing pET22(b)-derived plasmids as described elsewhere (2).
Computer analysis.
Phylogenetic tree constructions for the comQ and comX genes based on variation at synonymous sites were performed on eight sequences with the modified Nei-Gojobori method (19), estimating the number of differences (synonymous substitutions only) by pairwise comparison, with the MEGA 2.1 software (11) downloaded from http://www.megasoftware.net/. For this analysis, sequences representative of each pherotype (2, 27) were selected, including some from the same pherotype that were divergent. The GenBank accession numbers were NC000964 (168), AF456134 (RO-C-2), AF456130 (RO-FF-1), AY003901 (RO-H-1), AF456135 (RO-B-2), AF459919 (B. licheniformis), AF456137 (RO-F-3), and AF456138 (RO-E-2). We analyzed only a subset of the available sequences because within each pherotype the sequences are more similar than between pherotypes, and some pherotypes contain more sequences than others. Including all the sequences would therefore have biased some of the analyses, and for that reason only the most divergent sequences of each pherotype were chosen. We included the sequence of the quorum-sensing locus identified in a strain of B. licheniformis which is phylogenetically the most distant Bacillus strain containing the comQXPA quorum-sensing locus (12).
To test the fit of the sequences to the model of neutral evolution, the D test statistic proposed by Tajima (26) was computed for all eight sequences with the DnaSP program (24) downloaded from http://www.bio.ub.es/≈julio/DnaSP.html. Sequences were first aligned with ClustalW and formatted with MEGA 2.1 (11). Only the segregating sites were considered, with exclusion of sites with gaps. Results are expressed as the overall mean of Tajima's D value computed at each segregating site with a sliding window of 100 sites.
As a second method to test adherence to the neutral evolution model, the modified Nei-Gojobori method (19) was used to estimate the frequencies of synonymous and nonsynonymous differences between sequences, with standard errors estimated by the bootstrap method and 500 replications of the data. For this purpose MEGA 2.1 software (11) was used.
For computer analysis by both methods, only five complete comP sequences were available (B. subtilis 168, B. natto NAF4, B. mojavensis RO-H1, B. subtilis [W23 group] RS-B-1, and B. licheniformis ATCC 14580) and were divided in two parts. The first part encodes the more variable sensory region and consists of the first 1,200 nucleotides of the five genes analyzed, and the other corresponds to the transmitter domain (from position 1200 to the ends of the genes). For both methods, the comQ and comX sequences from each isolate were combined because the comX genes were too small to analyze separately.
In vivo pheromone activity.
β-Galactosidase assays, with a srfA-lacZ reporter, were performed as described (27) with tester strains grown in competence medium in the presence of purified pheromones. Samples were collected at 2 h past the end of exponential growth and assayed for β-galactosidase activity with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate. Each assay was repeated at least three times. Values represent the average of at last three determinations, which exhibited a variation from the mean of no more than 10%.
Analytical protein methods.
N-terminal sequencing by Edman degradation and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry were performed on purified pheromones at the Protein Core Facility, Columbia University.
RESULTS
Positive selection at the comQXP loci.
Significant polymorphism occurs at the comQXP loci in natural isolates of Bacillus compared with the variability of the housekeeping genes rpoB and gyrA (2). In each case, this polymorphism was more pronounced in the N-terminal sensor domain of ComP than in the conserved transmitter domain. When these strains were grouped by similarities at the quorum-sensing locus, a strong correlation was noted with specificity in the quorum-sensing response. These data permitted the classification of the strains into pherotype groups, defined as groups of strains that cross activate for the quorum-sensing response.
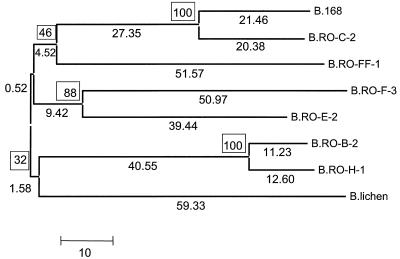
To pursue the relationship between polymorphism at the quorum-sensing locus and the emergence of different pherotypes, we constructed a tree from the comQ and comX sequences, based on the incidence of synonymous changes, inferred from the modified Nei-Gojobori method (19). This method is used to compute the numbers of synonymous and nonsynonymous substitutions and the numbers of potential synonymous and nonsynonymous sites. Diversifying selection results in an excess of nonsynonymous substitutions compared to the frequency expected from random (neutral) evolution. The comQ and comX sequences were combined and analyzed together because comX is a small gene. This simplification is appropriate, since previous phylogenetic analysis suggested that comQ and comX have similar evolutionary histories (2). A tree showing the phylogeny of the combined comQ and comX sequences of the eight strains analyzed is shown in Fig. 1.
FIG. 1.
Phylogenetic reconstruction of eight comQX sequences. The tree was drawn with the MEGA 2.1 software. The numbers at branches represent the number of synonymous differences, whereas the boxed numbers at internal branches represent the percent bootstrap probabilities of 500 bootstrap replications.
The extensive polymorphism exhibited by the comQ and comX genes may be due to positive selection. To test this hypothesis, we again used the modified Nei-Gojobori method to compute the frequencies of nonsynonymous mutations at nonsynonymous sites (dN) and the frequencies of synonymous substitutions at synonymous sites (dS), in pairwise comparisons of the eight sequences (19) (Table 1). Neutral theory predicts that the dN/dS ratio will equal 1, whereas positive selection will result in a ratio greater than 1. With our sequences, the average values obtained were dN = 230.9 and dS = 103.4, showing a strong bias toward nonsynonymous substitutions (Table 2). Each of the comparisons gave P values less than 0.004 with a Z test, rejecting the neutral evolution hypothesis, except for the comparison between RO-H-1 and RO-B-2, which gave a P value of 0.461 (data not shown). Figure 1 shows that RO-H-1 and RO-B-2 comprised the most similar pair. The Fisher exact test and the nonmodified (original) Nei-Gojobori method (19) yielded very similar results, attesting to the robustness of these results (data not shown).
TABLE 1.
comQX synonymous and nonsynonymous substitutionsa
| Nonsynonymous difference (dN) in strain: | Synonymous difference (dS) in strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 168 | RO-C-2 | RO-FF-1 | RO-H-1 | RO-B-2 | B. licheniformis | RO-F-3 | RO-E-2 | |
| 168 | 41.833 | 104.750 | 103.167 | 101.083 | 119.917 | 121.083 | 96.833 | |
| RO-C-2 | 85.167 | 94.917 | 111.250 | 106.833 | 117.417 | 113.833 | 96.08 | |
| RO-FF-1 | 181.250 | 185.083 | 118.417 | 110.083 | 108.500 | 117.167 | 106.083 | |
| RO-H-1 | 236.833 | 231.750 | 260.583 | 23.833 | 113.500 | 108.083 | 105.250 | |
| RO-B-2 | 249.917 | 240.167 | 258.917 | 74.167 | 110.083 | 110.333 | 113.000 | |
| B. licheniformis | 254.083 | 251.583 | 279.500 | 223.500 | 241.917 | 124.083 | 108.167 | |
| RO-F-3 | 254.917 | 261.167 | 261.833 | 245.917 | 239.667 | 252.917 | 90.417 | |
| RO-E-2 | 244.167 | 244.917 | 266.917 | 236.750 | 226.000 | 234.833 | 239.583 | |
dS and dN were computed with the modified Nei-Gojobori test and MEGA 2.0 software. The values represent the frequencies of differences at synonymous (right) and nonsynonymous (left) sites for each pair.
TABLE 2.
Statistic test summarya
| Parameter | Analyzed sequences
|
||
|---|---|---|---|
| comQX | comP SD | comP TD | |
| No. of sequences | 8 | 5 | 5 |
| No. of sites | 1,113 | 1,200 | 642 |
| No. of segregating sites | 713 | 1115 | 235 |
| Nucleotide diversity (Pi) | 0.405 | 0.598 | 0.179 |
| Nucleotide difference (avg) | 379.53 | 717.70 | 108.30 |
| Tajima's test | |||
| D value | 2.09 | 2.61 | −0.34 |
| P value | <0.05 | <0.001 | >0.05 |
| Modified Nei-Gojobori test | |||
| dN | 230.9 (9.0) | 558.2 (7.5) | 54.6 (4.7) |
| dS | 103.4 (4.3) | 159.4 (4.0) | 51.49 (3.8) |
| dN/dS | 2.232 | 3.502 | 1.061 |
SD, sensor domain; TD, transmitter domain. Tajima's test was performed with DnaSP software. Values correspond to the mean of Tajima's D value computed at each segregating site with exclusion of the sites with gaps. The modified Nei-Gojobori test was performed with MEGA 2.0 software. The values represent the overall mean of the differences at synonymous and nonsynonymous sites after pairwise distance evaluation. Standard errors (in parentheses) were computed by bootstrap replications (500 replicates).
Another test of neutral evolution is that of Tajima (26), which compares two measures of nucleotide diversity, one based on the number of segregating (variable) sites and the other on the average number of pairwise differences between sequences. Neutral evolution predicts that these two measures will be equal. However, diversifying selection will alter the population structure so that the second measure (based on the frequency of allelic differences) will produce a higher value. D, the difference between the two estimates, will therefore be positive. With this method, we obtained a D value of 2.09 with a total of 936 sites and 713 segregating sites (Table 2). Since the expected mean of D based on neutral evolution is 0 and the P value in this case for rejection of the neutral evolution hypothesis was less than 0.05, this result independently suggests that neutral evolution cannot explain the observed polymorphism. The large positive value of D strongly suggests that the sequences comprising comX and comQ have been subjected to diversifying selection.
This analysis was extended to comP. The ComP protein consists of a relatively conserved cytoplasmic transmitter domain and a polytopic, membrane-localized sensor domain of about 400 amino acid residues. Based on work with comP chimeras, we established that the sensor moiety, which is also the polymorphic portion of ComP, determines the specificity of the quorum-sensing response (unpublished data). We therefore restricted our analysis to the sensor domain of the comP genes, consisting of the promoter-proximal 1,200 nucleotide residues. The dN and dS values obtained for the receiver domain of ComP were 558.2 and 159.4, respectively (Table 2). Pairwise comparisons with a Z test gave P values that overall supported neither the positive nor neutral evolution hypothesis, but when the Tajima test was applied to the sensor domain of comP, we obtained a D value of 2.61 with 1,115 segregating sites and a P value of <0.001 (Table 2). Again, the results suggest that diversifying selection rather than drift was responsible for the marked polymorphism at this locus.
When we applied this analysis to 634 residues, including 235 segregating sites, encoding the conserved transmitter domain of comP as a control, we obtained a D value of −0.3 with a P value of >0.05 (Table 2). For this domain, in contrast to the sensor domain, we cannot exclude the possibility that the limited polymorphism is due to neutral evolution. Taken as a whole, our analysis strongly supports a role for positive Darwinian selection in the coevolution of the comX and comQ genes and of the comP sensor domains of these Bacillus species.
Our previous work with 13 sequences suggested that comX, comQ, and comP coevolved, since similarity trees constructed for these three genes are completely congruent and differ from trees constructed for the housekeeping genes rpoB and gyrA (2). Since the quorum-sensing genes coevolved under positive selection, they presumably did so in order to maintain critical interactions between proteins, e.g., between ComQ and pre-ComX and between ComX and the sensor domain of ComP. We have begun to explore these interactions and the factors that determine the specificity of the posttranslational modifications of ComX.
Amino acid sequence of mature ComX determines the nature of the posttranslational modification.
Each pheromone precursor must be processed and modified by its cognate ComQ enzyme. We previously showed that the isoprenoid modifications on ComX pheromones generally differ among the pherotypes (2). Based on analysis by mass spectrometry, there are three different modifications, which correspond in mass to geranyl or farnesyl groups and to an unknown isoprenoid derivative. We therefore asked whether the nature of the modification was determined by the sequence of pre-ComX or by ComQ. To accomplish this, we performed a comQ-comX exchange experiment between the genes of two phylogenetically related quorum-sensing loci, RO-H-1 and RO-B-2 (Fig. 1), which produce pheromones with different modifications (120 and 136 Da, respectively). Both combinations (comQRO-H-1comXRO-B-2 and comQRO-B-2 comXRO-H-1) were cloned into the pET22(b) vector and expressed in E. coli. Culture supernatants were subjected to the pheromone purification protocol, and fractions were tested for their abilities to induce srfA-lacZ expression in RO-H-1 and RO-B-2 tester strains (Table 3).
TABLE 3.
ComQ and pre-ComX exchange experimentsa
| Plasmid | comQ | comX | β-Galactosidase activity (U/mg) in tester strain:
|
||
|---|---|---|---|---|---|
| RO-H-1 | RO-B-2 | RO-E-2 | |||
| pED409 | RO-B-2 | RO-B-2 | 48 | 210 | ND |
| pED433 | RO-B-2 | RO-H-1 | 1 | 0 | ND |
| pED368 | RO-H-1 | RO-H-1 | 254 | 7 | ND |
| pED434 | RO-H-1 | RO-B-2 | 43 | 112 | ND |
| pED413 | RO-E-2 | RO-E-2 | 21 | ND | 270 |
| pET-QH1/XE2 | RO-H-1 | RO-E-2 | 23 | ND | 72 |
| pED622 | RO-H-1 | RO-H-1/RO-E-2 | 22 | ND | 122 |
RO-H-1 (BD2962), RO-B-2 (BD2983), and RO-E-2 (BD3020) tester strains were grown in competence medium in the presence of partially purified pheromones (1:1,000 dilutions) produced by the indicated constructs. β-Galactosidase assays were performed on samples collected 2 h after sporulation and expressed as units per milligram of protein. Values represent the average of at least three determinations with a variation of no more than 10% from the mean. ND, not done.
The comQRO-B-2 comXRO-H-1 combination produced no active pheromone, and no peak with absorbance at 214 nm corresponding to inactive pheromone eluted from the C18 column used for purification (not shown). In contrast, the comQRO-H-1 comXRO-B-2 combination produced an active pheromone exhibiting the RO-B-2 specificity (Table 3), although the in vivo activity was about half that obtained with the native combination (comQRO-B-2 comXRO-B-2), probably indicating a reduced efficiency in the maturation process. Mass spectrometry analysis of the purified comQRO-H-1 comXRO-B-2 pheromone revealed that it carried the RO-B-2 modification of 136 Da (data not shown), even though processed by a modifying enzyme that ordinarily introduces a modification with a mass of 120 Da. Thus, the nature of the posttranslational modification was determined by the pre-ComX sequence itself rather than by the modifying enzyme.
Based on these results, two other comX genes (comXRO-C-2 and comXRO-E-2) were individually combined with comQRO-H-1 in the same vector (see Materials and Methods). The comQRO-H-1 comXRO-C-2 combination produced no active pheromone detectable by biological activity (data not shown), whereas the comQRO-H-1 comXRO-E-2 combination produced some active pheromone with the RO-E-2 specificity, although again with reduced yield (Table 3). Clearly, productive heterologous ComQ-pre-ComX interactions occur with reduced efficiency or not at all, consistent with the idea that in each case, comQ and comX coevolved to ensure optimal protein-protein interactions.
In the ComX precursors, the N-terminal part is more conserved than the extreme C-terminal part, which corresponds to the mature peptide (Fig. 2). It is thus probable that the ComQ proteins interact at least in part with the precursor part of ComX, in order to cleave and modify pre-ComX. To address this hypothesis and to determine whether the extreme C-terminal sequence of pre-ComX is sufficient to direct the choice of modification, we constructed a pre-ComX chimera by combining the N-terminal domain from pre-ComXRO-H-1 and the C-terminal sequence of ComXRO-E-2. The joining site was chosen at a residue with conserved hydrophobicity close to the cleavage sites (Fig. 2). This chimera was coexpressed in E. coli with comQRO-H-1. Active pheromone was produced and was purified from the culture supernatant. This pheromone exhibited the RO-E-2 specificity (Table 3). Mass spectrometry identified an RO-E-2 peptide bearing a modification of 136 Da, corresponding to the expected ComXRO-E-2 modification of 136 Da (data not shown), which differs from the native RO-H-1 modification of 120 Da. This result shows that the mature part of the peptide encoded by comX was sufficient to determine the correct modification. The yield of pheromone activity with the chimeric pre-ComX molecule was intermediate between that obtained with the comQRO-H-1 comXRO-E-2 and comQRO-E-2 comXRO-E-2 combinations, suggesting that ComQ interacts with both the conserved N-terminal domain of pre-ComX and the less conserved C-terminal moiety (Table 3).
FIG. 2.
Pre-ComX sequence alignment. An arrow indicates the site chosen for the chimeric junction. In cases where the cleavage site is known, the mature peptide sequence is underlined.
DISCUSSION
The quorum-sensing locus has an average G+C content markedly lower than that of the B. subtilis chromosome (2) and was probably introduced by horizontal transmission into a common ancestor of the strains that we have studied. The results presented in this report suggest that since that event, the quorum-sensing locus has been subjected to strong positive selection, resulting in the evolution of new pherotype specificities and of dramatic sequence polymorphism. At least three protein-protein interactions must occur for the quorum-sensing system to work: ComQ with pre-ComX, ComX with the sensor domain of ComP, and ComP with ComA. The last of these interactions is between the conserved transmitter domain of ComP and the conserved receiver domain of ComA, and these interactions will probably not differ significantly among the pherotypes. The other two interactions are pherotype specific.
We have previously noted that similarity trees constructed for ComQ, pre-ComX, and the N-terminal domain of ComP are completely congruent. These trees do not correspond with those constructed by using housekeeping genes (2) or with the independently derived phylogeny of the cognate strains (22). These results indicate that comQ, comX, and comP coevolved, as expected from their protein-protein interactions, and did so independently of the rest of the genome. It appears likely that a change in one component which alters response specificity will be followed by changes in the other components in order to optimize the protein interactions. For instance, we may envisage that a mutational alteration in comP that decreases its interaction with a foreign (noncognate) ComX molecule might increase fitness by preventing inappropriate quorum-sensing responses. Since foreign pheromones sometimes interfere with the response to a cognate pheromone (2), change may protect against such interference. Such changes may carry a price, since they may also decrease responsiveness to the cognate ComX molecule. A change in comX which improves the fit between ComX and the altered ComP might mitigate this effect, but at the further price of decreased interaction between ComQ and pre-ComX. A mutational change in comQ might then result in a temporarily “optimized” system. Note that our evidence (Table 3) suggests that the efficiency of pheromone production depends on interactions between ComQ and both the N-terminal domain and the mature pheromone moieties of pre-ComX.
What fitness advantages are conferred by the quorum-sensing system in general and by the acquisition of new pherotypes? The quorum-sensing system of B. subtilis 168 has been identified as the first step in competence regulation (15). ComA phosphorylation leads to the synthesis of ComK, the transcriptional activator of the late competence genes. However, ComA is not devoted only to competence development, as shown by the computer search for ComA binding sites (13) and by microarray analysis (21). For example, ComA activates transcription of the sublancin lantibiotic precursor gene, the surfactin operon, the gene for pectate lyase, and several genes of unknown function. ComK itself induces the expression of many genes involved in the uptake and utilization of novel substrates, in growth arrest, and in DNA repair, which together comprise what has been described as the K-state (4, 8, 20).
It is not known which of the downstream genes of the quorum-sensing regulon might contribute to the fitness advantages of quorum-sensing specificity. However, the widespread presence of these quorum-sensing genes among the bacilli suggests that they do contribute to fitness, and the present demonstration of positive selection for specificity-determining change at the quorum-sensing locus suggests strongly that specificity itself contributes some advantage. What might this advantage be? The very existence of quorum sensing suggests that a given strain has evolved to respond when its population density is high. Presumably it will maximize its fitness when it responds to its own population density and not that of nearby but related strains. A promiscuous response would make a strain subject to fluctuations in the presence of related bacilli. If we consider the development of competence as a fitness-enhancing factor, specificity would be equivalent to a sexual isolation mechanism. The presence of related pheromones would raise the specter of genetic invasion, and therefore selection would drive changes in the quorum-sensing genes, as described above.
The polymorphism exhibited by the Bacillus quorum-sensing genes can be placed in a wider context. The competence pheromones of the streptococci exhibit a similar diversity (9), as do the pheromones of the staphylococcal Agr system (10). In those systems too, the trees built from sequence comparisons of the pheromones and of the receptors are congruent, indicating probable coevolution of the corresponding genes. Particularly interesting is the generalization that proteins mediating sexual reproduction in many eukaryotic species, including protozoa, fungi, insects, and numerous additional invertebrates as well as mammals, exhibit more divergence than do nonreproductive proteins, and several such systems have been shown to exhibit diversifying selection (reviewed in reference 25).
In fact, both the ciliate Euplotes and certain basidiomycete fungi secrete peptide pheromones that mediate mating. In Euplotes raikovi the 36- to 39-residue mature pheromones from seven mating types exhibit only seven conserved amino acids, and six of these are cysteines that form three disulfide bonds (14). In the fungal system, the sex pheromones bind to cell surface receptors and induce the expression of mating genes via a phosphorylation cascade (5), much as in the case of competence induction in the bacilli. It is remarkable that these fungal sex pheromones are also processed from preproteins and are farnesylated near their C termini, as is also true in the yeast Saccharomyces cerevisiae (5). If we regard the quorum-sensing systems of Bacillus and of Streptococcus spp. as primarily directed toward the regulation of competence, it is tempting to place these systems in the wider context of cell-cell communication mechanisms that regulate sexual behavior.
We do not know if the different isoprenoid modifications contribute to specific recognition by the ComP receptor. However, it is likely that they do, since the modifications generally differ between pherotypes (2) and unmodified peptides are completely inactive (2, 15). Our results suggest that the ComQ protein recognizes both the N-terminal and C-terminal parts of the ComX precursor. Perhaps the active modifying enzyme is a heteromultimer containing both pre-ComX and ComQ. This hypothesis is supported by the success of the ComX exchange experiment in the case of the closely related strains RO-H-1 and RO-B-2, while for RO-H-1 and RO-E-2, which are less phylogenetically related, modification of the precursor portion of pre-ComX improved the response (Table 3). The gene exchange experiments also revealed that the mature part of ComX was sufficient to determine the choice of modification (Table 3).
The studies reported here provide the basis for further exploration of the protein interactions that mediate competence induction and transmembrane signaling in the ComP/ComA two-component system as well as investigations of the evolution of quorum sensing in bacteria.
Acknowledgments
We thank all the members of our laboratory for frequent discussions. We also thank A. Sorokin for providing the B. licheniformis sequences.
This work was supported by NIH grant GM 57720.
REFERENCES
- 1.Albano, M., J. Hahn, and D. Dubnau. 1987. Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169:3110-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaldi, M., D. Marolt, T. Stebe, I. Mandic-Mulec, and D. Dubnau. 2002. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol Microbiol. 44:1561-1573. [DOI] [PubMed] [Google Scholar]
- 3.Bacon Schneider, K., T. M. Palmer, and A. D. Grossman. 2002. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 5.Casselton, L. A., and N. S. Olesnicky. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Souza, C., M. M. Nakano, and P. Zuber. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. 91:9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamoen, L. W., H. Eshuis, J. Jongbloed, G. Venema, and D. van Sinderen. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55-63. [DOI] [PubMed] [Google Scholar]
- 8.Hamoen, L. W., W. K. Smits, A. Jong Ad, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis with a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Håvarstein, L. S., and D. A. Morrison. 1999. Quorum-sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-26. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 10.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 12.Lapidus, A., N. Galleron, J. T. Andersen, P. L. Jorgensen, S. D. Ehrlich, and A. Sorokin. 2002. Co-linear scaffold of the Bacillus licheniformis and Bacillus subtilis genomes and its use to compare their competence genes. FEMS Microbiol. Lett. 209:23-30. [DOI] [PubMed] [Google Scholar]
- 13.Lazazzera, B. A., T. Palmer, J. Quisel, and A. D. Grossman. 1999. Cell density control of gene expression and development in Bacillus subtilis, p. 27-46. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 14.Luporini, P., A. Vallesi, C. Miceli, and R. A. Bradshaw. 1995. Chemical signaling in ciliates. J. Eukaryot. Microbiol. 42:208-212. [DOI] [PubMed] [Google Scholar]
- 15.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone. Cell 77:207-216. [DOI] [PubMed] [Google Scholar]
- 16.Mandic-Mulec, I., B. Kraigher, U. cepeon, and I. Mahne. 2003. Variability of the quorum-sensing system in natural isolates of Bacillus sp. Food Technol. Biotechnol. 41:23-28. [Google Scholar]
- 17.Nakano, M. M., L. Xia, and P. Zuber. 1991. Transcription initiation region of the srfA operon which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 173:5487-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, M. M., and P. Zuber. 1991. The primary role of ComA in establishment of the competent state in Bacillus subtilis is to activate the expression of srfA. J. Bacteriol. 173:7269-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, England.
- 20.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, M. S., and F. M. Cohan. 1995. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution 49:1081-1094. [DOI] [PubMed] [Google Scholar]
- 23.Roggiani, M., and D. Dubnau. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 175:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 25.Swanson, W. J., and V. D. Vacquier. 2002. The rapid evolution of reproductive proteins. Nat. Genet. Rev. 3:137-144. [DOI] [PubMed] [Google Scholar]
- 26.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortosa, P., L. Logsdon, B. Kraigher, Y. Itoh, I. Mandic-Mulec, and D. Dubnau. 2001. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 183:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran, L.-S. P., T. Nagai, and Y. Itoh. 2000. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 37:1159-1171. [DOI] [PubMed] [Google Scholar]
- 29.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:119-128. [DOI] [PubMed] [Google Scholar]
- 31.Weinrauch, Y., T. Msadek, F. Kunst, and D. Dubnau. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 173:5685-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]