The crystal structure of flap endonuclease 1 from the hyperthermophilic archaeon D. amylolyticus was determined at 2.00 Å resolution.
Keywords: FEN1, DNA repair, DNA replication
Abstract
Flap endonuclease 1 (FEN1) is a key enzyme in DNA repair and DNA replication. It is a structure-specific nuclease that removes 5′-overhanging flaps and the RNA/DNA primer during maturation of the Okazaki fragment. Homologues of FEN1 exist in a wide range of bacteria, archaea and eukaryotes. In order to further understand the structural basis of the DNA recognition, binding and cleavage mechanism of FEN1, the structure of FEN1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus (DaFEN1) was determined at 2.00 Å resolution. The overall fold of DaFEN1 was similar to those of other archaeal FEN1 proteins; however, the helical clamp and the flexible loop exhibited a putative substrate-binding pocket with a unique conformation.
1. Introduction
Flap endonuclease 1 (FEN1) is an enzyme that is involved in aspects of DNA metabolism such as replication, repair and recombination. It belongs to a structure-specific nuclease family that is evolutionarily conserved from bacteriophages to humans (Harrington & Lieber, 1994 ▶; Murante et al., 1995 ▶; Hosfield et al., 1998 ▶; Hwang et al., 1998 ▶). Owing to the importance of FEN1 in genome maintenance, defects in the gene encoding FEN1 give rise to a number of genetic diseases, several ataxias, fragile X syndrome and cancer (Tishkoff et al., 1997 ▶; Freudenreich et al., 1998 ▶; Schweitzer & Livingston, 1998 ▶). FEN1 recognizes 5′-flap structures formed by a branched DNA structure containing a variable length of 5′ single-stranded DNA (Storici et al., 2002 ▶; Kaiser et al., 1999 ▶; Chapados et al., 2004 ▶), one chain of which has an unannealed flapped 5′-end, and removes the flapped DNA strand (Murante et al., 1994 ▶). FEN1 plays an important role in the removal of an RNA primer during the maturation of the Okazaki fragment in DNA replication and in the removal of the 5′-flapped DNA structure of damaged nucleotides generated by an incision by an apurinic/apyrimidinic endonuclease. In addition, structural studies of FEN1 proteins have demonstrated the 3′-flap recognition mechanism of FEN1 and the structural basis of the interaction between FEN1 and proliferating cell nuclear antigen (PCNA) in DNA replication (Devos et al., 2007 ▶; Hwang et al., 1998 ▶; Hosfield et al., 1998 ▶; Chapados et al., 2004 ▶; Sakurai et al., 2005 ▶; Ceska et al., 1996 ▶; Mueser et al., 1996 ▶; Kim et al., 1995 ▶). However, the structural basis of the 5′-flap structure-recognition and cleavage mechanism of FEN1 enzymes remains unclear.
To elucidate this basis, we determined the three-dimensional structure of FEN1 from Desulfurococcus amylolyticus (DaFEN1), a hyperthermophilic archaeon with an optimum growth temperature range of 363–365 K (Kil et al., 2000 ▶), at 2.00 Å resolution using the molecular-replacement method. The overall structure of DaFEN1 is similar to those of other FEN1-family proteins, except that DaFEN1 has a clamp region with long and stable α-helices (α3–α4), unlike previous FEN1 structures, which displayed unstable helical clamp regions. The structure of DaFEN1 may represent a 5′-flap structure-recognition state of a FEN1-family protein.
2. Methods and results
2.1. Structure determination
The expression, purification and crystallization of DaFEN1, as well as the preliminary X-ray crystallographic study, were performed as described by Mase et al. (2009 ▶). The DaFEN1 crystal diffracted X-rays to 2.00 Å resolution. The structure of DaFEN1 was determined by the molecular-replacement method. Molecular replacement was performed using the program MOLREP from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶) using the coordinates of FEN1 from Pyrococcus horikoshii (PDB code 1mc8; 57% sequence identity; Matsui et al., 2002 ▶) as a search model. The initial structure was refined and rebuilt using the program ARP/wARP (Lamzin & Wilson, 1997 ▶). After automated model building and refinement, several cycles of manual model rebuilding, restrained refinement and TLS B-factor refinement were performed using the programs REFMAC5 (Murshudov et al., 1997 ▶) from the CCP4 suite and Coot (Emsley & Cowtan, 2004 ▶). Water molecules were picked up from the F o − F c map on the basis of peak height, distance and hydrogen-bonding criteria using the program Coot. The geometry of the final structure was evaluated with the program RAMPAGE (Lovell et al., 2003 ▶). The coordinates of DaFEN1 were deposited in the Protein Data Bank (PDB) with accession code 3ory. A summary of the refinement statistics is provided in Table 1 ▶.
Table 1. Summary of data-collection and refinement statistics for DaFEN1.
| Diffraction data collection | |
| Beamline | Photon Factory BL-5A |
| Space group | P321 |
| Unit-cell parameters () | a = b = 103.76, c = 84.58 |
| Wavelength () | 1.00000 |
| Resolution () | 50.02.00 (2.062.00) |
| No. of observations | 389978 (26362) |
| No. of unique reflections | 35852 (2607) |
| Data completeness (%) | 99.8 (100) |
| Multiplicity | 10.9 (10.1) |
| R merge | 0.037 (0.393) |
| I/(I) | 41.73 (6.06) |
| Refinement | |
| R (%) | 21.0 |
| R free (%) | 22.5 |
| No. of non-H atoms | |
| Protein | 2701 |
| Water | 101 |
| R.m.s. deviations | |
| Bond lengths () | 0.010 |
| Bond angles () | 1.119 |
| Ramachandran plot (%) | |
| Favoured | 98.8 |
| Allowed | 1.2 |
| Disallowed | 0 |
2.2. Structural analysis
Structural analysis was carried out using the following software programs: DALI (Holm et al., 2008 ▶) was used to search for similar structures from the database, 3D-Coffee (O’Sullivan et al., 2004 ▶) was used to align multiple sequences using structural information, ESpript (Gouet et al., 1999 ▶) was used to prepare alignment figures, MATRAS (Kawabata, 2003 ▶) was used to calculate multiple structural alignment, PyMOL (http://www.pymol.org) was used to depict the structure and DSSP (Kabsch & Sander, 1983 ▶) was used to calculate secondary-structure assignments.
3. Results and discussion
3.1. Overall structure of DaFEN1
The crystal structure of DaFEN1 was determined at 2.00 Å resolution with R-factor and R free values of 21.0% and 22.5%, respectively. The final model contained one DaFEN1 molecule and 101 water molecules in the asymmetric unit. Owing to the poor electron density, we could not build the coordinates of residues −10 to −4 and 340–353. In the Ramachandran plot, 98.8% of the residues were included in the favoured region and the remainder of the residues were in the allowed region. The refinement statistics of DaFEN1 are summarized in Table 1 ▶.
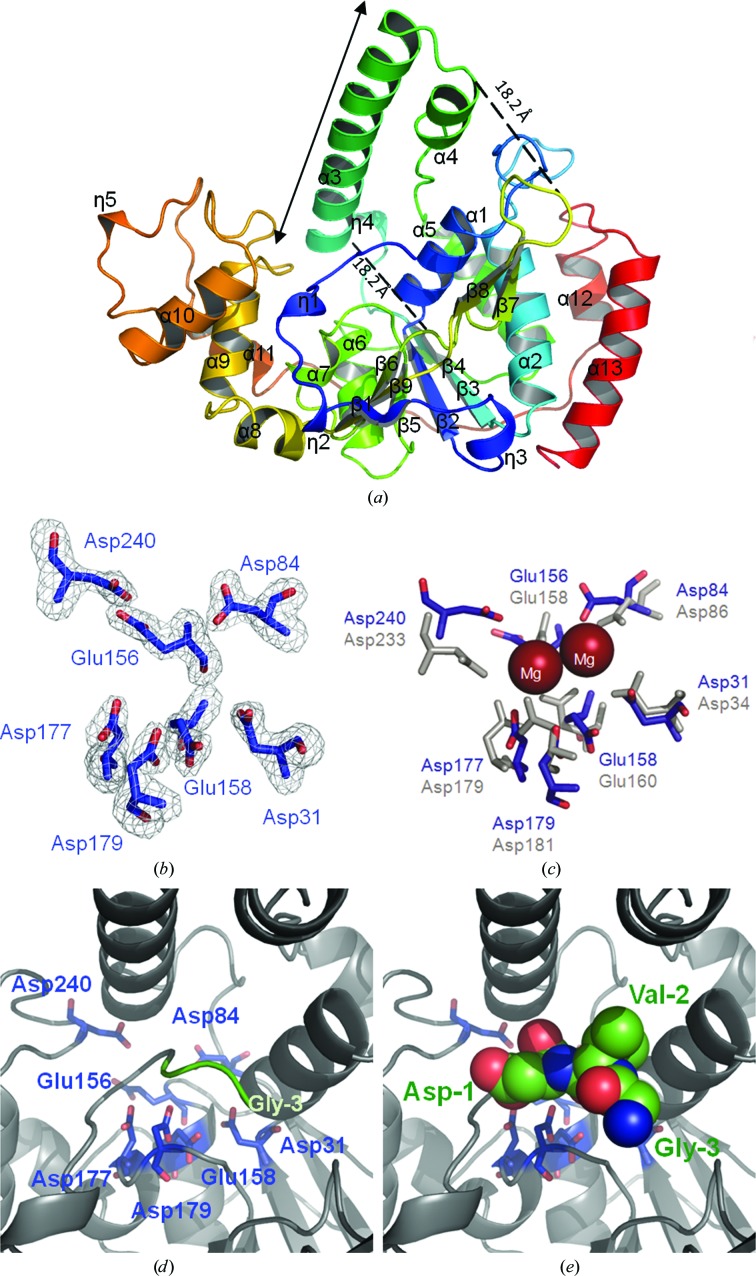
The structure of DaFEN1 is presented as a ribbon model in Fig. 1 ▶(a). DaFEN1 is composed of 13 α-helices (α1–α13) and nine β-strands (β1–β9), with topology η1-η2-β1-η3-β2-α1-α2-β3-η4-α3-α4-α5-β4-α6-β5-α7-β6-β7-β8-β9-α8-α9-α10-η5-α11-α12-α13, where η represents a 310-helix. The structure of DaFEN1 consists of a central six-stranded parallel β-sheet surrounded by α-helices, along with two additional small regions, which in FEN1-family proteins are called the ‘helical clamp’ (the region including α3 and α4 of DaFEN1) and the ‘flexible loop’ (the region including β7, the loop and β8 of DaFEN1), that extend out of the main body of the protein. The flat surface of the glycine-rich loop connecting the α9 and α10 helices has structural and sequence similarity to the DNA-binding helix–turn–helix (HTH) motif. In FEN1 proteins the HTH motif interacts with the phosphodiester backbone of the double-stranded DNA of the flap substrate (Hosfield et al., 1998 ▶). In DaFEN1, 13 residues between the α9 and α10 helices form three turn structures, which may thus bind to substrate double-stranded DNA.
Figure 1.
Structure of DaFEN1. (a) Ribbon diagram of DaFEN1. Colour-coding runs from blue at the N-terminal region to red at the C-terminal region. Secondary-structure assignments are labelled on the ribbon model. (b) σA-weighted F o − F c OMIT map of the putative active site of DaFEN1. The map is contoured at 4σ. (c) Proposed Mg2+ positions in the DaFEN1 active site based on superposition of the DaFEN1 (blue) and hFEN1 (grey) active-site residues. Mg2+ ions are shown as red spheres. (d) Active-site structure of DaFEN1. Active-site acidic residues are shown as blue stick models. (e) Structure of the additional N-terminal residues of DaFEN1. These residues (Gly−3, Val−2 and Asp−1) are shown as sphere models.
3.2. Comparison with other structures
A database search using the DALI server revealed that the overall structure of DaFEN1 is particularly similar to those of archaeal FEN1 proteins. Examples include the chain A structure of FEN1 from P. horikoshii (PhFEN1; Matsui et al., 2002 ▶; PDB entry 1mc8), which was used as a search model for structure determination by molecular replacement in the present study (Z score = 33.7, sequence identity = 59%, r.m.s.d. for 274 aligned Cα atoms = 2.0 Å), the chain A structure of FEN1 from P. furiosus (PfFEN1; Hosfield et al., 1998 ▶; PDB entry 1b43; Z score = 35.8, sequence identity = 60%, r.m.s.d. for 287 aligned Cα atoms = 2.2 Å) and the chain A structure of FEN1 from Archaeoglobus fulgidus (AfFEN1; Chapados et al., 2004 ▶; PDB entry 1rxv; Z score = 34.4, sequence identity = 52%, r.m.s.d. for 301 aligned Cα atoms = 3.3 Å). In addition, the chain X structure of human FEN1 (hFEN1; Sakurai et al., 2005 ▶; PDB entry 1ul1; Z score = 31.8, sequence identity = 38%, r.m.s.d. for 276 aligned Cα atoms = 2.6 Å) has a similar structure.
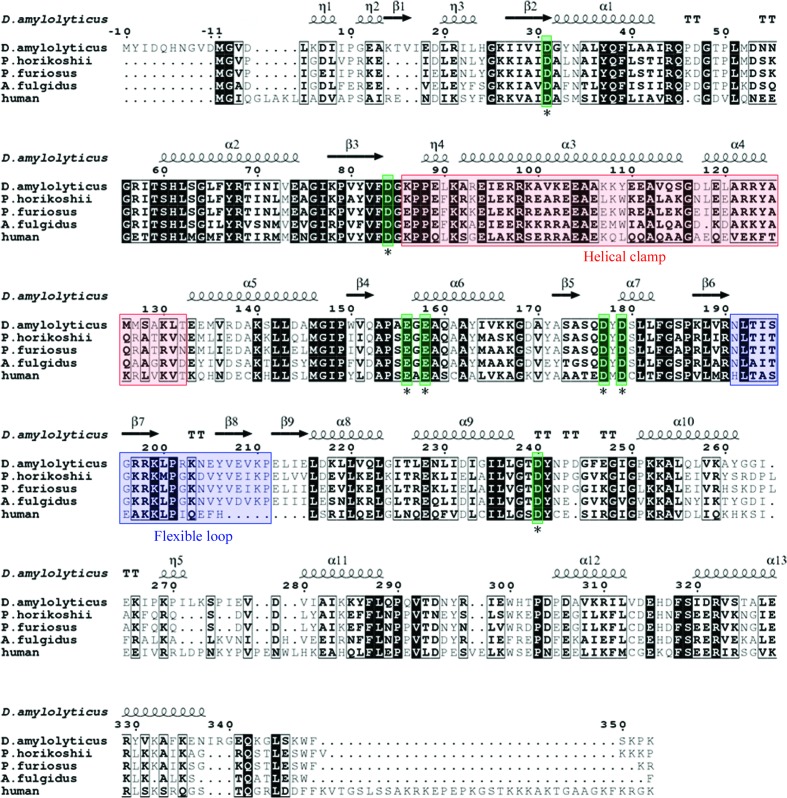
A multiple-sequence alignment shows a high degree of similarity in the active-site pocket residues between DaFEN1 and other FEN1-family proteins (Fig. 2 ▶). In particular, the sequence and structure of the Mg2+-coordinating residues in the active-site pocket are highly conserved. In hFEN1 seven acidic residues (Asp34, Asp86, Glu158, Glu160, Asp179, Asp181 and Asp233) are assigned as active-site residues that tightly coordinate to two Mg2+ ions (Shen et al., 1997 ▶). In DaFEN1 seven acidic residues (Asp31, Asp84, Glu156, Glu158, Asp177, Asp179 and Asp240) correspond to the acidic active-site residues of hFEN1 (Figs. 1 ▶ b and 1 ▶ c). These residues would form the active site of DaFEN1 and coordinate to the two Mg2+ ions which are necessary for enzymatic activity (Fig. 1 ▶ c).
Figure 2.
Amino-acid sequence alignment of DaFEN1 and other FEN1-family proteins [P. horikoshii FEN1 (PDB entry 1mc8), P. furiosus FEN1 (PDB entry 1b43), A. fulgidus FEN1 (PDB entry 1rxv) and human FEN1 (PDB entry 1ul1)]. The secondary-structure assignment of DaFEN1 is indicated by helices (α-helices and 310-helices) and arrows (β-strands). Highly conserved acidic residues in the active site are indicated by asterisks.
3.3. Additional residues in the N-terminal region
For structural analysis, we used a construct that has ten additional amino acids at its N-terminus (M−10YIDQHNGVD−1-DaFEN1) to obtain crystals that were suitable for X-ray diffraction measurements (Mase et al., 2009 ▶), based on the gene arrangement of DaFEN1 preceding the initiation methionine. In the crystal structure of DaFEN1 we built a structural model of the three amino-acid residues on the C-terminal side (GVD−1; Figs. 1 ▶ d and 1 ▶ e). These three residues fill the active-site pocket to prevent substrates from accessing and coordinating to the Mg2+ ions; this prevention inhibits the enzymatic activity of DaFEN1. The approximate distances between the Cα atoms of Asp−1 and Glu158 and between those of Val−2 and Asp31 were 13.7 and 13.9 Å, respectively. DaFEN1 with the ten additional N-terminal residues was found to have a lower enzymatic activity than wild-type DaFEN1 (data not shown). The observation that the N-terminal methionine residue was not observed in FEN1 proteins for which structures have been determined indicates that the decrease in enzymatic activity was not influenced by the positive charge of the N-terminus but simply by the existence of other residues. In addition, the sequence between Met−10 and Asp−1 of DaFEN1 is not conserved in FEN1-family proteins. According to this evidence, we presume that the starting methionine of DaFEN1 is Met1.
3.4. Unique structure of DaFEN1
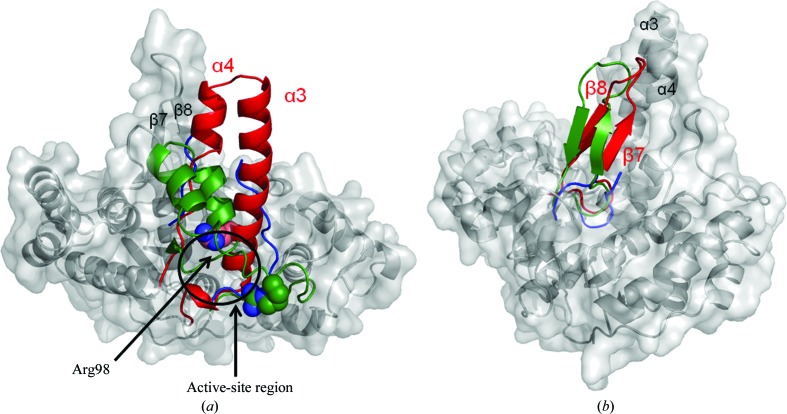
A structural comparison of DaFEN1 with homologous proteins shows that the structure of DaFEN1 contains a unique helical clamp structure (Fig. 3 ▶ a). DaFEN1 is the first FEN1 to show a well ordered helical clamp structure with a long α-helix (α3). Although this region has structural diversity among the FEN1-family proteins, the structure of the helical clamp region probably changes when it binds to the 5′-flap based on the results of mutation analysis (Matsui et al., 2004 ▶; Storici et al., 2002 ▶). One of the residues in the helical clamp region (Arg94) of PfFEN1 has been predicted to bind the phosphate group at the cleavage site of substrate DNA after a substrate-induced conformational change in the helical clamp (Allawi et al., 2003 ▶). The helical clamp region of DaFEN1 is mostly α-helical (the approximate length of the α3 helix is 34 Å) and differs from those of other FEN1 proteins, which are rich in loop structures or are disordered. Interestingly, Arg98 of DaFEN1, which corresponds to Arg94 of PfFEN1, is located in the N-terminal part of the α3 helix near the putative active site (Fig. 3 ▶ a).
Figure 3.
Comparison of DaFEN1 with other FEN1 proteins. (a) Comparison of the helical clamp structures of DaFEN1 (red) with PfFEN1 and hFEN1 (green and blue, respectively). Arg98 of DaFEN1 and Arg94 of PfFEN1 are shown as sphere models. (b) Comparison of the flexible-loop structure of DaFEN1 (red) with those of PfFEN1 and hFEN1 (green and blue, respectively).
DaFEN1 has an ‘antiparallel ribbon’ in the flexible-loop region which consists of antiparallel β-strands with a relatively long loop structure. A flexible loop containing such a ribbon has previously been reported in the structure of PfFEN1 (Hosfield et al., 1998 ▶) and this ribbon supports tighter binding to a flapped DNA than in eukaryotic FEN1 enzymes (Fig. 3 ▶ b). However, the β-sheet is not present in PhFEN1 (Matsui et al., 2002 ▶). It has been reported that most eukaryotic FEN1 enzymes show a deletion of amino acids in this region and in the case of hFEN1 this deletion forms a shorter loop structure (Sakurai et al., 2005 ▶).
In the structure of DaFEN1 the distances between the helical clamp and the flexible loop are estimated to be 18.2 and 18.2 Å between the Cα atoms of Glu94 and Asn191 and those of Glu119 and Lys203, respectively (Fig. 1 ▶ a). The helical clamp and flexible loop form a hole that is sufficiently large to accommodate single-stranded DNA at the kinked position of the flap substrate. The results described in the present study may indicate that the helical clamp of FEN1 becomes α-helical as in the structure of DaFEN1 when the proteins bind and digest DNA with a flapped structure.
Supplementary Material
PDB reference: flap endonuclease 1, 3ory
Acknowledgments
We are grateful to the staff members of beamline BL5A at the Photon Factory (Tsukuba, Japan; Proposal No. 2003S2-002). This work was supported in part by the National Project on Protein Structural and Functional Analyses and the Targeted Proteins Research Program (TPRP) of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Allawi, H., Kaiser, M., Onufriev, A., Ma, W., Brogaard, A., Case, D., Neri, B. & Lyamichev, V. (2003). J. Mol. Biol. 328, 537–554. [DOI] [PubMed]
- Ceska, T., Sayers, J., Stier, G. & Suck, D. (1996). Nature (London), 382, 90–93. [DOI] [PubMed]
- Chapados, B., Hosfield, D., Han, S., Qiu, J., Yelent, B., Shen, B. & Tainer, J. (2004). Cell, 116, 39–50. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Devos, J., Tomanicek, S., Jones, C., Nossal, N. & Mueser, T. (2007). J. Biol. Chem. 282, 31713–31724. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Freudenreich, C., Kantrow, S. & Zakian, V. (1998). Science, 279, 853–856. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Harrington, J. & Lieber, M. (1994). EMBO J. 13, 1235–1246. [DOI] [PMC free article] [PubMed]
- Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Hosfield, D., Mol, C., Shen, B. & Tainer, J. (1998). Cell, 95, 135–146. [DOI] [PubMed]
- Hwang, K., Baek, K., Kim, H. & Cho, Y. (1998). Nature Struct. Biol. 5, 707–713. [DOI] [PubMed]
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Kaiser, M., Lyamicheva, N., Ma, W., Miller, C., Neri, B., Fors, L. & Lyamichev, V. (1999). J. Biol. Chem. 274, 21387–21394. [DOI] [PubMed]
- Kawabata, T. (2003). Nucleic Acids Res. 31, 3367–3369. [DOI] [PMC free article] [PubMed]
- Kil, Y., Baitin, D., Masui, R., Bonch-Osmolovskaya, E., Kuramitsu, S. & Lanzov, V. (2000). J. Bacteriol. 182, 130–134. [DOI] [PMC free article] [PubMed]
- Kim, Y., Eom, S., Wang, J., Lee, D., Suh, S. & Steitz, T. (1995). Nature (London), 376, 612–616. [DOI] [PubMed]
- Lamzin, V. & Wilson, K. (1997). Methods Enzymol. 277, 269–305. [DOI] [PubMed]
- Lovell, S., Davis, I., Arendall, W. III, de Bakker, P., Word, J., Prisant, M., Richardson, J. & Richardson, D. (2003). Proteins, 50, 437–450. [DOI] [PubMed]
- Mase, T., Kubota, K., Miyazono, K., Kawarabayasi, Y. & Tanokura, M. (2009). Acta Cryst. F65, 923–925. [DOI] [PMC free article] [PubMed]
- Matsui, E., Abe, J., Yokoyama, H. & Matsui, I. (2004). J. Biol. Chem. 279, 16687–16696. [DOI] [PubMed]
- Matsui, E., Musti, K., Abe, J., Yamasaki, K., Matsui, I. & Harata, K. (2002). J. Biol. Chem. 277, 37840–37847. [DOI] [PubMed]
- Mueser, T., Nossal, N. & Hyde, C. (1996). Cell, 85, 1101–1112. [DOI] [PubMed]
- Murante, R., Huang, L., Turchi, J. & Bambara, R. (1994). J. Biol. Chem. 269, 1191–1196. [PubMed]
- Murante, R., Rust, L. & Bambara, R. (1995). J. Biol. Chem. 270, 30377–30383. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- O’Sullivan, O., Suhre, K., Abergel, C., Higgins, D. G. & Notredame, C. (2004). J. Mol. Biol. 340, 385–395. [DOI] [PubMed]
- Sakurai, S., Kitano, K., Yamaguchi, H., Hamada, K., Okada, K., Fukuda, K., Uchida, M., Ohtsuka, E., Morioka, H. & Hakoshima, T. (2005). EMBO J. 24, 683–693. [DOI] [PMC free article] [PubMed]
- Schweitzer, J. & Livingston, D. (1998). Hum. Mol. Genet. 7, 69–74. [DOI] [PubMed]
- Shen, B., Nolan, J., Sklar, L. & Park, M. (1997). Nucleic Acids Res. 25, 3332–3338. [DOI] [PMC free article] [PubMed]
- Storici, F., Henneke, G., Ferrari, E., Gordenin, D., Hübscher, U. & Resnick, M. (2002). EMBO J. 21, 5930–5942. [DOI] [PMC free article] [PubMed]
- Tishkoff, D., Filosi, N., Gaida, G. & Kolodner, R. (1997). Cell, 88, 253–263. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: flap endonuclease 1, 3ory