Abstract
Mammalian imprinted genes are associated with differentially methylated regions (DMRs) that are CpG methylated on one of the two parental chromosomes. In mice, at least 21 DMRs acquire differential methylation in the germline and many of them act as imprint centres. We previously reported the physical extents of differential methylation at 15 DMRs in mouse embryos at 12.5 days postcoitum. To reveal the ontogeny of differential methylation, we determined and compared methylation patterns of the corresponding regions in sperm and oocytes. We found that the extent of the gametic DMRs differs significantly from that of the embryonic DMRs, especially in the case of paternal gametic DMRs. These results suggest that the gametic DMR sequences should be used to extract the features specifying methylation imprint establishment in the germline: from this analysis, we noted that the maternal gametic DMRs appear as unmethylated islands in male germ cells, which suggests a novel component in the mechanism of gamete-specific marking. Analysis of selected DMRs in blastocysts revealed dynamic changes in allelic methylation in early development, indicating that DMRs are not fully protected from the major epigenetic reprogramming events occurring during preimplantation development. Furthermore, we observed non-CpG methylation in oocytes, but not in sperm, which disappeared by the blastocyst stage. Non-CpG methylation was frequently found at maternally methylated DMRs as well as non-DMR regions, suggesting its prevalence in the oocyte genome. These results provide evidence for a unique methylation profile in oocytes and reveal the surprisingly dynamic nature of DMRs in the early embryo.
Keywords: DNA methylation, Differentially methylated region (DMR), Genomic imprinting, Non-CpG methylation, Oocyte, Sperm, Mouse
INTRODUCTION
Genomic imprinting is a parental origin-specific gene-marking phenomenon crucial for normal mammalian development. The marks, or imprints, render certain genes as expressed from only one of the two inherited chromosomes. At the molecular level, imprints are characterised by epigenetic modifications (Reik and Walter, 2001; Sasaki and Matsui, 2008), which include DNA methylation: imprinted genes are associated with differentially methylated regions (DMRs) that are methylated on either the paternal or maternal allele. In the mouse, 21 DMRs (of which four are paternally methylated and 17 are maternally methylated) are known to acquire methylation in the gametes (primary DMRs) (Chotalia et al., 2009; Kobayashi et al., 2009), and many of them have been shown to constitute imprint centres (ICs) that regulate nearby imprinted genes in cis (Wutz et al., 1997; Thorvaldsen et al., 1998; Fitzpatrick et al., 2002; Yoon et al., 2002; Lin et al., 2003; Williamson et al., 2004; Shiura et al., 2009). The methylation marks of such primary DMRs are presumed to be maintained throughout embryonic development, including during the preimplantation stages at which extensive demethylation of the genome takes place, but few have been examined in detail throughout development (Reik et al., 2001).
The primary DMRs are methylated by a de novo methyltransferase, Dnmt3a, and its co-factor Dnmt3L in both male and female germlines (Hata et al., 2002; Bourc'his and Bestor, 2004; Kaneda et al., 2004). However, how the Dnmt3a-Dnmt3L complex finds and distinguishes DMRs in the two germlines remains unclear. Sequence studies have shown that short interspersed nuclear elements (SINEs) are less enriched in imprinted regions than in non-imprinted regions (Greally, 2002; Ke et al., 2002), and tandem repeats are frequently found within or adjacent to DMRs (Neumann et al., 1995; Hutter et al., 2006; Reinhart et al., 2006; Walter et al., 2006). Furthermore, an 8-10 bp spacing of CpGs and unmodified histone H3 lysine 4 (H3K4) may be preferred features for interaction of the Dnmt3a-Dnmt3L complex (Jia et al., 2007; Ooi et al., 2007). Indeed, it was recently reported that a histone H3K4 demethylase is required for methylation of some maternally methylated DMRs in oocytes (Ciccone et al., 2009). Other work has suggested that transcription across the DMRs during oocyte growth is required for methylation at maternally imprinted regions (Chotalia et al., 2009). Despite these advances, we still lack a full explanation of the locus- and parent-specific methylation of DMRs (Walter et al., 2006; Ferguson-Smith and Greally, 2007).
Previously in our laboratory, the physical extent of allelic differential methylation was determined for 15 primary DMRs (three paternally methylated and 12 maternally methylated DMRs) in mouse embryos at 12.5 days postcoitum (dpc) by bisulphite sequencing (Kobayashi et al., 2006). However, very little systematic analysis has yet been performed on the extents of DMRs in gametes, and this would be essential information in fully defining mechanisms involved in locus- and parent-specific methylation in the germline. We have now determined the extents of this set of DMRs in the gametes (gametic DMRs) and find that they differ significantly from the allelic DMRs in the embryos (embryonic DMRs), indicating that the gametic DMR sequences should be used to identify the features that determine methylation imprint establishment. Interestingly, most maternally methylated DMRs are flanked by regions methylated in both oocytes and sperm, so that the DMRs can be viewed as unmethylated islands in sperm. Together with results obtained for selected DMRs in blastocysts, we found that the extent and methylation landscape of DMRs change dynamically during early development. Furthermore, we unexpectedly identified oocyte-specific non-CpG methylation, similar to that recently reported in human embryonic stem (ES) cells (Lister et al., 2009). The presence of non-CpG methylation was evident within CpG-methylated regions not only at DMRs, but also at other regions, suggesting its prevalence in the oocyte genome.
MATERIALS AND METHODS
Mice
C57BL/6 mice were purchased from Clea Japan or bred at the Babraham Institute, and JF1 mice (Koide et al., 1998) were obtained from the Genetics Strains Research Center at the National Institute of Genetics, Japan. C57BL/6 females were crossed with JF1 males to obtain F1 hybrid mice to isolate sperm, oocytes and blastocysts.
Preparation of sperm, oocytes and blastocysts
Sperm was isolated from the epididymis of adult mice. Fully grown oocytes were isolated from ovarian follicles of adult mice as follows: ovaries were placed in M2 medium (Sigma-Aldrich) and slashed with a needle to release oocytes. Only oocytes with a diameter over ~65 μm were collected and carefully washed several times by pipetting in a drop of M2 medium to eliminate somatic cells. Blastocysts were flushed out from the uteri at 3.5 dpc.
DNA isolation and bisulphite methylation analysis
Genomic DNA was prepared from oocytes and blastocysts as described previously (Hirasawa et al., 2008), except that lambda DNA (300 ng) was added before ethanol precipitation. Sperm DNA was prepared by a standard method. Bisulphite treatment of genomic DNA was carried out as described (Hirasawa et al., 2008). We used bisulphite-treated genomic DNA from sperm (~5 ng) or from oocytes and blastocysts (~1.4-2.8 ng) for each PCR amplification. The result for each oocyte amplicon was obtained from single PCR amplifications using at least 100 oocytes. For sperm DNA, the following PCR program was used: 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. For oocyte and blastocyst DNA, the program consisted of eight cycles of 95°C for 30 seconds, 60-56°C (with a 0.5°C decrement per cycle) for 1 minute, and 72°C for 1 minute, followed by 32 cycles of 95°C for 30 seconds, 56°C for 1 minute, and 72°C for 1 minute. The PCR primers are listed in Table S1 in the supplementary material. The PCR products were cloned and sequenced.
To determine the extent of each DMR, we applied the criteria described previously (Kobayashi et al., 2006). Briefly, if none or only one of four consecutive CpGs showed a methylation level below 70% in the allele where the DMR was more methylated than in the other allele, or over 30% in the allele where the DMR was less methylated than in the other allele, these four CpGs were judged to be a part of the DMR. The genomic region between the two outmost CpG sites was defined as a DMR. In cases in which single nucleotide polymorphisms (SNPs) were not available at potential DMR endpoints in embryos, the following criterion was applied: if none or only one of four consecutive CpGs showed a methylation level below 85% or over 15%, these four CpGs were judged to be a part of the DMR. However, when the methylation status of such a region was highly mosaic, this criterion was not applied and thus the precise DMR end was not defined. Prior to application of the above criteria, all bisulphite-PCR clones potentially derived from the same genomic DNA molecule were removed using the strict criteria defined by BISMA (Rohde et al., 2010). The genomic coordinates of the DMRs so determined are given in Table S2 in the supplementary material.
Sequence analyses of DMRs
We used in-house scripts to analyse the contents of CpG, G+C, oligonucleotides, repetitive elements and CpG spacing of the gametic DMR sequences determined in this study. For those DMRs for which the precise extents could not be determined, tentative endpoints of differential methylation were used. The sequence properties of the DMRs were compared with those of all mouse CpG islands (Illingworth et al., 2010) and with mouse chromosome 19 as an approximation of the whole genome. Genomic positions of repetitive elements were extracted from the EnsEMBL dataset to analyse their coverage in the DMRs and CpG islands.
Non-CpG methylation analysis
To verify the presence of non-CpG methylation in oocytes, 100 ng unmethylated lambda DNA (Promega) was added as a conversion control to 7 ng of DNA from oocytes, bisulphite treated in duplicate and representative regions of lambda DNA and DMRs were analysed by sequencing. An equivalent amount of DNA from blastocysts mixed with lambda DNA was treated in parallel for comparison. The data obtained by bisulphite sequencing were analysed using the CyMATE program (Hetzl et al., 2007) to determine the positions and levels of non-CpG methylation. The rate of experimental errors caused by PCR and sequencing was estimated using the observed frequency of thymine to cytosine transition (oocytes, 0.22%; sperm, 0.18%; blastocysts, 0.19%; 12.5 dpc embryos, 0.20%), and was used for data normalisation. The Logo plot images were generated with WebLogo (Crooks et al., 2004). To exclude cytosines that likely resulted from experimental errors, including failed conversion, cytosine sites represented by at least two clones were used to extract proximal sequences for the generation of logos.
RESULTS
Determination of the extents of DMRs in oocytes and sperm
We collected fully grown oocytes and epididymal sperm, both of which have fully established methylation imprints (Hiura et al., 2006; Kato et al., 2007), for bisulphite sequencing. To distinguish between the parental alleles we used SNPs identified between C57BL/6 and JF1 strains at 86 out of 109 amplicons and 110 out of 128 amplicons in oocytes and sperm, respectively. DNA corresponding to at least 100 oocytes was used for each assay. The methylation status in oocytes was compared with that in sperm and the extents of gametic DMRs were determined using the criteria described previously (Kobayashi et al., 2006). An average of 18 and 16 unique sequences were present in each amplicon at the boundaries of the DMRs so defined in oocytes and sperm, respectively, suggesting that the results were not skewed by possible clonal amplification events.
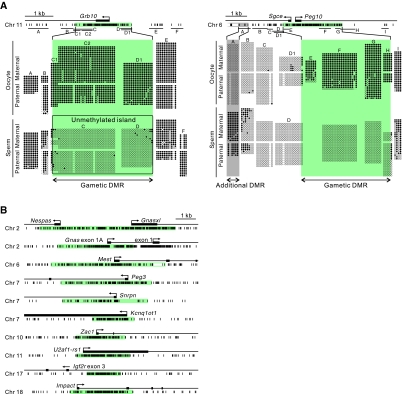
We determined the CpG methylation status of 12 maternally methylated DMRs (Fig. 1) and three paternally methylated DMRs (Fig. 2). PCR amplicons were designed to close the gaps of undetermined regions toward the endpoint of each DMR. Within each gamete, most regions showed no significant difference in CpG methylation between the parental alleles, confirming that gamete-specific methylation was already established. Most of the regions flanking the maternally methylated DMRs were consistently methylated in both oocytes and sperm, and thus the DMRs were viewed as ‘unmethylated islands’ on sperm DNA [see Fig. 1A for the example at the Grb10 (also known as Meg1) DMR]. Exceptions to this generality were the Peg10 and Gnas 1A DMRs, which were flanked by an unmethylated region in both sperm and oocytes (Figs 1 and 3). Note, however, that these are both complex transcription units: directly upstream of Peg10 is the promoter for Sgce, which is transcribed in the opposite direction, and 3′ to the Gnas 1A DMR is the Gnas promoter, which is a constitutively unmethylated CpG island (Chotalia et al., 2009). Interestingly, the Peg10 locus had an additional DMR, with a 1.3 kb intervening unmethylated region (Fig. 1A). The additional DMR was methylated only in sperm, the opposite of the primary DMR, and lost differential methylation after fertilisation (see later).
Fig. 1.
DNA methylation patterns and the extents of differential methylation obtained by bisulphite sequence analysis at the twelve maternally methylated DMRs in oocytes and sperm. (A) Examples of primary bisulphite sequencing results (before the removal of potential clonal amplification events) showing methylated (filled circles) and unmethylated (open circles) CpG sites of two representative DMRs at the mouse Grb10 and Peg10 loci. CpG sites at the DMRs are shown by short vertical lines below the gene organisation schemes; PCR amplicons are indicated by horizontal bars. In the bisulphite sequencing profiles shown, hyphens represent missing or undetermined CpG sites due to SNPs or sequencing problems. Paternal and maternal alleles were discriminated by SNPs if available. The extents of the gametic DMRs determined are shown by the green areas. The Grb10 DMR can be seen as an unmethylated island on the sperm DNA (see text). An additional DMR found at the Peg10 locus is shown in grey. The endpoint of the additional DMR was not determined and therefore the 5′ end is shown by an open box. (B) Extents of the remainder of the maternally methylated DMRs analysed in this study. The DMR in the gametes determined at each locus is shown in green. The precise 3′ endpoint of the Mest DMR was not determined due to repetitive sequences overlapping this region.
Fig. 2.
DNA methylation patterns obtained and extents of differential methylation determined at the three paternally methylated DMRs in oocytes and sperm. (A) CpG methylation profile of the mouse H19 (top) and Rasgrf1 (bottom) DMRs in the gametes. Additional DMRs (grey) were found separate from the gametic DMRs containing the imprint centres (ICs, green) at both loci. The gametic and additional DMRs extended over long distances and the endpoints were not determined (denoted by open boxes). Tandem repeat regions are represented by triangles. (B) The extent of the gametic DMR determined at the Dlk1-Gtl2 locus in oocytes and sperm.
Fig. 3.
Developmental changes in CpG methylation and DMR extent. DNA methylation status in the gametes (oocyte, red; sperm, blue) compared with that in 12.5 dpc mouse embryos (maternal allele, red; paternal allele, blue). The extents of the DMRs shown were determined after the removal of potential clonal amplification events. Undetermined endpoints of methylation are indicated by gradient shading. Gametic DMRs containing ICs are centred and the changes in DMR extent that have occurred in embryos by 12.5 dpc are represented by pink (maternal DMRs) and light blue (paternal DMRs) areas. Major transcription start sites are indicated by arrows. DMRs were classified into four groups (enlarged, shifted, contracted and fused) according to the pattern of change in extent.
The regions containing the three paternally methylated DMRs were methylated in sperm and unmethylated in oocytes, and this differential methylation extended over long distances (>10 kb). Thus, we were not able to determine the precise endpoints of these DMRs (for the subsequent feature analyses, we tentatively used the endpoints to which differential methylation was confirmed by sequencing). At the H19 DMR, there was a 1 kb region unmethylated in both sperm and oocytes, which divided the DMR into two (Fig. 2A). Of the two parts, that located 5′ to the H19 gene was the previously reported primary DMR containing the IC (Thorvaldsen et al., 1998), and the other was defined as an additional DMR. Similarly, the Rasgrf1 DMR was divided by a 2.8-3.9 kb region that was unmethylated in both sperm and oocytes (Fig. 2A), and here the more 5′ part was the IC (Yoon et al., 2002) and the other region constituted an additional DMR.
Gametic DMRs differ from embryonic DMRs
The gametic DMRs were compared with the corresponding embryonic DMRs previously determined at 12.5 dpc (Kobayashi et al., 2006), for which additional bisulphite analysis in this study provided more accurate extents (Fig. 3). We found that they differed significantly, and the DMRs were classified into four groups according to the pattern of change (i.e. enlargement, shift, contraction and fusion). Thus, the extents of all DMRs changed to a greater or lesser degree during development.
For example, while one end of the maternally methylated Nespas-Gnasxl, Peg10, Mest (also known as Peg1), Kcnq1ot1 (also known as Lit1), Grb10, U2af1-rs1 (also known as Zrsr1) and Igf2r DMRs remained unchanged, the other end either extended (enlarged DMRs) or regressed (contracted DMRs). The Gnas 1A, Peg3 and Snrpn DMRs extended at one end and regressed at the other end (shifted DMRs). The rest of the maternally methylated DMRs, i.e. the Zac1 (also known as Plagl1) and Impact DMRs, regressed at both ends and thus contracted. Almost all these changes were attributable to changes that occurred in the ‘unmethylated islands’ on the sperm-derived chromosomes. The Peg10 and Gnas 1A DMRs, which were not seen as unmethylated islands, were exceptions: at the Peg10 DMR, an unmethylated flanking region on the oocyte-derived chromosome acquired methylation, which contributed not only to the enlargement of the primary DMR but also to the loss of differential methylation at the additional DMR; and the 3′ endpoint of the Gnas 1A DMR showed a subtle change and the flanking region remained unmethylated on both paternal and maternal alleles.
The paternally methylated H19 and Rasgrf1 DMRs were divided into two in the gametes by a region unmethylated in both sperm and oocytes (see above). The unmethylated region of both loci became methylated on the sperm-derived chromosomes by 12.5 dpc and thus the two DMRs were fused into one (Fig. 3). This is consistent with previous, but more limited, reports that the unmethylated H19 promoter in sperm becomes methylated later in embryos (Ferguson-Smith et al., 1993), and that an unmethylated region is present at the Rasgrf1 locus in sperm (Lindroth et al., 2008). Both fused DMRs, and also the paternally methylated Dlk1-Gtl2 (also known as Meg3) DMR, were contracted substantially in 12.5 dpc embryos by regressions at both ends. This was caused by the acquisition of methylation at the flanking regions on the oocyte-derived chromosomes. Interestingly, the overall changes of the paternally methylated DMRs were far more extensive than for the maternally methylated DMRs.
Analysis of DMRs in blastocysts suggests dynamic changes in CpG methylation
Having established that the gametic DMRs differ from the embryonic DMRs determined at 12.5 dpc, we were interested to know whether the extent of the embryonic DMRs is established at the preimplantation stage or later in development. We collected 3.5 dpc blastocysts and analysed the methylation status of representative DMRs from each group as defined by the patterns of change (Fig. 3), i.e. the maternally methylated Peg10 (enlarged), Snrpn (shifted) and Impact (contracted) DMRs and the paternally methylated Dlk1-Gtl2 (contracted) and H19 (fused) DMRs.
All five allelic DMRs analysed in blastocysts had significantly different methylation profiles from the corresponding gametic DMRs and also from the postimplantation embryonic DMRs (Fig. 4). Most blastocyst DMRs showed contraction (except for the Snrpn DMR, the precise 3′ endpoint was not determined), resulting from demethylation of parts of the DMRs and their flanking regions. The maternally methylated DMRs showed more subtle changes than the paternally methylated DMRs between gametes and blastocysts. Of particular note, however, was the substantial decrease in methylation of flanking regions on the sperm-derived chromosomes. The Peg10, Snrpn and Impact DMRs, together with their flanking regions including the additional Peg10 DMR, were almost completely demethylated on the sperm-derived chromosomes in blastocysts. Therefore, the Snrpn and Impact DMRs can no longer be viewed as unmethylated islands on the sperm-derived chromosome. However, demethylation also occurred on the oocyte-derived chromosomes at the regions flanking their gametic DMRs, and this contributed to the contraction of the Peg10 and Impact DMRs.
Fig. 4.
Developmental changes of DMRs in preimplantation mouse embryos. CpG methylation at three maternally methylated DMRs (Peg10, enlarged DMR; Snrpn, shifted DMR; Impact, contracted DMR) and two paternally methylated DMRs (Dlk1-Gtl2, contracted DMR; H19, fused DMR) were analysed in blastocysts and compared with those in the gametes and 12.5 dpc embryos. CpG sites are indicated by short vertical bars below gene organisations. Horizontal bars indicate positions of PCR amplicons used. Methylation level at each CpG site is shown by vertical bars, with the height of each bar representing the degree of methylation (red, oocyte and maternal allele; blue, sperm and paternal allele). Potential clonal amplification events were removed. Where there was no SNP for allele discrimination, methylation level is indicated by black bars. DMRs are highlighted in pink (maternally methylated) and light blue (paternally methylated). Additional DMRs found at the Peg10 and the H19 loci in the gametes are shown in grey. Striped DMR ends denote undetermined endpoints. ND, not determined.
Concomitant with the demethylation on the sperm-derived chromosomes, the paternally methylated DMRs showed more extensive changes in their extents. The additional H19 DMR disappeared in blastocysts and both the H19 and Dlk1-Gtl2 DMRs showed substantial contraction. This is consistent with the previous, more limited, observations for the H19 locus, which showed that the region overlapping the H19 gene body (corresponding to amplicons C and E) is unmethylated in early embryos (Brandeis et al., 1993) and that the methylation level is reduced at the promoter-proximal region in blastocysts (corresponding to amplicon F) (Tremblay et al., 1995; Tremblay et al., 1997). Interestingly, the Dlk1-Gtl2 DMR in blastocysts did not overlap with that in 12.5 dpc embryos.
These studies revealed surprisingly dynamic changes in CpG methylation at the DMRs in early embryos. Most changes observed in blastocysts were associated with demethylation, which is most likely part of the genome-wide demethylation that occurs in preimplantation development (Reik et al., 2001).
Sequence feature analyses of gametic DMRs
To date, a number of attempts have been made to identify sequence features that may distinguish DMRs from the rest of the genome with the aim of establishing the properties that specifically direct the acquisition of gametic methylation imprints (Walter et al., 2006; Jia et al., 2007). However, the only available comprehensive sequence information on DMRs has been obtained from 12.5 dpc embryos (Kobayashi et al., 2006), the extents of which we have now shown to be significantly different in the gametes. Here, we analysed features of the gametic DMR sequences obtained in this study.
We previously reported that the embryonic maternally methylated DMRs were more CpG-rich than the paternally methylated DMRs (Kobayashi et al., 2006). To determine whether this is also true for gametic DMRs, we compared their CpG and G+C contents with 23,021 mouse CpG islands that were biochemically defined (Illingworth et al., 2010) and with the mouse genome (chromosome 19) as references. Consistent with the previous study, the maternally methylated gametic DMRs showed higher contents of CpG and G+C (see Fig. S1A,B in the supplementary material) than the paternally methylated gametic DMRs. The CpG and G+C contents dropped within the 1 kb flanking regions of the maternally methylated DMRs. When compared with CpG islands and the mouse genome, the CpG and G+C contents of the maternally methylated DMRs and their flanking regions were very close to those of CpG islands, whereas the paternally methylated DMRs were more similar to the genome average.
To explore the possibility that the DMRs have a distinct sequence composition from the rest of the genome by which they might be recognised by the DNA methylation machinery in the gametes, we searched for short sequence motifs that were over-represented in the gametic DMRs. However, similar to their CpG and G+C contents, the maternally methylated DMRs had very similar patterns of di-, tri- and tetranucleotide enrichment to that of CpG islands, and the paternally methylated DMRs were similar to the genome average with no apparent over-representation of specific sequence contents (Fig. S2 in the supplementary material). Analysis of repetitive elements showed that the maternally and paternally methylated DMRs have a higher mean coverage of tandem repeats than CpG islands (see Fig. S3A in the supplementary material), which is consistent with previous reports (Hutter et al., 2006). Nevertheless, there was still a substantial number of CpG islands that had higher coverage of tandem repeats than the DMRs, suggesting that DMRs cannot be distinguished from such CpG islands simply by this difference (see Fig. S3B in the supplementary material). The paternally methylated DMRs showed a higher mean coverage of SINEs, long interspersed nuclear elements (LINEs) and long terminal repeats (LTRs) than CpG islands; however, this mainly reflected the exceptionally repeat-rich sequences at the Rasgrf1 DMR (Hirasawa et al., 2008). Also, in contrast to a previous study that showed that maternally methylated DMRs (as determined by their extents in 12.5 dpc embryos) have a characteristic 8-10 bp CpG periodicity, our analysis did not find this feature in the gametic DMRs (see Fig. S4 in the supplementary material). We also examined whether CpG sites in the gametic DMRs have the specific sequence contexts that have been reported to be preferred by human DNMT3A or DNMT3B (Wienholz et al., 2010), but we did not observe this, consistent with the proposal that DNMT3L attenuates the flanking sequence preference of these enzymes (data not shown). Thus, these results indicate that the sequence compositions of the maternally and paternally methylated gametic DMRs were distinct, but did not differ from those of CpG islands or the genome average, respectively.
Non-CpG methylation in oocytes
During this study, we noticed a high incidence of unconverted cytosines at non-CpG sites at the maternally methylated DMRs in fully grown oocytes, but not in sperm, blastocysts or 12.5 dpc embryos. The presence of unconverted cytosines at non-CpG sites was not due to failed bisulphite reactions as lambda DNA, which was treated together with oocyte DNA, was almost fully bisulphite converted (99.9%) at two representative regions analysed (see Fig. S5 in the supplementary material). These regions had CpG dinucleotide densities of 9.4-12.2% and G+C contents of 48.1-50.2%, which are comparable to those of the maternally methylated DMRs (see Fig. S1 in the supplementary material). In addition, blastocyst DNA treated in parallel also showed very few unconverted cytosines at non-CpG sites (see Fig. S5 in the supplementary material).
Examples of frequent non-CpG methylation in fully grown oocytes are shown for regions of the Igf2r, Snrpn and Kcnq1ot1 DMRs (Fig. 5A and see Fig. S5 in the supplementary material). In the region of the Igf2r DMR shown, methylation levels at non-CpG sites were not as high as those at CpG sites (which showed nearly 100% methylation), but some non-CpG sites showed in excess of 50% methylation. Within the Snrpn DMR, a number of non-CpG sites were methylated and some sites showed nearly 100% methylation. By contrast, the corresponding regions in sperm and 12.5 dpc embryos had very little non-CpG methylation. The average methylation levels in oocytes at methylated sites of all maternally methylated DMRs examined were: CpG, 91.9%; CpHpG, 22.2%; and CpHpH, 22.0% (where H=A, C or T). Analysis of all DMRs also showed that non-CpG methylation in oocytes tended to occur within or near regions with high CpG methylation, but was absent from CpG-unmethylated regions such as those DMRs methylated on the paternal allele (Fig. 5B and see Fig. S6 in the supplementary material for representative regions). When all data were combined, 37.0% of 5-methylcytosines occurred in non-CpG contexts in oocytes (Fig. 5C). Interestingly, blastocysts had a slightly higher proportion of non-CpG methylation (1.9%) than sperm (1.0%) or 12.5 dpc embryos (0.7%).
Fig. 5.
Non-CpG methylation in oocytes. (A) Distribution and levels of CpG and non-CpG methylation in representative regions of the maternally methylated Igf2r and Snrpn DMRs in fully grown oocytes, sperm and 12.5 dpc mouse embryos. The regions correspond to PCR amplicons C of the Igf2r DMR and D of the Snrpn DMR (Fig. 4 and see Fig. S5 in the supplementary material for the positions of the amplicons). Positions of cytosines are shown at the bottom. (B) Number of methylcytosines in each sequence context in oocytes at the maternally and paternally methylated DMRs. The number of cytosines sequenced was divided by the total length of amplicons sequenced. (C) Percentage of methylcytosines in each sequence context in oocytes, sperm, blastocysts and 12.5 dpc embryos. Values were normalised against the expected experimental error rates (see Materials and methods). (D) Percentage of all possible cytosine sites at which methylcytosine was observed at the maternally methylated DMRs in oocytes. (E) Logo plots for the sequences proximal to the methylated CpHpG and CpHpH sites in oocytes. (F) CpG and non-CpG methylation status at non-DMR regions within the Snrnp70 and Pard6b loci in oocytes and sperm.
It is also interesting to note that non-CpG methylation in oocytes did not appear to occur at all possible sites, in contrast to CpG methylation (Fig. 5D). Furthermore, there was a good concordance in the sites of non-CpG methylation between replicate bisulphite-treated oocyte samples (Fig. 5A and see Fig. S5 in the supplementary material). Sequence context analysis showed that at non-CpG sites, the nucleotide immediately 3′ to 5-methylcytosine was most likely to be adenine (CpA) (Fig. 5E), which is also a feature of non-CpG methylation observed previously at one locus in oocytes (Ramsahoye et al., 2000; Haines et al., 2001; Lister et al., 2009) and more widely in ES cells (Ramsahoye et al., 2000; Haines et al., 2001; Lister et al., 2009). However, unlike human ES cells (Lister et al., 2009), mouse oocytes had 5-methylcytosine more frequently in a CpHpH context than in a CpHpG context (Fig. 5C). We also examined whether there was an 8-10 bp periodicity between methylated sites when methylcytosines in all sequence contexts were included in the analysis, but this was not found to be the case (data not shown).
To assess whether non-CpG methylation is a feature specific to maternally methylated DMRs, we performed bisulphite analysis at non-DMR CpG-rich elements (S. Smallwood, personal communication), at which both oocytes and sperm are CpG-methylated (Fig. 5F). The results showed a similar high frequency of non-CpG methylation at the CpG islands of the Snrnp70 and Pard6b loci in oocytes, suggesting that it is not restricted to the DMRs, but prevalent in the CpG-methylated compartment of the oocyte genome. The absence of non-CpG methylation in these CpG-methylated regions in sperm contrasts markedly with the methylation status in oocytes. Non-CpG methylation was previously reported at the Nf1 locus and at the Mest DMR in mouse oocytes (Haines et al., 2001; Imamura et al., 2005), as well as in mouse and human ES cells (Ramsahoye et al., 2000; Lister et al., 2009), but the data in oocytes and early embryos were limited and the information in sperm was lacking. Here, for the first time, we have obtained solid evidence for the prevalence of non-CpG methylation in mammalian oocytes.
DISCUSSION
As an essential step to understanding the mechanisms of methylation imprint establishment in the germline, we have determined the extents of the DMRs in sperm and oocytes. Our data provide first access to detailed information on the methylation and sequence extents of gametic DMRs and begin to provide new insights into how we view the mechanism of methylation imprint establishment and its parental germline specificity. Moreover, we find dynamic developmental changes in differential CpG methylation and non-CpG methylation in oocytes (summarised in Fig. 6).
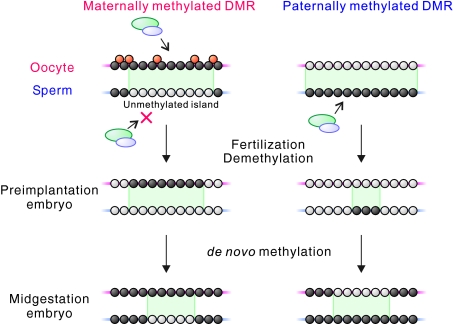
Fig. 6.
Developmental changes in methylation landscapes at typical DMRs. (Top) At a maternally methylated DMR, the de novo methyltransferase complex (green and blue ovals) methylates both CpG (black circles) and non-CpG (orange circles) sites in oocytes, whereas in sperm it cannot methylate the region, which consequently remains as an unmethylated island (unmethylated CpG sites shown by white circles), establishing the gametic DMR (green region). (Middle) After fertilisation, the peripheral regions of the DMR are affected by global demethylation, resulting in a change in the extent of the DMR, and the unmethylated island can no longer be seen on the sperm-derived allele. By this stage, non-CpG methylation on the oocyte-derived genome disappears. (Bottom) Then, an unmethylated island on the paternal allele is reformed by de novo methylation of flanking sequences by the midgestation stage; however, the extent of the DMR is distinct from that in the gametes. At a paternally methylated DMR, demethylation on the sperm-derived allele results in more extensive changes in DMR extent. At the midgestation stage, the DMR is seen as an unmethylated island on the maternal genome.
Recent publications have suggested several factors, such as transcription (Chotalia et al., 2009), H3K4 methylation status (Ooi et al., 2007; Ciccone et al., 2009) and CpG spacing (Jia et al., 2007), as being required for the establishment of maternal methylation imprints in oocytes by the DNA methyltransferase complex. Our methylation analysis revealed that most maternally methylated DMRs are flanked by regions that are methylated in both oocytes and sperm, and the DMRs can be seen as unmethylated islands in the sperm genome. This observation adds another possible component to the mechanism of formation of maternally methylated DMRs, suggesting that DMRs could be methylated by default and concomitant with flanking sequences in oocytes, but might have to be specifically protected from methylation in male germ cells. It was recently reported that CpG islands recruit the CXXC-domain protein Cfp1 (also known as Cxxc1), which determines their levels of H3K4me3 and protects them from CpG methylation (Thomson et al., 2010), and the H3K36me2 demethylase Kdm2a (Blackledge et al., 2010). By altering the histone modification profile at CpG islands, both factors could influence the interaction of the Dnmt3a-Dnmt3L complex (Dhayalan et al., 2010). It is possible that a similar mechanism might apply to unmethylated islands in sperm. Indeed, the CpG richness of the maternally methylated DMRs is close to that of CpG islands as defined by the regions bound to the CXXC domain of Cfp1 (Illingworth et al., 2010). By contrast, the paternally methylated H19 and the Rasgrf1 DMRs do not appear to be viewed as unmethylated islands in the gametes, suggesting that this model does not apply for these regions.
We also found that the methylation landscapes and the extents of gametic DMRs differ significantly from the DMRs as they exist in preimplantation and postimplantation embryos. The extent of the three paternally methylated DMRs, together with the additional DMRs at the H19 and Rasgrf1 loci, showed striking differences between the gametes and 12.5 dpc embryos. The maternally methylated DMRs also change, but vary less in size between the gametes and 12.5 dpc embryos. In blastocysts, the maternally methylated DMRs can no longer be viewed as unmethylated islands on the paternal allele owing to extensive loss of methylation in the peripheral regions and, in some cases, more internal regions, which also results in changes in the extent of the DMR. Thus, it appears that the DMRs, which are considered to be resistant to developmental reprogramming of methylation, are also affected, at least in part, by the genome-wide demethylation that occurs during preimplantation development (Reik et al., 2001; Morgan et al., 2005). In other words, the DMRs appear to be demethylated and remethylated to some extent during early embryonic development. The more prominent changes occurring at the paternally methylated DMRs might be linked to the difference in the mechanism of global demethylation between the oocyte- and sperm-derived genomes (Reik et al., 2001). After subsequent de novo methylation, which occurs by the midgestation stage, the DMRs appear again as unmethylated islands, but the extent of the maternally methylated DMRs is distinct from that in the gametes. It is notable that the paternally methylated DMRs can also be seen as unmethylated islands on the maternal allele at this stage.
Given how DMRs change during development, it is logical that the gametic DMR sequences themselves should be used to extract features that might dictate the establishment of methylation in the germline. However, our bioinformatic analyses of the gametic DMRs suggested that the sequence composition of the maternally methylated DMRs is indistinguishable from that of CpG islands. Similarly, the paternally methylated DMRs did not appear to have significant sequence properties that distinguish them from the remainder of the genome. It is possible that there might not be common sequence features required for imprint acquisition in the germline. Instead, it might be the ‘context’ of the DMRs that is important; for example, their presence within transcription units (Chotalia et al., 2009) or their histone modification state not driven primarily by underlying sequence. Alternatively, it might be that the gametic DMRs possess characteristic sequence features that could not be detected by our analyses which, in combination with other factors, could be important for imprint acquisition.
Finally, we observed abundant non-CpG methylation in oocytes, which contrasts with sperm in which non-CpG methylation was absent. Non-CpG methylation was previously reported at the Nf1 locus and the Mest DMR in oocytes (Haines et al., 2001; Imamura et al., 2005), but our study is far more extensive and provides the first solid evidence for the existence of abundant non-CpG methylation in vivo in mammals and suggests that it might be a general property of sequences undergoing CpG methylation. Previous studies showed that mouse and human ES cell lines contain abundant non-CpG methylation (Ramsahoye et al., 2000; Lister et al., 2009). Our data indicate that CpA is a preferred site for methylation in oocytes, consistent with the previous, albeit limited, observations; however, unlike human ES cells (Lister et al., 2009), mouse oocytes do not have a preference for CpHpG or, more specifically, CpApG. The non-CpG methylation in oocytes disappears after several rounds of DNA replication in preimplantation development, as its content in blastocysts is already close to that in postimplantation embryos. This is consistent with previous observations at the Nf1 locus and at the Mest DMR (Haines et al., 2001; Imamura et al., 2005) and is most likely because non-CpG methylation is not maintained through DNA replication during cleavage: it is known that the maintenance methyltransferase Dnmt1 is highly specific to CpG.
The previous report on the Nf1 gene (Haines et al., 2001) indicated that non-CpG methylation in oocytes is not limited to DMRs. It is noteworthy that our study on non-DMR regions, which were CpG-methylated in both oocytes and sperm, also identified non-CpG methylation specifically in oocytes. In addition, our re-examination of previous results from oocytes (Kaneda et al., 2010) also identified non-CpG methylation in retroelement sequences (data not shown). Thus, the existence of non-CpG methylation might be global in oocytes. We speculate that the high incidence of non-CpG methylation in oocytes and ES cells is a consequence of high expression of de novo methyltransferases (Okano et al., 1998; Vassena et al., 2005), as these enzymes act not only on CpG sites but also on CpA and CpT sites (Ramsahoye et al., 2000; Aoki et al., 2001). Non-CpG methylation introduced by these enzymes should accumulate in oocytes in particular, as they are non-replicating. Consistently high expression of de novo methyltransferases, or the expression of other factors that target these enzymes to DNA, might explain the presence of non-CpG methylation in ES cells despite replication.
In conclusion, our study illustrates for the first time a detailed picture of the DMRs in the gametes and reveals the dynamic developmental changes that occur in methylation, underscoring the importance of using gametic DMR sequences for the study of imprint establishment. This study provides a basis for understanding the mechanism of imprint establishment in the germline.
Supplementary Material
Acknowledgements
We thank T. Sado and K. Ichiyanagi for helpful discussion; H. Horiike for oocyte collection; S. Smallwood for providing information on non-DMR methylated CpG islands prior to publication; H. Furuumi and K. Sakai for mouse maintenance; J. Kitayama and K. Takada for technical assistance; T. Horiike for computational analysis; and all members of the H.S. laboratory for discussion and support. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Area from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.S., and by a grant from the United Kingdom Medical Research Council to G.K. Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.061416/-/DC1
References
- Aoki A., Suetake I., Miyagawa J., Fujio T., Chijiwa T., Sasaki H., Tajima S. (2001). Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 29, 3506-3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N. P., Zhou J. C., Tolstorukov M. Y., Farcas A. M., Park P. J., Klose R. J. (2010). CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell 38, 179-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D., Bestor T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96-99 [DOI] [PubMed] [Google Scholar]
- Brandeis M., Kafri T., Ariel M., Chaillet J. R., McCarrey J., Razin A., Cedar H. (1993). The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 12, 3669-3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotalia M., Smallwood S. A., Ruf N., Dawson C., Lucifero D., Frontera M., James K., Dean W., Kelsey G. (2009). Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 23, 105-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone D. N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T. (2009). KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415-418 [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R. Z., Ragozin S., Jeltsch A. (2010). The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114-26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith A. C., Greally J. M. (2007). Epigenetics: perceptive enzymes. Nature 449, 148-149 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith A. C., Sasaki H., Cattanach B. M., Surani M. A. (1993). Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751-755 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick G. V., Soloway P. D., Higgins M. J. (2002). Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32, 426-431 [DOI] [PubMed] [Google Scholar]
- Greally J. M. (2002). Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. USA 99, 327-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines T. R., Rodenhiser D. I., Ainsworth P. J. (2001). Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev. Biol. 240, 585-598 [DOI] [PubMed] [Google Scholar]
- Hata K., Okano M., Lei H., Li E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983-1993 [DOI] [PubMed] [Google Scholar]
- Hetzl J., Foerster A. M., Raidl G., Mittelsten Scheid O. (2007). CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51, 526-536 [DOI] [PubMed] [Google Scholar]
- Hirasawa R., Chiba H., Kaneda M., Tajima S., Li E., Jaenisch R., Sasaki H. (2008). Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 22, 1607-1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H., Obata Y., Komiyama J., Shirai M., Kono T. (2006). Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells 11, 353-361 [DOI] [PubMed] [Google Scholar]
- Hutter B., Helms V., Paulsen M. (2006). Tandem repeats in the CpG islands of imprinted genes. Genomics 88, 323-332 [DOI] [PubMed] [Google Scholar]
- Illingworth R. S., Gruenewald-Schneider U., Webb S., Kerr A. R., James K. D., Turner D. J., Smith C., Harrison D. J., Andrews R., Bird A. P. (2010). Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 6, e1001134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T., Kerjean A., Heams T., Kupiec J. J., Thenevin C., Paldi A. (2005). Dynamic CpG and non-CpG methylation of the Peg1/Mest gene in the mouse oocyte and preimplantation embryo. J. Biol. Chem. 280, 20171-20175 [DOI] [PubMed] [Google Scholar]
- Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. (2007). Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900-903 [DOI] [PubMed] [Google Scholar]
- Kaneda M., Hirasawa R., Chiba H., Okano M., Li E., Sasaki H. (2010). Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells 15, 169-179 [DOI] [PubMed] [Google Scholar]
- Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. (2007). Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 16, 2272-2280 [DOI] [PubMed] [Google Scholar]
- Ke X., Thomas N. S., Robinson D. O., Collins A. (2002). The distinguishing sequence characteristics of mouse imprinted genes. Mamm. Genome 13, 639-645 [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Suda C., Abe T., Kohara Y., Ikemura T., Sasaki H. (2006). Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 113, 130-137 [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Yamada K., Morita S., Hiura H., Fukuda A., Kagami M., Ogata T., Hata K., Sotomaru Y., Kono T. (2009). Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics 93, 461-472 [DOI] [PubMed] [Google Scholar]
- Koide T., Moriwaki K., Uchida K., Mita A., Sagai T., Yonekawa H., Katoh H., Miyashita N., Tsuchiya K., Nielsen T. J., et al. (1998). A new inbred strain JF1 established from Japanese fancy mouse carrying the classic piebald allele. Mamm. Genome 9, 15-19 [DOI] [PubMed] [Google Scholar]
- Lin S. P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J., Ferguson-Smith A. C. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97-102 [DOI] [PubMed] [Google Scholar]
- Lindroth A. M., Park Y. J., McLean C. M., Dokshin G. A., Persson J. M., Herman H., Pasini D., Miro X., Donohoe M. E., Lee J. T., et al. (2008). Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 4, e1000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H. D., Santos F., Green K., Dean W., Reik W. (2005). Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14 Spec No 1, R47-R58 [DOI] [PubMed] [Google Scholar]
- Neumann B., Kubicka P., Barlow D. P. (1995). Characteristics of imprinted genes. Nat. Genet. 9, 12-13 [DOI] [PubMed] [Google Scholar]
- Okano M., Xie S., Li E. (1998). Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219-220 [DOI] [PubMed] [Google Scholar]
- Ooi S. K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S. P., Allis C. D., et al. (2007). DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714-717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye B. H., Biniszkiewicz D., Lyko F., Clark V., Bird A. P., Jaenisch R. (2000). Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 97, 5237-5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Walter J. (2001). Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21-32 [DOI] [PubMed] [Google Scholar]
- Reik W., Dean W., Walter J. (2001). Epigenetic reprogramming in mammalian development. Science 293, 1089-1093 [DOI] [PubMed] [Google Scholar]
- Reinhart B., Paoloni-Giacobino A., Chaillet J. R. (2006). Specific differentially methylated domain sequences direct the maintenance of methylation at imprinted genes. Mol. Cell. Biol. 26, 8347-8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde C., Zhang Y., Reinhardt R., Jeltsch A. (2010). BISMA-fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics 11, 230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Matsui Y. (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129-140 [DOI] [PubMed] [Google Scholar]
- Shiura H., Nakamura K., Hikichi T., Hino T., Oda K., Suzuki-Migishima R., Kohda T., Kaneko-ishino T., Ishino F. (2009). Paternal deletion of Meg1/Grb10 DMR causes maternalization of the Meg1/Grb10 cluster in mouse proximal chromosome 11 leading to severe pre- and postnatal growth retardation. Hum. Mol. Genet. 18, 1424-1438 [DOI] [PubMed] [Google Scholar]
- Thomson J. P., Skene P. J., Selfridge J., Clouaire T., Guy J., Webb S., Kerr A. R., Deaton A., Andrews R., James K. D., et al. (2010). CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464, 1082-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen J. L., Duran K. L., Bartolomei M. S. (1998). Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12, 3693-3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay K. D., Saam J. R., Ingram R. S., Tilghman S. M., Bartolomei M. S. (1995). A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9, 407-413 [DOI] [PubMed] [Google Scholar]
- Tremblay K. D., Duran K. L., Bartolomei M. S. (1997). A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17, 4322-4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R., Dee Schramm R., Latham K. E. (2005). Species-dependent expression patterns of DNA methyltransferase genes in mammalian oocytes and preimplantation embryos. Mol. Reprod. Dev. 72, 430-436 [DOI] [PubMed] [Google Scholar]
- Walter J., Hutter B., Khare T., Paulsen M. (2006). Repetitive elements in imprinted genes. Cytogenet. Genome Res. 113, 109-115 [DOI] [PubMed] [Google Scholar]
- Wienholz B. L., Kareta M. S., Moarefi A. H., Gordon C. A., Ginno P. A., Chedin F. (2010). DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 6, e1001106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson C. M., Ball S. T., Nottingham W. T., Skinner J. A., Plagge A., Turner M. D., Powles N., Hough T., Papworth D., Fraser W. D., et al. (2004). A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat. Genet. 36, 894-899 [DOI] [PubMed] [Google Scholar]
- Wutz A., Smrzka O. W., Schweifer N., Schellander K., Wagner E. F., Barlow D. P. (1997). Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389, 745-749 [DOI] [PubMed] [Google Scholar]
- Yoon B. J., Herman H., Sikora A., Smith L. T., Plass C., Soloway P. D. (2002). Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30, 92-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.