Abstract
The Cajal body (CB) is an evolutionarily conserved nuclear subcompartment, enriched in components of the RNA processing machinery. The composition and dynamics of CBs in cells of living organisms is not well understood. Here we establish the zebrafish embryo as a model system to investigate the properties of CBs during rapid growth and cell division, taking advantage of the ease of live-cell imaging. We show that zebrafish embryo CBs contain coilin and multiple components of the pre-mRNA splicing machinery. Histone mRNA 3′ end processing factors, present in CBs in some systems, were instead concentrated in a distinct nuclear body. CBs were present in embryos before and after activation of zygotic gene expression, indicating a maternal contribution of CB components. During the first 24 hours of development, embryonic cells displayed up to 30 CBs per nucleus; these dispersed prior to mitosis and reassembled within minutes upon daughter cell nucleus formation. Following zygotic genome activation, snRNP biogenesis was required for CB assembly and maintenance, suggesting a self-assembly process that determines CB numbers in embryos. Differentiation into muscle, neurons and epidermis was associated with the achievement of a steady state number of 2 CBs per nucleus. We propose that CB number is regulated during development to respond to the demands of gene expression in a rapidly growing embryo.
Key words: Cajal body, zygotic gene expression, pre-mRNA splicing, snRNA, scaRNA, snRNP
Introduction
The Cajal body (CB) is a nuclear compartment initially described by Ramon y Cajal in silver-stained sections of vertebrate cerebral cortex.1 Like other cellular inclusions, e.g., nucleoli, polar granules, stress granules and PML bodies, the CB is a discrete body that is not enclosed by a lipid bilayer. Because the CB is uniquely marked by the protein coilin,2–7 it has been possible to characterize the components of CBs in multiple species at the molecular level. Numerous factors essential for pre-mRNA splicing, histone mRNA 3′-end processing, telomere maintenance and rRNA processing are concentrated in CBs, suggesting a role for CBs in RNP-dependent processes.8–10 Because both the coilin protein and the CB as a morphological entity are well conserved in evolution, CBs have served as a general model for the study of the assembly and function of subnuclear compartments. Two basic questions about CBs warrant further investigation. First, are CBs universal nuclear compartments with a defined composition?11 Second, how is CB number regulated in the somatic cells of living organisms?
Much of what is known about the molecular composition of CBs comes from immortalized, often aneuploid tissue culture cells, on the one hand, and from the CB-like “spheres” of amphibian germinal vesicles (GVs), on the other. It is clear that neither system provides an accurate or general view of CBs in diploid somatic nuclei. For example, the Xenopus GV contains up to 100 CBs ranging in diameter from 1–10 µm, whereas nuclei of mammalian, plant and Drosophila cells contain on average 1–2 CBs that range in diameter from 0.5–1 µm.1,12,13 While CBs in all of these organisms contain components of the pre-mRNA splicing machinery, other machineries vary in CB localization. For example, CBs in the GV are present at histone loci and contain massive amounts of the U7 snRNP, a factor required for histone mRNA 3′ end formation.14–16 CBs in mammalian tissue culture cells also contain the U7 snRNP and partially colocalize with histone gene loci.1,17 Both observations suggest that CBs may deliver vital RNA processing machinery to the sites of histone mRNA synthesis. However, recent studies in Drosophila showed that the U7 snRNP is not concentrated in CBs but rather in a distinct nuclear body named the Histone Locus Body (HLB), due to its precise colocalization with the histone gene locus.13 Similar observations in human ES cells suggests that the presence of this particular machinery in the CBs of transformed human cells may reflect the cancerous state.10,18,19 These disparate observations raise the question of whether the RNA processing machineries previously assigned to CBs in selected systems are generally present in all cells and organisms.
CB components exchange rapidly with nucleoplasm, where they engage in numerous molecular processes.20,21 CBs in most somatic cells can move, split and arise de novo within nucleoplasm.12,22 Their size and number vary throughout the cell cycle and can be influenced by post-translational modifications of the coilin protein.23–27 This dynamic behavior suggests that CBs respond to the physiological requirements of living cells; the system that provides such a variety of changing cellular environments is a developing multicellular organism. Differences in CB numbers between various cell types in the context of an organism have been reported for Arabidopsis and Drosophila. In Arabidopsis, the number of CBs per nucleus was shown to depend on cell type and developmental stage, similar to nurse cells in Drosophila; however, somatic cells in Drosophila appear to have a constant number of 1 CB per nucleus.12,13 It is still an open question whether this is the case for all animal systems or if, alternatively, CB numbers vary during embryogenesis and/or cellular differentiation.
Here, we take advantage of the rapid, external development of the zebrafish embryo to investigate the CB. Because the embryo is amenable to imaging from fertilization onwards, this model system permits a dynamic description and detailed analysis of CBs in a living vertebrate system. In all species examined to date, the spliceosomal snRNPs—essential components of the premRNA splicing machinery—are concentrated in CBs.1 Indeed, prior to the identification of coilin in some species, snRNPs were considered bona fide markers of CBs.12,13 Therefore, we focus on zebrafish coilin, the spliceosomal snRNPs and the U7 snRNP. Our study reveals the molecular composition and dynamic characteristics of CBs during vertebrate embryogenesis and establishes the zebrafish system for future exploration of the role of nuclear architecture in a living animal.
Results
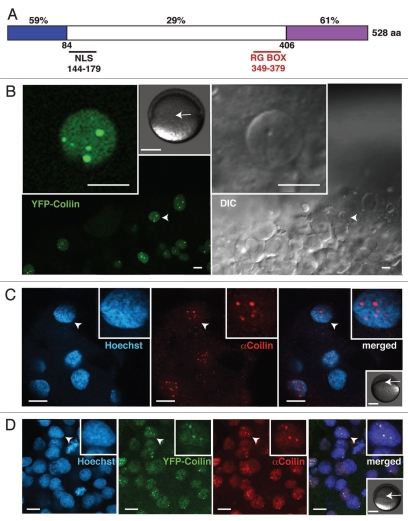
Zebrafish coilin is 34% identical to the human protein (Fig. 1A), with conserved N- and C-termini that have been shown to mediate coilin-coilin self-association and Sm-protein binding, respectively.5,28,29 In vivo imaging performed after injection of mRNA encoding YFP-tagged zebrafish coilin revealed YFP-positive nuclear foci in the early zebrafish embryo (Fig. 1B). We also raised an antibody reactive with zebrafish coilin (Fig. S1). Whole-mount immunohistochemistry revealed nuclear foci similar in size, number and distribution to those observed in vivo (Figs. 1C and S2). When combined, the two methods detected the same coilin-positive nuclear compartment (Fig. 1D). This indicates that zebrafish coilin is concentrated in nuclear foci in the embryo, and that our anti-serum recognizes coilin in situ.
Figure 1.
Detection of zebrafish coilin in nuclear bodies. (A) Schematic representation of zebrafish coilin indicating the percentage identity to human coilin, the position of the nuclear localization signal (NLS) and the arginine-glycine-rich (RG) box. Coilin self-association occurs via the N-terminal conserved domain (blue), whereas binding of Sm proteins has been mapped to the C-terminal domain (purple). (B) Expression of YFP-tagged zebrafish coilin following injection of mRNA into 1-cell embryos assayed by live-cell confocal imaging at early gastrula stage (5.5 hpf, hours post fertilization), single confocal section in YFP channel and corresponding DIC images. (C) Labeling of fixed late blastula stage embryos (4.5 hpf) with anti-zebrafish coilin (AB 9EA2, Fig. S1), DNA counterstained with Hoechst. (D) Colocalization of endogenous coilin with YFP-tagged zebrafish coilin expressed from in vitro transcribed and injected mRNA is shown by immunostaining of fixed embryos at early gastrula stage (5.5 hpf). Single confocal sections are shown, and arrowheads indicate nuclei magnified in the insets (two-fold magnification in C and D). Scale bars: 10 µm. Additional grayscale insets in (B–D) show embryonic stage at the time of imaging, and the approximate imaged areas (arrows). Scale bars: 250 µm.
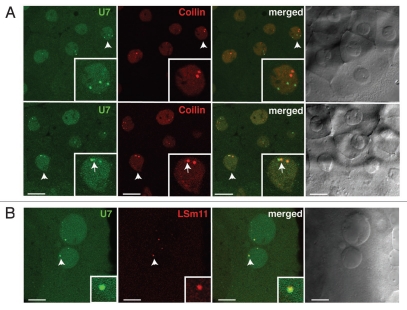
In human tissue culture cells and the GVs of Xenopus laevis, coilin-positive CBs are enriched in components of the histone mRNA 3′ end processing machinery, particularly the U7 snRNP.16,17,30 However, in Drosophila melanogaster embryos, this machinery is instead concentrated in a separate compartment termed the Histone Locus Body (HLB), which is coincident with the histone gene cluster.13 To determine whether coilin-positive nuclear bodies in zebrafish embryos contain the U7 snRNP, in vitro transcribed Alexa488-U7 snRNA and mRNA encoding mRFP-Lsm11 (a U7 snRNP-specific protein) were injected into 1-cell embryos and imaged at 3 hours post-fertilization (hpf). U7 snRNA was detected in nuclear bodies distinct from those marked by mRFP-coilin (Fig. 2A). In some cases, low levels of mRFP-coilin were detected in U7-positive bodies, which were sometimes adjacent to major sites of coilin concentration. In contrast, U7 snRNA always colocalized strongly with mRFP-Lsm11 protein (Fig. 2B), indicating that U7 snRNP is concentrated in nuclear bodies distinct from the most intense coilin-positive nuclear bodies. We conclude that, as in Drosophila, zebrafish nuclei contain distinct nuclear compartments enriched in the histone mRNA 3′ end processing machinery.
Figure 2.
Zebrafish nuclear bodies enriched in U7 snRNP are deficient in coilin. Embryos were injected with mRNAs encoding either mRFP-coilin (A) or mRFP-LSm11 (B) and fluorescently labeled U7 snRNA (A and B). Representative confocal sections taken at the blastula stage are shown. Insets show two-fold magnifications of the nuclei (A) or three-fold magnifications of the foci (B) marked by arrowheads. Arrows point to the adjacent coilin- and U7 snRNA-positive foci. Scale bars: 10 µm.
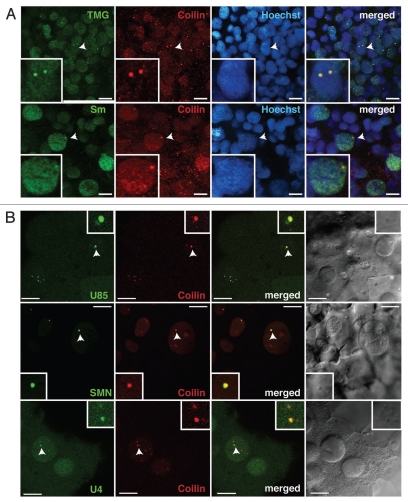
To deduce whether coilin-positive nuclear bodies in zebrafish embryos represent bona fide CBs, the presence of components of the pre-mRNA splicing machinery was tested. First, double immunostaining revealed that shared epitopes on spliceosomal snRNPs (namely, Sm proteins and the tri-methylguanosine or TMG cap) are concentrated in coilin-positive nuclear bodies (Fig. 3A). Second, additional representative factors—the U4 snRNA, U85 scaRNA and SMN protein—were visualized following RNA injection in living zebrafish embryos together with mRNA encoding mRFP-coilin. Figure 3B shows that all three components, characteristic of CBs in all systems studied,1,31,32 were highly concentrated in coilin-positive nuclear bodies in the zebrafish embryo. Consistent colocalization was observed at all stages of development and embryonic regions examined. Taken together, the colocalization of coilin with endogenous snRNP proteins and TMG caps, SMN, U4 snRNA and U85 scaRNA confirms the identity of the coilin-positive nuclear bodies as the zebrafish counterparts of CBs described in other organisms.
Figure 3.
Spliceosomal snRNPs and scaRNAs are CB components in zebrafish embryos. (A) Embryos were fixed at the onset of segmentation (∼10 hpf) and double-stained with antibodies specific for coilin (red) and either tri-methylguanosine cap (TMG) or Sm proteins (green) present on spliceosomal snRNPs. Insets show two-fold magnification of the nucleus indicated with arrowheads. (B) Embryos were injected with either mRNAs encoding mRFP-coilin (red) and SMN-CFP (green) or mRFP-coilin (red) and Alexa488-labeled U85 scaRNA or U4 snRNA, as indicated in the green channel. Representative confocal sections for various stages of development are shown: blastula (upper), segmentation (middle), gastrulation (bottom). Insets show three-fold magnification of the CBs marked by arrowheads. Scale bars: 10 µm.
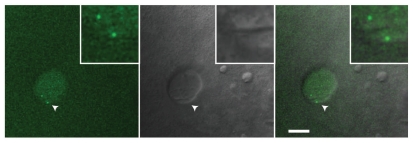
How early in embryogenesis are CBs detectable? Are CBs maternally provided and maintained in embryonic cells before the onset of zygotic gene transcription? Zygotic transcription starts at the 512-cell stage at ∼2.75 hours post-fertilization (hpf),33 and numerous coilin-positive nuclear foci could be observed by immunostaining from the 8-cell (∼1.25 hpf) to 512-cell stage, indicating that zygotic gene transcription is not necessary for the assembly or maintenance of CBs (Fig. S2). The difficulty of immunostaining in early embryos prevented unequivocal identification of nuclear substructures before the 8-cell stage. However, the clear presence of coilin immunostaining even at the 2-cell stage (Fig. S2) suggests a maternal contribution of coilin protein and perhaps CBs. Therefore, we turned to in vivo imaging of early embryos, taking advantage of injected, fluorescently labeled U4 snRNA as a marker of CBs. It was previously shown in tissue culture cells and amphibian GVs that fluorescently labeled U4 snRNA rapidly assembles with Sm proteins, leading to nuclear import and CB localization within 30 minutes.32,34 Remarkably, Alexa-488 U4 snRNA was detectable in discrete foci in 4-cell embryonic nuclei (∼1 hpf), even though endogenous snRNAs are not yet being transcribed (Fig. 4). U4 snRNA is likely able to assemble with maternally contributed Sm proteins.35 We conclude that CBs containing coilin and capable of concentrating U4 snRNPs are present at least from the 4-cell stage onward, indicating that CB components are maternally deposited and that newly assembled zygotic components are able to integrate into CBs.
Figure 4.
U4 snRNA positive CBs are present in the nuclei of 4-cell embryos. Embryos were injected at the 1-cell stage with 2.5 femtomole of Alexa-488-labeled, in vitro synthesized U4 snRNA. This embryo was allowed to develop for 1 hour at 28°C and mounted for in vivo imaging in 2% methylcellulose. A single confocal section is shown. Arrowheads indicate regions magnified three-fold in the insets. Scale bar: 10 µm.
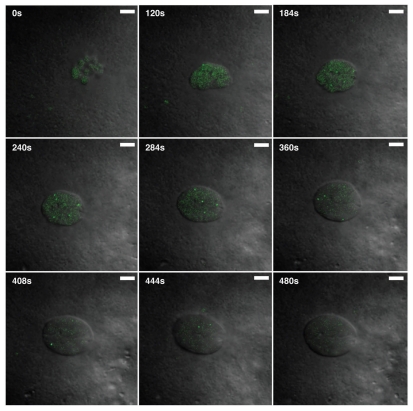
The cells of the early zebrafish embryo divide synchronously every 15 minutes. Studies in tissue culture cells indicate that CBs disperse at mitosis and reform after cell division, taking ∼20 minutes for reassembly.36 If these reassembly kinetics were similar in zebrafish embryos, it seems unlikely that CBs would be observed at all. To determine whether CBs of the zebrafish embryo disperse at mitosis, time-lapse microscopy was performed on embryos injected with Alexa488-labeled U4 snRNA to mark CBs and DIC imaging to detect the assembly and function of the mitotic spindle. Figure 5 shows still images from a movie (Suppl. material), focusing on a daughter cell at the 32-cell stage shortly after cytokinesis; the nucleus has not yet completely reformed, and U4 snRNA is distributed evenly throughout the mass of chromatin. After ∼3 minutes, U4 snRNA is visible at higher concentrations in larger round bodies, consistent with CBs. These CBs move extensively throughout the interphase nucleus, becoming progressively more prominent. Toward the end of interphase, CBs disappear and the U4 signal disperses throughout the nucleoplasm before mitosis. This analysis shows that zebrafish embryonic CBs are highly dynamic, disassemble at mitosis, and reform within minutes thereafter. Thus, the time-scale for CB assembly is not fixed at ∼20 min, but may be subject to regulation in systems with differing temporal requirements.
Figure 5.
Rapid assembly and disassembly of CBs at mitosis. Embryos injected with Alexa488-labeled U4 snRNA were subjected to live cell imaging at the 32-cell stage. Still images of a time-lapse movie (see Suppl. Material) show that U4 snRNA concentrates in nuclear bodies in the early blastula when cell cycles are only ∼15 minutes long. U4-positive nuclear bodies disperse immediately prior to mitosis (after frame: 444 s) and reform in daughter cells within 3 minutes (frame: 184 s). Scale bar: 5 µm.
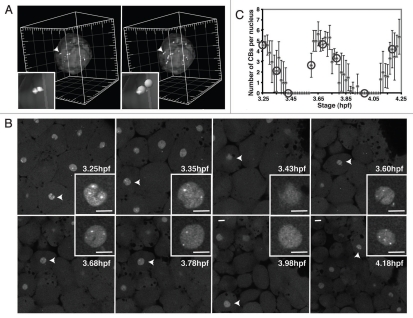
We sought to validate and quantify CB dynamics detected before and after mitosis. To this end, we used 3D rendering of imaged nuclei labeled with YFP-coilin, followed by automatic detection and counting in order to determine the numbers of CBs in embryonic zebrafish nuclei (Fig. 6A). CBs were defined by a size threshold of 0.5 µm and an intensity threshold of threefold above the average nucleoplasmic intensity (see Materials and Methods for details). Live imaging of individual cells dividing every ∼30 minutes confirmed the rapid dispersal of CBs at mitosis and reformation of up to 5 CBs within ∼10 minutes (Fig. 6B and C). Interestingly, mother and daughter cells had similar numbers of CBs. A two-tailed Student's t-test revealed that differences in the peak average number of CBs per nucleus measured at different time points were not significant (p = 0.22 and p = 0.47 for maximal CB numbers per cell in the first peak versus the second peak and the second peak versus the third peak, respectively). This indicates a remarkable control of CB number before and after cell division.
Figure 6.
Regulation of CB number during the cell cycle. (A) Two perspectives of a representative volume-rendered nucleus containing CBs marked by YFP-coilin for quantitation of CB number. Arrowheads point to region magnified in the inset. Major tick: 5 µm. (B) Time series of YFP-coilin expressing embryo from time-lapse confocal imaging. Embryos expressing YFP-coilin imaged at 28°C from 3.25 to 4.25 hpf, confocal sections 1.5 µm apart collected every 50 seconds. Arrowheads: nuclei in the insets. (c) Plot of CB number per nucleus (mean ± SD) from the time-lapse movie shown in (B), quantified as in (A). Each point represents an average number of CBs in the nuclei (N = 4–10 nuclei) present in the imaged volume for a particular time point. No CBs were detected in mitosis. Circles mark the time points shown in (B). Note that single confocal sections (B) do not show all the CBs from each respective nucleus (A). The maximal number of CBs per nucleus in the consecutive interphases is not statistically different (p = 0.22 for the first two interphases, and p = 0.47 for the second and third interphase). Scale bar: 10 µm.
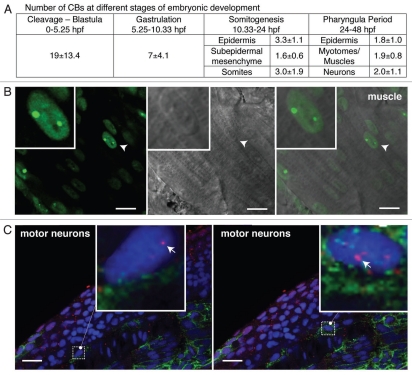
To further explore the regulation of CB number throughout zebrafish embryogenesis, we applied the same imaging and counting approach to different developmental stages and tissues from 0–48 hpf. At cleavage and blastula stages—before YFP-coilin (∼3 hpf) was expressed—anti-coilin immunostaining was used to detect and count CBs. Following translation and maturation of injected YFP-coilin mRNA (after ∼3.5 hpf), both methods were used. The average number of CBs per nucleus in early blastula stages was strikingly high (19 ± 13 per nucleus); we assume that variation in these values is due to the short cell cycle times and is likely skewed to smaller numbers by the analysis of datasets in which cells are approaching or have just finished mitosis. CB number gradually decreased as development progressed (Fig. 7A), such that nuclei of muscle and neuronal cells in embryos at 30 hpf contain on average 2 CBs (Fig. 7B and C). Interestingly, CBs were maintained at 2 per nucleus from early somitogenesis until 48 hours (when the experiment was terminated), suggesting CB number reaches a steady state. Thus, although daughter cells tended to have a similar number of CBs to the mother cell, an overall decrease in CB number is observed during the first 10 hours of development, in which thousands of cells are produced and the zygotic genome is activated.
Figure 7.
Regulation of CB number during development and differentiation. (A) Number of CBs per nucleus at different stages of embryonic development, mean ± SD (N = 3–9 independent experiments). (B and C) Single confocal sections of striated muscle and motor neurons in live zebrafish embryos at 30 hpf injected with YFP-coilin mRNA (B) or fixed and labeled with an antibody against zebrafish coilin (9EA2; red) and neurolin (zn-8; green) a cell surface molecule on secondary motor neurons (C); hoechst counterstained nuclei are shown in blue. Arrowheads in (B) point to the nuclei magnified in the insets. Magnification is three-fold in (B) and seven-fold in (C). Arrows in the insets point to selected CBs. Scale bars: 10 µm in (B) and 20 µm in (c).
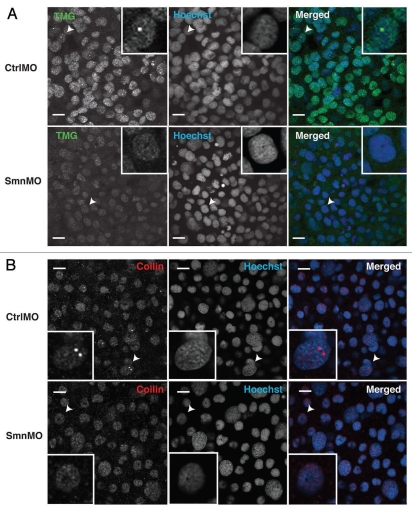
The dramatic decrease in the number of CBs per nucleus during embryogenesis might reflect a dilution of maternal stores. Yet, the observation of CB disassembly and reassembly at mitosis indicates the involvement of an active process. To address the possibility that ongoing snRNP biogenesis contributes to CB number in the embryo, we sought to disrupt snRNP biogenesis. SMN protein is required for the assembly of Sm rings on spliceosomal snRNAs; subsequently 2,2′-methylation of the 7-methylguanosine cap generates tri-methylguanosine (TMG) at the 5′ end of snRNAs, and nuclear import occurs.37–39 In mammalian tissue culture cells, SMN knockdown reduces levels of Sm ring assembly and leads to coilin protein dispersal.40,41 SMN depletion with translation-blocking morpholinos (SmnMO) has been previously accomplished in zebrafish, establishing an animal model for spinal muscular atrophy;42,43 however, these previous studies in zebrafish did not examine nuclear morphology. To determine whether SMN depletion in embryos affected CB number, SmnMO or CtrlMO was injected into 1-cell embryos, which were left to develop for 10 hours at 28°C. Immunostaining and immunoblotting verified the loss of SMN protein in SmnMO but not CtrlMO injected embryos (Fig. S3). The resulting loss of anti-TMG immunoreactivity in cells of the embryos injected with SmnMO indicates a robust defect in Sm ring formation on snRNAs and confirms the expected effect on snRNP biogenesis (Fig. 8A). Strikingly, SmnMO injection nearly abolished TMG- and coilin-positive nuclear foci (Fig. 8A and B): quantifying CB numbers in single confocal sections, 50% +/− 11% of nuclei had CBs in CtrlMO-injected embryos, whereas only 9% +/− 2% of nuclei had CBs following SMN depletion. We conclude that SMN protein and active snRNP biogenesis is required for CB integrity in zebrafish embryos.
Figure 8.
Depletion of SMN leads to dispersal of embryonic CBs. Zebrafish embryos were injected with control morpholino (CtrlMO) or morpholino targeting the 5′-UTR of Smn mRNA (SmnMO) and fixed at the onset of segmentation (∼10 hpf). Reduction in snRNA trimethylation is evident from the overall loss of signal in samples immunostained with anti-TMG (a). Loss of snRNP-containing CBs is also shown by immunostaining with anti-TMG (A) as well as anti-coilin (B). Single confocal sections are shown, and arrowheads indicate nuclei magnified three-fold in the insets. Scale bars: 10 µm.
Discussion
The present study establishes the zebrafish, Danio rerio, as a powerful model system for the field of nuclear architecture. The embryo's accessibility to imaging and morpholino knockdown of key proteins provides opportunities for analysis of nuclear organization before and after activation of the zygotic genome. Here we show that the nuclei of the zebrafish embryo contain numerous Cajal bodies (CBs) of the expected composition, which display rapid assembly and disassembly dynamics at mitosis as well as specific changes in number according to developmental stage. The application of additional tools, such as injection of fluorescently labeled in vitro transcribed RNAs, and 3D reconstruction of nuclei facilitated several key findings. First, CBs in the zebrafish do not contain the histone mRNA 3′ end processing machinery, which is instead concentrated in distinct nuclear bodies. Second, CB number per nucleus is highly variable during embryogenesis, asymptotically approaching two per nucleus in differentiated cells. Third, CBs in zebrafish embryonic cells with extremely short cell cycle times (15 minutes) are highly dynamic, dispersing and reassembling at each mitosis. The conservation of CB numbers before and after cell division suggests that an active process regulates CB assembly and/or maintenance. We provide evidence that snRNP biogenesis is required for CB integrity in the embryo.
We initially identified the zebrafish CB by immunofluorescence with an antiserum specific for zebrafish coilin, as well as through expression of injected mRNAs encoding fluorescently tagged versions of zebrafish coilin. These coilin-positive bodies, appropriately sized in the range of 0.5–1 µm in diameter, were shown to contain numerous components of the splicing machinery, including Sm proteins and the tri-methylguanosine cap on snRNAs. In addition, fluorescently labeled small RNAs, namely U4 snRNA and U85 scaRNA, concentrated in the CBs of living embryonic cells when injected into 1-cell stage embryos. Finally, the SMN protein, visualized by injection of mRNA encoding fluorescently-tagged SMN, was also concentrated in the CBs of embryonic cells. In contrast, components of the U7 snRNP involved in histone mRNA 3′ end processing, namely U7 snRNA and the Lsm11 protein, were concentrated in a distinct nuclear body reminiscent of the Histone Locus Body observed in Drosophila cells.13 The specific concentration of coilin with components of the splicing machinery definitively identifies CBs in zebrafish embryos for the first time.
It is currently thought that CBs play a role in the assembly of spliceosomal snRNPs. Each snRNP contains a 100–200 nucleotide-long snRNA that is extensively modified post-transcriptionally and assembled with numerous snRNP-specific proteins.9,31 After transcription, the U1, U2, U4 and U5 snRNAs are exported to the cytoplasm, where a hetero-heptameric ring of Sm proteins is loaded onto each snRNA by the SMN complex and the 5′ end is hypermethylated.37,44 Subsequently, these still immature snRNPs are re-imported to the nucleus together with SMN.45,46 ScaRNAs, such as U85 studied here, then guide site-specific modification of the snRNAs in CBs.47,48 Importantly, transient intermediates in final assembly steps involving RNA structural rearrangements and association of additional snRNP-specific proteins are concentrated in CBs.49–51 During splicing, snRNPs are partially disassembled, and the final assembly steps leading to regeneration of active snRNPs also occur in CBs.52 These latter steps in snRNP assembly are predicted to occur approximately 10-fold more quickly in cells that have four CBs/nucleus—the average number detected in HeLa cell nuclei—due to the increased likelihood of collisions between the components.32 Model nuclei with fewer CBs were predicted to support slower rates of snRNP assembly, providing a framework for considering the significance of CB number.
The striking change in the number of CBs per nucleus observed during the course of development suggests that the physiological requirement for CBs in embryonic cells varies. In the early blastula, numerous CBs were observed (∼19/cell) despite extremely short cell cycle times (15 minutes), which require disassembly and reassembly of CBs within minutes. This was surprising, because pre-mRNA splicing per se is not required until ∼4.5 hpf, after the onset of zygotic transcription.53 Indeed, mammalian CBs only appear after zygotic transcription begins, and are disrupted when transcription is blocked in tissue culture cells.54–56 However, unassembled snRNP components are abundant in the oocyte nuclei of insects and amphibians,1,35 suggesting that CB components are maternally supplied and that CBs may arise in the early embryo to support ongoing snRNP biogenesis even in the absence of zygotic transcription. Consistent with this, CBs were present in zebrafish embryonic nuclei at the 4-cell stage (see Fig. 4), well before the 512-cell stage when zygotic gene activation occurs. These observations point to species-specific differences in the assembly of the gene expression machinery.
One possibility is that at least some of the CBs in the developing embryo are maternally contributed and diluted out over progressive cell divisions. However, CB integrity in the late gastrula was dependent on SMN and ongoing snRNP biogenesis, because depletion of the SMN protein, required for assembly of the Sm ring on snRNAs, led to coilin dispersal. This is consistent with prior observations in tissue culture cells.40,41 In the embryo, this observation demonstrates that, although maternally contributed CBs and/or CB components may persist in the blastula and early gastrula, they do not account for the CBs present at later stages, after transcription of the zygotic genome is activated. Prior studies indicate that coilin is required for CB assembly and maintenance,57,58 yet even though coilin is still present in the SMN morphants, it is not sufficient for CB assembly. Instead, SMN and snRNP biogenesis in the embryo clearly contributes to CB morphology, likely by providing multivalent binding partners for the coilin protein that can lead to a network of low affinity interactions necessary for CB formation.9,28,29 It is therefore likely that the profile of CB number in the first 48 hours of zebrafish development represents assembly from a combination of maternal stores and newly synthesized CB components.
A correlation between cell metabolic activity and CB number has been suggested.1,56,59 High metabolic activity can be associated with either accelerated cell division or cell growth, which may not be strictly linked. In zebrafish embryos, the cells with the highest CB numbers have the shortest cell cycles. Because cells become smaller during early cell divisions in the embryo,33 it follows that CB number correlates directly with the cell size. This is in agreement with an interesting study reporting that neuronal cell size correlates with CB number.60 However, neurons are postmitotic and may attain a homeostatic state, relating metabolism to CB number. In contrast, the cells of the early zebrafish embryo divide rapidly; CBs disassemble at mitosis and reform quickly in the nuclei of daughter cells, and the number of CBs generated in consecutive generations is statistically identical to the number of CBs present in the mother cells (see Fig. 6). Prior studies link dispersal of CBs at mitosis with coilin hyper-phosphorylation, and phosphorylation of residues at the coilin C-terminus can control CB number.23,27,61 Interestingly, we observed a doublet of coilin in zebrafish embryos, with the upper—presumably hyperphosphorylated band—being most abundant (Fig. S1); the prevalence of this high molecular weight form of coilin in embryos is consistent with the high frequency of cell divisions.23 The maintenance of a constant number of CBs per nucleus after cell division might suggest a correspondence between DNA content and CB number. However, given that the numbers of CBs do not generally correlate with cell ploidy and that SMN morphants lack CBs altogether, it seems that DNA content is not the only determinant of CB number.62
As cell division cycles proceed and the zygotic genome is activated, the average CB number decreases, asymptotically approaching two per nucleus upon cellular differentiation. Similar to the zebrafish, differentiated cells in developing Arabidopsis, Drosophila and human fetal tissues also tend to have one to two CBs per nucleus,12,13,25 suggesting that this is the optimal number of CBs for differentiated cells. We speculate that CB number may reflect general transcription and splicing levels at these later stages. Based on the modeling results correlating the rate of spliceosomal snRNP assembly with CB number (see above), it follows that two CBs per nucleus must be sufficient to carry out vital functions in post-mitotic cells that have reached a steady state in overall gene expression. We propose that active snRNP biogenesis, which likely reflects overall gene expression levels, defines a given cell's facility for CB self-assembly. As embryonic cells grow and divide, the production of new coilin and snRNP molecules—initially due to translation of maternal mRNAs and eventually due to zygotic transcription and translation—leads to homeostasis in CB assembly and maintenance, in which two CBs per nucleus are produced in differentiated cells of the organism.
Materials and Methods
Fish maintenance and cell culture.
Zebrafish AB strain adults and embryos were maintained and harvested as previously described.63 Embryos were grown at ∼28°C, and staged in hours post fertilization.64 When necessary (e.g., western blot), both chorion and yolk were removed as previously described.63,65 PAC2 cells (gift from Nicholas S. Foulkes, MPI for Developmental Biology, Tuebingen) were grown in L15 culture medium (Gibco), supplemented with 15% fetal calf serum, 100 U/ml Penicillin, 100 µg/ml Streptomycin (Gibco) at 28°C, and atmospheric CO2.
Cloning and vectors.
To generate zebrafish cDNA, total RNA was isolated from 48 hpf embryos using TRIzol® Reagent (Invitrogen) and treated with DNA-free kit (Ambion). Oligo-dT18 primer and SuperScript III Reverse Transcriptase (Invitrogen) were used to generate cDNA. Fluorescent protein tags were constructed in pCS2+ vectors (Dave Turner, Ralph Rupp, Jackie Lee, University of Michigan, MI, USA; unpublished). pCS2+N-mRFP was made by inserting mRFP (AF506027) from DsRed-C3 (Clontech), into pCS2+. Likewise, pCS2+N-YFP and pCS2+CFPC were made by inserting eYFP from pEYFPC1 (Clontech) and eCFP from pECFP-N1 (Clontech), respectively, into pCS2+. The zebrafish coilin orthologue (BC045858) was cloned as a N-terminally tagged fusion in pCS2+N-YFP and pCS2+N-mRFP. The zebrafish SMN orthologue (NM_131191) was cloned as a C-terminally tagged fusion into pCS2+CFP-C. LSm11 (ENSG00000022555) was cloned as a N-terminally tagged fusion into pCS2+N-mRFP. Human U85 scaRNA plasmid was a gift of Arnold M. Kiss. Human U4 snRNA cloned into pSP65 and zebrafish U4 snRNA cloned into a TOPO vector were kind gifts of Albrecht Bindereif.66,67
In vitro transcription.
In vitro transcription was carried out with mMESSAGE mMACHINE®SP6 Kit (Ambion) for mRNA synthesis, and MEGAshortscript™ T7 Kit for snRNA synthesis (Ambion). Run-off transcription of snRNAs and scaRNA and incorporation of Alexa488-UTP was carried out as previously described.32
For in vitro synthesis of mouse U7 snRNA, an antisense U7 oligo: AGG GGT TTT CCG ACC GAA GTC AGA AAA CCT GCT AGA CAA ATT CTA AAA GAG CTG TAA CAC TTC CCT ATA GTG AGT CGT ATT A, was annealed to a T7 promoter oligo: TAA TAC GAC TCA CTA TAG GG, and used at 500 nM as template for in vitro transcription.
Microinjections.
Embryos were injected into the yolk or the 1-cell blastoderm with 1 nl of material following standard methods.68 Morpholinos (Gene Tools LLC, Philomath, Oregon) had the following sequences (5′→3′):
Smn-MO: CGA CAT CTT CTG CAC CAT TGG C42
Standard control (CtrlMO): CCT CTT ACC TCA GTT ACA ATT TAT A.
Morpholinos were dissolved and injected in Danieu's buffer. Prior to each injection, morpholino stock solution was incubated at 65°C for 5 min, snap-cooled on ice and an aliquot taken to measure concentration, which was then adjusted to 1 mM. In vitro transcribed mRNAs and fluorescently labeled RNAs were injected in 100 mM KCl, 5 mM Tris-HCl pH 7.4, 0.25 mM EDTA.
Antibodies.
A screen for the maximum length, soluble GST-tagged fragment of zebrafish coilin expressed in E. coli was performed by ELISA at the MPI-CBG Protein Expression Facility. This fragment covering 82% of the peptide (aa 96–528) was used to immunize rabbits. Immunoreactivity of sera (9EA2) was tested by immunocytochemistry and by western blot. The following antibodies were also used: anti-Sm (Y12) at 1:1;69 anti-TMG (K121, Calbiochem) at 1:300;70 anti-SMN (1F1) at 1:100;71 antineurolin/DM-GRASP (zn-8) at 1:200.72 Secondary antibodies were anti-mouse conjugated with FITC and/or anti-rabbit conjugated with TRITC (Jackson ImmunoResearch Laboratories).
Western blot.
Dechorionated and deyolked embryos were resuspended in cold SDS sample buffer.65 For western analysis, 40 µg of total protein extract was analyzed with anti-coilin (9EA2) antiserum at 1:3,000 and anti-SMN antibody at 1:1,000. As secondary antibody HRP-coupled anti-rabbit IgG, (1:10,000, Amersham) and anti-mouse (1:80,000, Sigma) were used.
Indirect immunofluorescence.
Embryos were fixed at RT in 4% paraformaldehyde in 100 mM PIPES, pH 6.9, 2 mM MgCl2, 1.25 mM EGTA for 15–30 min, permeabilized for 15–30 min at room temperature with 0.2% Triton X-100, and blocked in 5% Normal Goat Serum (Jackson ImmunoResearch Laboratories). Subsequently, embryos were incubated in a humidified chamber for 1 h at RT with primary antibody in PBS plus 3% BSA, 0.2% β-glycerophosphate and 10 mM MgCl2. Secondary antibody was used at 1:200 dilution in the dark. The samples were washed with PBS with 10 mM MgCl2, rinsed with water, mounted on glass slides, and left to dry. 80% glycerol (spectrophotometric grade; Fischer Scientific) supplemented with 25 mg/ml DABCO as an anti-fading reagent (Sigma) and 2 µg/ml Hoechst 33342 was applied.
Microscopy.
In vivo imaging was performed in 2–3% methyl cellulose in E3 medium.63 Fixed specimens were mounted in 80% glycerol (spectrophotometric grade; Fischer Scientific) with 25 mg/ml DABCO (Sigma) and 2 µg/ml Hoechst 33342 (Sigma). Imaging was carried out on an Olympus FluoView 1000 confocal microscope, using UPlanApo 10×/NA = 0.4, PlanApo 60×/NA = 1.4 Oil, UPlanApo 60×/NA = 1.3 Oil objectives.
Image processing.
Images were processed using ImageJ and Bitplane Imaris (Bitplane AG, Zurich, Switzerland). Using ImageJ, an average cytoplasmic and nucleoplasmic intensity was determined for every experimental time-lapse sequence or z-stack that was subsequently used for spot counting. For the early developmental stages, spot counting was done using Imaris software. Briefly, on the basis of the collected z-stacks with slices taken every 1 µm to minimize double appearance of the same spot in two subsequent z-slices, a 3D volume was rendered. It was cropped in 3D to accommodate each nucleus in a new volume. Subsequently, to define the nucleus, an isosurface was created using the average cytoplasmic and nucleoplasmic intensities previously determined in ImageJ. Usually, 3× average cytoplasmic intensity was used to set the threshold for isosurface. Information from outside the nucleus was removed from the image. For spot counting, the size threshold was set to 0.5 µm and the intensity threshold to three-fold the average nucleoplasmic intensity calculated by ImageJ. Due to the tissue compaction at segmentation and pharyngula stages, counting was done manually. Figures were prepared using Adobe Illustrator and Adobe Photoshop.
Acknowledgements
We thank David Stanek, Patricia Heyn and Martin Machyna for helpful discussions and comments on the manuscript. We are grateful to Jan Peychl and Mike Tipsword for expert technical assistance as well as the MPI-CBG fish facility. This work was supported by the Max Planck Society (to K.M.N. and A.O.) and a grant from the German Research Foundation (NE909/2-1 to K.M.N.).
Abbreviations
- snRNP
small nuclear ribonucleoprotein
- snRNA
small nuclear RNA
- scaRNA
small Cajal body-associated RNA
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/10680
Supplementary Material
References
- 1.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:14–16. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 2.Tuma RS, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, et al. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- 4.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker KE, Massello LK, Gao L, Barber TJ, Hebert MD, Chan EK, et al. Structure and characterization of the murine p80 coilin gene, Coil. J Struct Biol. 2000;129:269–277. doi: 10.1006/jsbi.2000.4234. [DOI] [PubMed] [Google Scholar]
- 6.Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell. 2006;17:2942–2951. doi: 10.1091/mbc.E05-12-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 9.Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- 10.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- 12.Boudonck K, Dolan L, Shaw PJ. Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cell cycle parameters. J Cell Sci. 1998;111:3687–3694. doi: 10.1242/jcs.111.24.3687. [DOI] [PubMed] [Google Scholar]
- 13.Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- 15.Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, et al. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. Rna. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, et al. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handwerger KE, Murphy C, Gall JG. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J Cell Biol. 2003;160:495–504. doi: 10.1083/jcb.200212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, et al. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis—evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudonck K, Dolan L, Shaw PJ. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- 26.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 27.Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, et al. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;122:1872–1881. doi: 10.1242/jcs.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, et al. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005;114:155–166. doi: 10.1007/s00412-005-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbott J, Marzluff WF, Gall JG. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 32.Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 34.Gerbi SA, Borovjagin AV, Odreman FE, Lange TS. U4 snRNA nucleolar localization requires the NHPX/15.5-kD protein binding site but not Sm protein or U6 snRNA association. J Cell Biol. 2003;162:821–832. doi: 10.1083/jcb.200301071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes DJ, Kornberg TB, Kirschner MW. Small nuclear RNA transcription and ribonucleoprotein assembly in early Xenopus development. J Cell Biol. 1983;97:62–72. doi: 10.1083/jcb.97.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 38.Narayanan U, Achsel T, Luhrmann R, Matera AG. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 39.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum Mol Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, et al. Ongoing U snRNP Biogenesis Is Required for the Integrity of Cajal Bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, et al. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 45.Fischer U, Luhrmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 46.Fischer U, Sumpter V, Sekine M, Satoh T, Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- 52.Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, et al. Spliceosomal Small Nuclear Ribonucleoprotein Particles Repeatedly Cycle through Cajal Bodies. Mol Biol Cell. 2008;19:2534–2543. doi: 10.1091/mbc.E07-12-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konig H, Matter N, Bader R, Thiele W, Muller F. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell. 2007;131:718–729. doi: 10.1016/j.cell.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira J, Carmo-Fonseca M. Nuclear morphogenesis and the onset of transcriptional activity in early hamster embryos. Chromosoma. 1996;105:1–11. doi: 10.1007/BF02510033. [DOI] [PubMed] [Google Scholar]
- 55.Ferreira J, Carmo-Fonseca M. The biogenesis of the coiled body during early mouse development. Development. 1995;121:601–612. doi: 10.1242/dev.121.2.601. [DOI] [PubMed] [Google Scholar]
- 56.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6 and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, et al. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 59.Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pena E, Berciano MT, Fernandez R, Ojeda JL, Lafarga M. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comp Neurol. 2001;430:250–263. doi: 10.1002/1096-9861(20010205)430:2<250::aid-cne1029>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 61.Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci. 2003;116:303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- 62.Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, et al. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- 63.Westerfield M. The Zebrafish Book: a guide for the laboratory use of zebrafish (Danio rerio) Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- 64.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 65.Link V, Shevchenko A, Heisenberg CP. Proteomics of early zebrafish embryos. BMC Dev Biol. 2006;6:1. doi: 10.1186/1471-213X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wersig C, Bindereif A. Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res. 1990;18:6223–6229. doi: 10.1093/nar/18.21.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trede NS, Medenbach J, Damianov A, Hung LH, Weber GJ, Paw BH, et al. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci USA. 2007;104:6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nüsslein-Volhard CaRD. Zebrafish: A Practical Approach. Oxford, UK: Oxford University Press; 1990. [Google Scholar]
- 69.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krainer AR. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16:9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young PJ, Man NT, Lorson CL, Le TT, Androphy EJ, Burghes AH, et al. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum Mol Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- 72.Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.