SUMMARY
The heart has the ability to grow in size in response to exercise, but little is known about the transcriptional mechanisms underlying physiological hypertrophy. Adult cardiomyocytes have also recently been proven to hold the potential for proliferation, a process which could be of great importance for regenerative medicine. Using a unique RT-PCR based screen against all transcriptional components, we showed that C/EBPβ was down-regulated with exercise, while the expression of CITED4 was increased. Reduction of C/EBPβ in vitro and in vivo resulted in a phenocopy of endurance exercise with cardiomyocyte hypertrophy and proliferation. This proliferation was mediated, at least in part by the increased CITED4. Importantly, mice with reduced cardiac C/EBPβ levels displayed substantial resistance to cardiac failure upon pressure overload. These data indicate that C/EBPβ represses cardiomyocyte growth and proliferation in the adult mammalian heart and that reduction in C/EBPβ is a central signal in physiologic hypertrophy and proliferation.
HIGHLIGHTS.
Endurance exercise of mice induces cardiomyocyte proliferation
Endurance exercise causes a reduction of C/EBPβ expression and an increase of CITED4
Reduction of C/EBPβ in vitro induces cellular hypertrophy and proliferation via CITED4
Genetic reduction of C/EBPβ in vivo results in a phenocopy of the exercised heart
INTRODUCTION
Cardiac muscle adapts to increased pressure and/or volume with hypertrophy. This holds true both for physiological stimuli, such as exercise, as well as for pathological provocations, including hypertension. While the earliest stages appear quite similar at a morphological level, pathological cardiac hypertrophy leads to cardiovascular diseases such as heart failure and arrhythmia, while physiological hypertrophy does not. Thus, understanding the mechanistic distinction between these two types of cardiac growth has very important clinical implications. Numerous prior reports document transcription factors involved in pathological hypertrophy (Akazawa and Komuro, 2003; Frey and Olson, 2003), but relatively little is known about the molecular mechanisms controlling physiological hypertrophy.
Cardiomyocytes in adult mammals retain a limited ability to proliferate in response to specific stimuli (Bergmann et al., 2009; Bersell et al., 2009; Kajstura et al., 2010). This has potential importance because cardiomyocyte proliferation could serve as a basis for regeneration of an injured heart, if it could be enhanced and harnessed. However, whether this process can be modulated through physiological interventions remains unclear. Moreover, while recent studies have implicated some signal transduction pathways involved in adult cardiomyocyte proliferation (Bersell et al., 2009), little is known about the detailed molecular mechanisms and particularly the transcriptional components that are decisive in this process. Furthermore, there is little data available on factors that control cardiomyocyte proliferation in the adult heart.
One of few known regulators of cardiomyocyte proliferation is Gata4, which was recently reported to mark proliferative cells during cardiac regeneration in the zebrafish (Kikuchi et al., 2010). This factor is also a well known regulator of cardiomyocyte differentiation in adult hypertrophy, together with SRF, Nkx2.5, Tbx5 and Gata6 (Akazawa and Komuro, 2003).
Here we studied a murine model of physiological hypertrophy induced by endurance exercise, using a method termed Quanttrx, that we recently developed for genome-wide, quantitative measurement of the expression of all transcriptional components (Gupta et al., 2010). The database underlying this method includes all known transcription factors, transcriptional co-regulatory proteins, and all proteins that contain a motif that has been associated with transcriptional components, whether their function is known or not. We report here that endurance exercise effected a hypertrophic and proliferative program in cardiomyocytes that was dependent on a reduction in the expression of the transcription factor C/EBPβ, and a linked increase in the expression of CITED4. Furthermore, mice with genetically reduced C/EBPβ levels recapitulated many of the cardiac phenotypes seen in exercised mice. Importantly, mice with reduced C/EBPβ were resistant to the development of cardiac dysfunction in response to pressure overload on the heart.
RESULTS
Cardiac hypertrophy and cardiomyocyte proliferation are induced following endurance exercise
To study transcriptional regulators of physiological cardiac hypertrophy, a ramp swimming exercise model was used (Taniike et al., 2008). As shown in Supplemental Figure I, mild cardiac hypertrophy was induced without alterations in the expression of pathological gene markers such as ANP and BNP. There was also a 45% increase in cardiomyocyte cell size (Supplemental Figure I) without any detectable difference in fibrosis or angiogenesis (Supplemental Figure I). Hence, we may conclude that healthy physiological cardiac hypertrophy occurred in these animals.
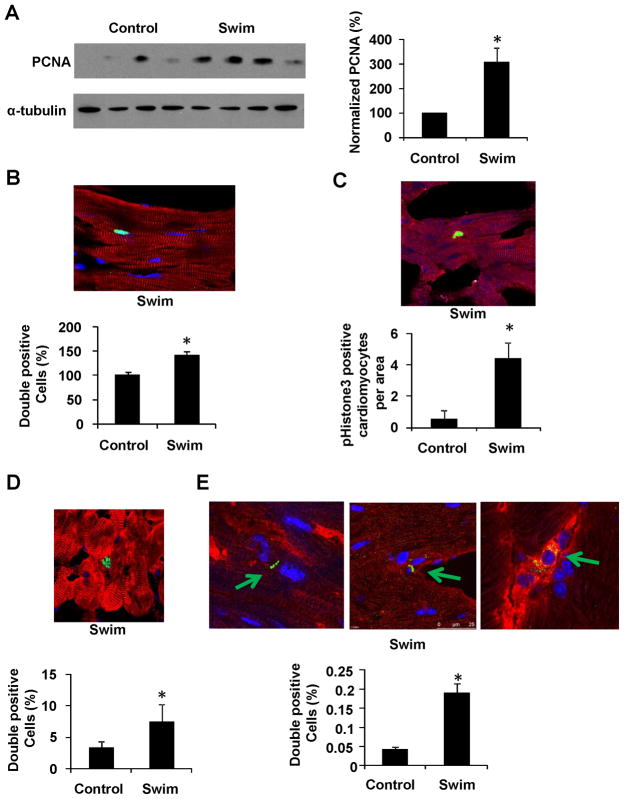
Further characterization revealed that expression of the cell proliferation marker PCNA was increased in the hearts from exercised mice (Figure 1A). To establish the cellular origin of the increased proliferation markers, confocal microscopy was employed in cardiac tissue stained for both the cardiomyocyte-selective protein α-actinin and the proliferation marker ki67. Robust ki67 staining was seen in 1-5% of cardiomyocytes (Figure 1B), Importantly, there was a significant increase in cardiomyocyte ki67 staining in the exercised mice compared to the non-exercised controls, as judged from blinded analyses of histological sections using digital software. The more specific mitosis marker, phospho-Histone3 (pH3), was also elevated (Figure 1C), though the absolute number of positive cells was relatively low. Supplemental Figure I shows pH3-positive cardiomyocytes in the different stages of mitosis. Thus, markers of both cell size and proliferation are elevated in the cardiomyocytes of mice in this model of physiological hypertrophy.
Figure 1. A swim model induces physiological cardiac hypertrophy and cardiomyocyte proliferation.
A. Mice swam using a ramp protocol for 2 weeks with respective controls (n=4), and were assayed for PCNA protein levels with subsequent quantification. Data is presented as PCNA per α-tubulin after subtraction of background. Data is representative for two independent mouse cohorts. B-C. Representative image and quantification of Ki67 (B) and phosphoHistone3 (C) staining from exercised mice (green) counterstained with α-actinin (red) and DAPI (blue) in cardiac tissue. D. Exercised and control mice were injected with BrdU 3 days prior to the final exercise day. The image shows a representative section from the exercised cohort. BrdU is labeled in green, α-actinin in red and DAPI in blue. E. Representative image and quantification of Aurora B kinase (green) staining from exercised mice counterstained with α-actinin (red) and DAPI (blue) in cardiac tissue. All quantifications were based on at least 80 (blinded) images from each group (from at least 4 mice). Any uncertainty concerning whether a ki67, AuroraB or BrdU positive cell was completely within an α-actinin positive area was resolved using confocal z-stacks. * indicates p<0.05 vs respective control using students t-test. Se also Supplemental Figure S1.
Previous studies have utilized the density of cardiomyocyte nuclei as an indicator of cardiomyocyte proliferation (Bersell et al., 2009). We could detect a small increase in nuclear density (100 and 132% in controls versus exercised mice respectively, p=0.07), but this did not reach statistically significance. However, it is worth noting that the observed increase in cardiomyocyte size would be expected to confound/dilute the calculated changes in nuclear density. DNA biosynthesis was therefore assayed using bromodeoxyuridine (BrdU) injections in mice undergoing this exercise protocol. We could detect double α-actinin and BrdU positive cardiomyocytes at a frequency of 1-6% in exercised mice. Figure 1D shows a substantial increase in BrdU positive cardiomyocytes in the hearts of exercised mice compared to the controls. Importantly, z-stack visualization of positive cells clearly demonstrates a distinct cardiomyocyte localization of the BrdU (Supplemental Figure I). Finally, staing for the cytokinesis marker Aurora B kinase was used to demonstrate that the exercise protocol did indeed promote completed cardiomyocyte cell division (Figure 1E). Conversely, we could not detect any of these changes in proliferation markers in a murine model of pathological pathological hypertrophy (data not shown). Taken together these data strongly suggest that adult cardiomyocytes increase in both size and proliferation rate in response to this form of endurance exercise.
C/EBPβ is down-regulated during physiological hypertrophy
To investigate the transcriptional basis of physiological cardiac hypertrophy, we utilized Quanttrx, a qPCR based screen we recently developed against all known and predicted transcriptional components (Gupta et al., 2010). We used cardiac samples from the mice characterized (above) together with cardiac samples from mice subjected to transaortic constriction (TAC) for two weeks. As shown in Supplemental Figure I, the TAC procedure induced a similar degree of hypertrophy as seen in the exercised animals, but with elevation of ANP/BNP, thus indicating pathological hypertrophy. There were no signs of heart failure or other pathological changes in the mice subjected to TAC at this time point.
The Quanttrx screen identified 175 genes significantly regulated in the exercise model, and 96 in the pathological (TAC) model (Supplemental Table I and II). Importantly, there was little overlap in the expression profile between the two models. In fact, there was a significant, negative correlation between the changes induced in respective model (Supplemental Figure II and r=−0.32, p<0.001). Thus, these models of pathological and physiological hypertrophy express distinct programs of transcriptional components.
Of the 175 transcription factors regulated by physiological hypertrophy, 47 (27%) had no known function (no PubMed annotation). However, 10 transcription factors with known functions in cardiac differentiation and/or adult hypertrophy were differentially regulated in the swim model. These included well known genes such as Nkx2.5, Gata4, Tbx5, Mef2c and Gata6. Furthermore, 13% of the differentially expressed factors had known or suggested roles in cellular proliferation. Less than 1% of the genes regulated in the pathological model had any known role in cellular proliferation.
Based on the magnitude of expression changes and absolute cardiac expression, we chose 30 genes out of this preliminary list of 272, and performed qPCR analyses in completely new samples of either physiological (endurance exercise) or pathological (TAC) hypertrophy.
Eight out of these 30 showed robust validation (Supplemental Figure II and Figure 2A) and these were further analyzed for cardiomyocyte-specific expression. Supplemental Figure II show expression of these genes in cardiomyocytes and non-cardiomyocytes, after density gradient separation. Five out of 8 were expressed primarily in the cardiomyocyte fraction, and these were sub sequentially analyzed using adenoviral-mediated forced expression and/or siRNA-mediated knockdown, using changes in cell size of rat neonatal cardiomyocytes as our end-point. Gbx2, Cited4, Mlx and Meox1 failed to demonstrate any effect on cardiomyocyte sizes (Supplemental Figure II), but reduction of C/EBPβ caused a clear increase in cell size (Figure 3A).
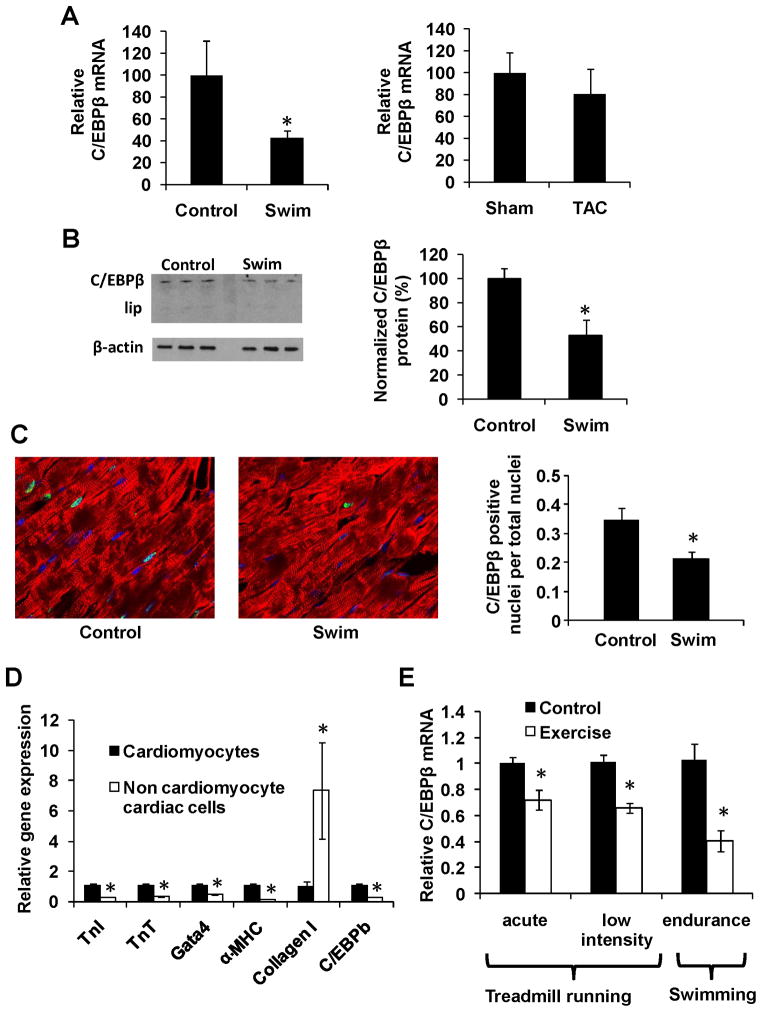
Figure 2. C/EBPβ is expressed in cardiomyocytes and down regulated with endurance exercise.
A. Cardiac C/EBPβ mRNA levels in a model for physiological (swim) and pathological (TAC) hypertrophy with respective controls (n=4) as described in methods. B. Western blot analysis of C/EBPβ and β-actin in the swim cohort (above) with subsequent quantification. Data is presented as percent of control with C/EBPβ relative to β-actin after background subtraction. C. Confocal microscopy after immunohistochemistry against C/EBPβ (green), α-actinin (red) and DAPI (blue) in cardiac tissue with subsequent quantification in indicated groups. Data is presented as the ratio of C/EBPβ positive cardiomyocyte nuclei. D. Gradient fractionation of primary rat neonatal cardiac cells followed by expression analysis of cardiomyocyte and fibroblast markers. E. Cardiac C/EBPβ mRNA levels from three different exercise regimens; “acute” = 40 minutes treadmill running (n=6), “low intensity” = 2 weeks of 30 minutes daily running (n=6) and “endurance” = the swimming protocol as described in methods (n=4). * indicates p<0.05 vs respective control using students t-test. Se also Supplemental Figure S2.
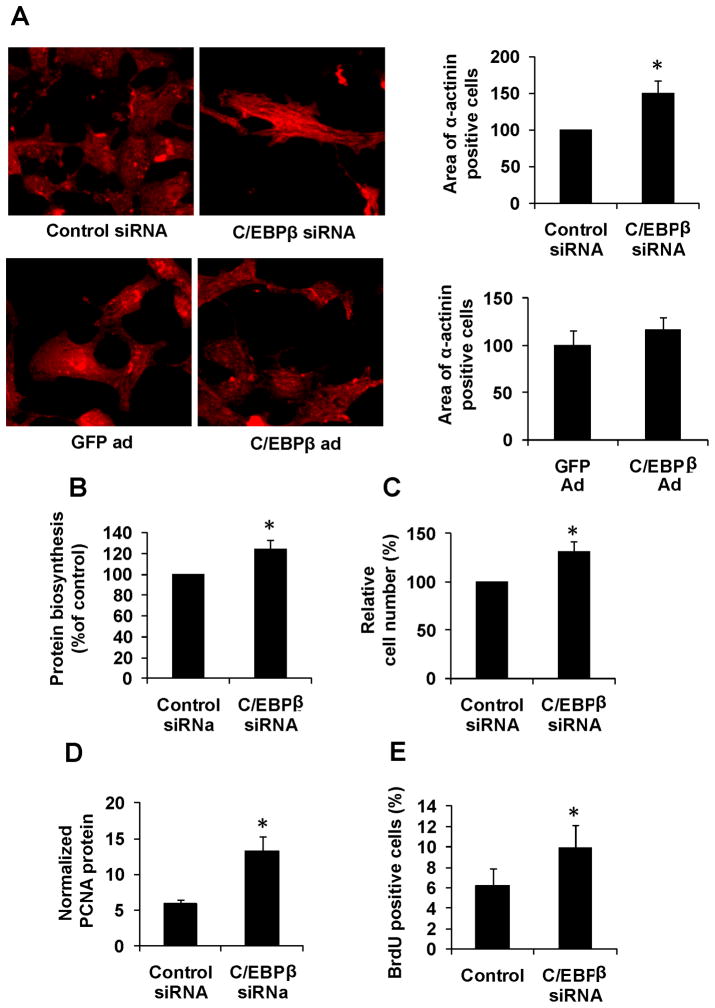
Figure 3. Reduction of C/EBPβ in primary cardiomyocytes results in hypertrophic cell growth and proliferation.
Primary rat neonatal cardiomyocytes treated with either a C/EBPβ siRNA or a C/EBPβ expressing adenovirus with respective controls. All experiments were performed 48 hours after transfection / transduction. A. Immunohistochemistry against α-actinin followed by cell area quantifications as described in methods. At least 100 cells were quantified in all groups. B. Protein biosynthesis measured as 35S-Met incorporation into the protein pool after a 1h pulse. Data is presented as percent of control. C. Quantification of cell numbers in primary after transfection with indicated siRNA constructs. D. Western blot analysis of PCNA followed by quantification (n=4). Data is presented as PCNA relative to β-actin after background subtraction. E. Primary rat neonatal cardiomyocytes were treated with BrdU 24 hours after transfection with indicated siRNA. Cells were then stained against BrdU and α-actinin and BrdU-positive cardiomyocytes were counted normalized to total number of cardiomyocytes. * indicates p<0.05 vs respective control using students t-test. Se also Supplemental Figure S3.
C/EBPβ is a member of the bHLH gene family of DNA-binding transcription factors, and has known roles in cell proliferation and differentiation in other tissues (Sebastian and Johnson, 2006), including adipose cells and liver. However, it has not been studied in the context of the cardiomyocyte. C/EBPβ mRNA expression was reduced by 61% in hearts in the exercise model when assayed in new samples, but this effect was not observed in a model of pathological hypertrophy using trans-aortic banding (TAC), performed in parallel (Figure 2A). Down-regulation of C/EBPβ was also seen in response to the exercise training at the protein level (Figure 2B). Expression of the short LIP C/EBPβ isoform that is inhibitory toward C/EBPβ function was undetectable in these cardiac samples (Figure 2B). Confocal microscopy illustrated that it was indeed cardiomyocyte-specific expression of C/EBPβ that was reduced with exercise (Figure 2C). Consistent with this, C/EBPβ expression was enriched in the cardiomyocyte fraction, versus non-cardiomyocyte cells, following gradient purification of rat neonatal cardiomyocytes (Figure 2D and Supplemental Figure II).
Two additional mouse models of exercise were tested for effects on C/EBPβ expression; acute and low intensity treadmill running. Neither treatment resulted in cardiac hypertrophy (data not shown), but reduction of C/EBPβ expression occurred in the acute setting (40 minutes of treadmill running), as seen in Figure 2E. Thus, C/EBPβ expression is reduced relatively early in endurance exercise.
Reduction in C/EBPβ expression induces hypertrophy and proliferation in cultured cardiomyocytes
The functional consequences of experimental reduction of C/EBPβ were assessed in detail in primary rat cardiomyocytes. A siRNA was used to reduce C/EBPβ levels to approximately the levels of mRNA observed after endurance exercise (43%, Supplemental Figure III). As demonstrated in Figure 3A-B, the forced decrease in C/EBPβ mRNA was sufficient to drive an increase in cell size and in the rate of protein biosynthesis. No statistically significant change in cell size or protein biosynthesis was seen with forced C/EBPβ expression (p=0.42 and p=0.054 respectively). The reduction of C/EBPβ expression was also sufficient to ablate the increase in ANP levels seen with pathological provocation (Supplemental Figure III).
Interestingly, the C/EBPβ reduction via siRNA also led to an increase in cell number (Figure 3C). This was accompanied by increased PCNA expression and increased BrdU incorporation into DNA, highly suggestive of cardiomyocyte proliferation (Figure 3D-E). These data indicate that a reduction in C/EBPβ expression is sufficient to induce hypertrophy and proliferation in cultured cardiomyocytes.
C/EBPβ regulates expression of a gene set characteristic of physiological hypertrophy by sequestering Serum Response Factor (SRF)
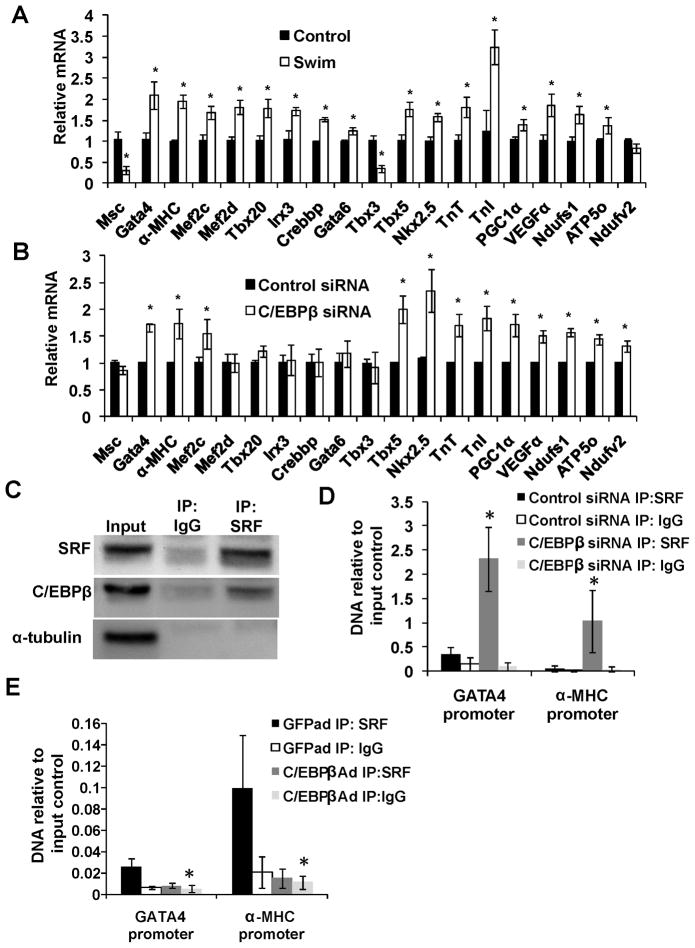
The initial Quanttrx screen revealed a specific set of regulated genes with known functions in cardiomyocyte differentiation and hypertrophy (Figure 4A and Supplemental Table I). Examples of such genes were Gata4, Tbx5 and Nkx2.5 which all induce hypertrophy(Akazawa and Komuro, 2003). Beyond these 11 genes, we also investigated expression levels of the genes well-known to be altered in cardiac hypertrophy or differentiation, including α-MHC, TnI and TnT, which were all up regulated in the hearts of exercised mice (Figure 4A). The transcriptional co-regulator PGC-1α has previously been demonstrated to have an important role in preventing cardiac dysfunction after the development of hypertrophy (Arany et al., 2006), and exercised mice indeed displayed increased cardiac expression levels of PGC-1α together with its downstream targets VEGF, Ndufs1, Ndufv2 and ATP5o (Figure 4A). Thus, by combining the Quanttrx screen hits with markers of hypertrophy and the PGC-1α as well as its downstream targets, we defined an exercise-induced gene set of 19 genes (Figure 4A). The changes in mRNA expression for Nkx2.5, Tbx5, Mef2c, Gata4 and PGC-1α following TAC surgery is shown in supplemental figure IV.
Figure 4. C/EBPβ controls a exercise –induced gene set by inhibiting SRF DNA binding.
QPCR analysis of indicated genes in hearts from endurance exercised mice (A) and in primary cardiomyocytes treated with C/EBPβ or control siRNA for 48 hours (B). * indicates p<0.05 vs respective control using students t-test. C, Immunoprecipitation against SRF or a non-immune IgG in primary cardiomyocytes followed by western blot against SRF, C/EBPβ and the control protein α-tubulin as indicated. D-E. Chromatin immunoprecipitation in primary cardiomyocytes treated with either C/EBPβ siRNA (D) or C/EBPβ adenoviral over expression (E) with respective controls. Precipitated DNA was then analyzed with RT-PCR using primers directed against promoter regions in the Gata4 and myh6 gene. ChIP experiments were repeated at least three times with similar results. * indicates p<0.05 vs respective control using one-way ANOVA statistics. Se also Supplemental Figure S4.
As shown in Figure 4B, reduction in C/EBPβ levels in primary cardiomyocytes resulted in a strikingly similar expression pattern. In fact, 11/19 of these genes were regulated in the same manner as in the exercise model (p<0.001 using Chi-Square statistics). Furthermore, adenoviral expression of C/EBPβ induced essentially the reverse expression pattern (Supplemental Figure IV). These data strongly suggest that C/EBPβ acts upstream of other transcriptional components that are also regulated in physiological hypertrophy. Especially interesting was the C/EBPβ-dependent regulation of Gata4, which has recently been demonstrated to play a major role in zebrafish cardiomyocyte proliferation during myocardial regeneration (Kikuchi et al., 2010).
Many of the genes regulated by both endurance exercise and the siRNA-mediated C/EBPβ reduction are known to be transcriptional targets of the transcription factor SRF, with SRE elements in their promoters (Akazawa and Komuro, 2003). C/EBPβ has been demonstrated to physically interact with SRF (Hanlon and Sealy, 1999), but the functional consequences of this interaction are not clear, especially in cardiomyocytes where the role of C/EBPβ is unknown. Previous reports on the C/EBPβ-SRF interaction have suggested a positive effect on SRE-dependent elements, but the increased expression levels of Gata4 and α-MHC (Figure 4A) following C/EBPβ siRNA here suggested a negative regulation of SRE-dependent genes in cardiomyocytes.
A physical interaction between the SRF and C/EBPβ proteins was indeed observed, using co-immunoprecipitation from primary cardiomyocytes (Figure 4C). Next, chromatin immunoprecipitation (ChIP) assays were performed, examining the binding of SRF to the Gata4 and α-MHC promoters, following gain- and loss of the C/EBPβ protein. As shown in Figure 4D, CEBPβ reduction using siRNA dramatically increased SRF binding to the promoters of both α-MHC and Gata4, where SRF is known to play a positive regulatory role. Conversely, adenoviral expression of C/EBPβ reduced SRF binding to these promoters. Thus, C/EBPβ interferes with SRF binding to the promoters of critical cardiac genes (Figure 4E).
The recent discovery of the ErbB4 pathway in cardiomyocyte proliferation led us to investigate whether C/EBPβ could be affected by such signaling. This was especially interesting as ErbB4 signals via AKT1, a kinase known to function in physiological hypertrophy (DeBosch et al., 2006). We thus investigated the connection of this pathway to C/EBPβ, using both forced expression of wild type AKT1 and a dominant negative AKT1 (Nagoshi et al., 2005). As demonstrated in Supplemental Figure IV, increasing AKT1 activity did indeed reduce C/EBPβ expression levels together with an expression profile resembling what was observed with exercise with increased expression of Tbx5, Gata4, TnI and α-MHC. Conversely, the dominant negative AKT1 mutant increased C/EBPβ paralleled with decreased mRNA levels of Tbx5, Gata4, TnI, TnT and α-MHC. We could thus conclude that AKT1 activity evoke the same gene expression profile as seen in exercise. We next used C/EBPβ adenovirus together with AKT1 over expression to establish whether C/EBPβ indeed act downstream on AKT1 in the regulation of these target genes. As demonstrated in Supplemental Figure IV, C/EBPβ over expression ablated the positive expression effects of AKT1 on Tbx5, Gata4, TnI, TnT and α-MHC. Taken together, these data suggest that AKT1 regulates C/EBPβ expression, connecting C/EBPβ to a previously characterized physiological growth pathway in the heart.
C/EBPβ negatively regulates CITED4, a transcription factor promoting cardiomyocyte proliferation
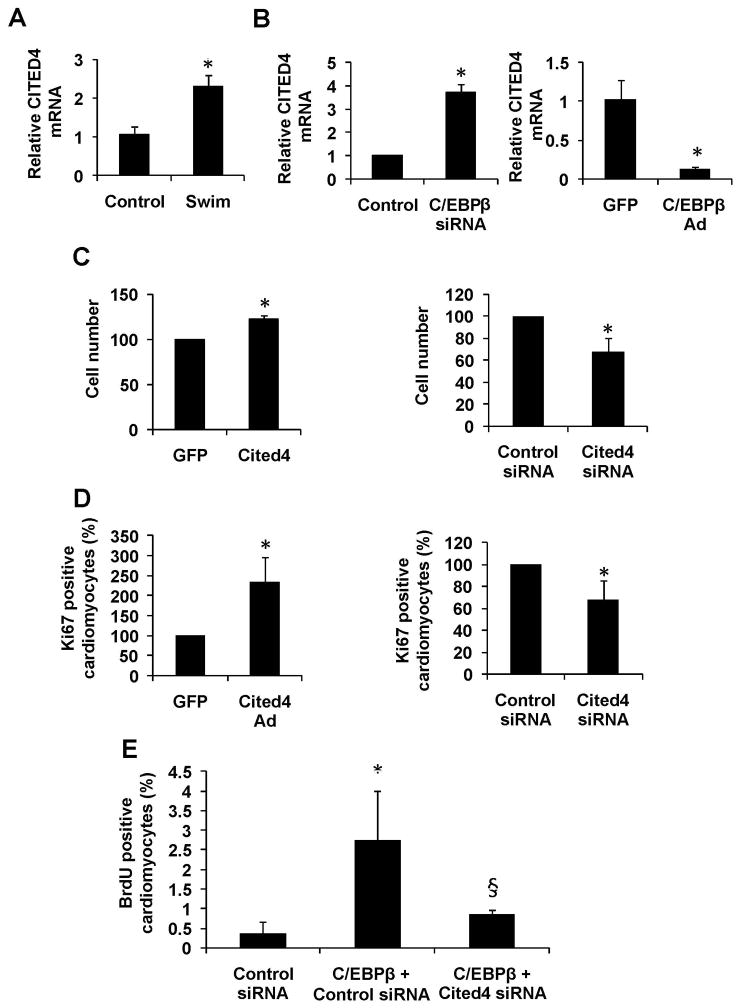
The SRF-dependent gene set described above includes well-known regulators of cardiomyocyte hypertrophy, but apart from Gata4, they are not known to participate in a proliferative phenotype, such as we see with C/EBPβ reduction. We thus analyzed the list of exercise-induced cardiac genes (Supplemental Figure II) for those controlled by C/EBPβ that might influence or control cardiomyocyte proliferation. Preliminary studies using gain and loss of function for 6 factors apparently downstream of CEBPβ led us to focus on Cbp/p300-interacting transactivator with ED-rich carboxy-terminal domain 4 (CITED4). Indeed, CITED4 showed a robust increase in exercised hearts (Figure 5A) and its expression level was markedly altered with C/EBPβ gain- or loss-of-function in vitro (Figure 5B).
Figure 5. CITED4 is up regulated with endurance exercise, regulated by C/EBPβ, and its forced expression induced cardiomyocyte proliferation in vitro.
A. mRNA levels of cardiac CITED4 in control and endurance exercised (swim) mice normalized to 18S expression. B. mRNA expression of CITED4 in primary rat neonatal cardiomyocytes after siRNA knockdown (left panel) or adenoviral over expression (right panel) of CITED4. Expression levels is normalized to 18S. C-D. Rat neonatal cardiomyocytes treated with either an adenovirus over expressing CITED4 (left) or a siRNA directed against CITED4 (right). Cells were then stained against α-actinin and ki67, and positive cells were counted in 20x view fields from 15 random images per group. E. Rat neonatal cardiomyocytes treated with control siRNA, C/EBPβ siRNA + control siRNA or C/EBPβ siRNA + Cited4 siRNA, followed by assay of BrdU incorporation as previously described. Data is pooled from three experiments and presented as percent of control. * indicates p<0.05 vs respective control, and § versus C/EBPβ siRNA using one-way ANOVA statistics. Se also Supplemental Figure S5.
Strikingly, forced CITED4 expression (Supplemental Figure V) increased cardiomyocyte cell numbers, together with an increased percentage of ki67 positive cells (Figure 5C-D). Conversely, siRNA directed against CITED4 (Supplemental Figure VC) reduced both cell numbers and ki67 staining (Figure 5C-D). Gene expression analysis also revealed a gene set of pro-proliferative genes increased in endurance exercise (Supplemental Figure V), where the important cell cycle/proliferation factors CyclinD1 and n-myc also displayed increased expression with forced CITED4 expression together with SRF (Supplemental Figure V).
To investigate the functional relationship between C/EBPβ and CITED4, we used siRNA directed against CITED4, together with C/EBPβ siRNA. As shown in Figure 5E, CITED4 knockdown could completely abolish the effect of cellular proliferation seen with the C/EBPβ reduction. Thus, C/EBPβ is likely to mediate its anti-proliferative effects, at least in part, via CITED4 expression.
C/EBPβ controls cardiomyocyte proliferation in the zebrafish embryo
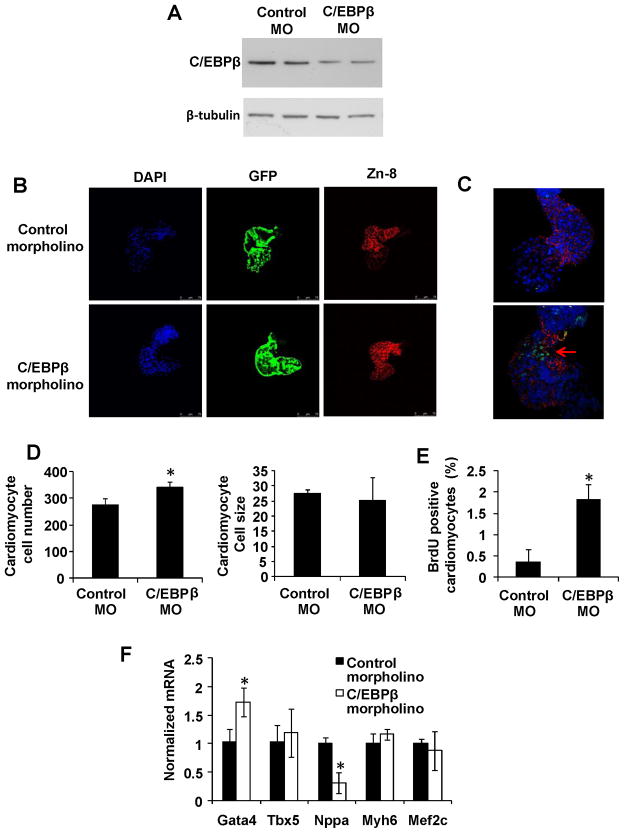
We next tested the role of C/EBPβ reduction in zebrafish, a simple and experimentally accessible in vivo system. Anti-C/EBPβ morpholino oligonucleotides were injected into zebrafish embryos transgenic for cmlc2-GFP at the one-cell stage (Figure 6A). Figure 6B shows zebrafish hearts dissected at 36h and stained for nuclei (DAPI–blue), and plasma membrane (integrin - red). Quantification of these images revealed increased cardiomyocyte cell number, but did not change cardiomyocyte size, up to 72 hours post fertilization (Figure 6D). Furthermore, 20 minutes BrdU labeling of 48h hpf embryos demonstrated increased DNA-synthesis (Figure 6C and quantification in Figure 6E). Of note, Gata4 mRNA expression was significantly increased, consistent with results shown above (Figure 6F). Thus, reduction of C/EBPβ induces cardiomyocyte proliferation in zebrafish hearts in vivo.
Figure 6. C/EBPβ reduction in the Zebrafish embryo results in cardiomyocyte proliferation.
An anti-C/EBPβ morpholino was injected into one-cell stage cmlc-GFP transgenic Zebrafish embryos. A. Western blot analysis against indicated proteins in zebrafish embryos 48 hpf. B. Dissection of beating hearts at 36 hours followed by integrin staining (blue = DAPI, green = GFP and red = zn-8). C. 20 minutes BrdU labeling prior to dissection of beating hearts and BrdU (green) / zn-8 (red) staining. D. Quantification (B) from totally 30 hearts per group. E. Quantification of integrin and BrdU positive cardiomyocytes (C) from n=6 embryos per group. Data is presented as percent of control and representative of two independent experiments. F, RT-PCR analysis of indicated transcripts. * indicates p<0.05 vs respective control using students t-test. Se also Supplemental Figure S6.
Heterozygosity for C/EBPβ in mice mimics cellular aspects of endurance exercise and is cardioprotective against pressure overload
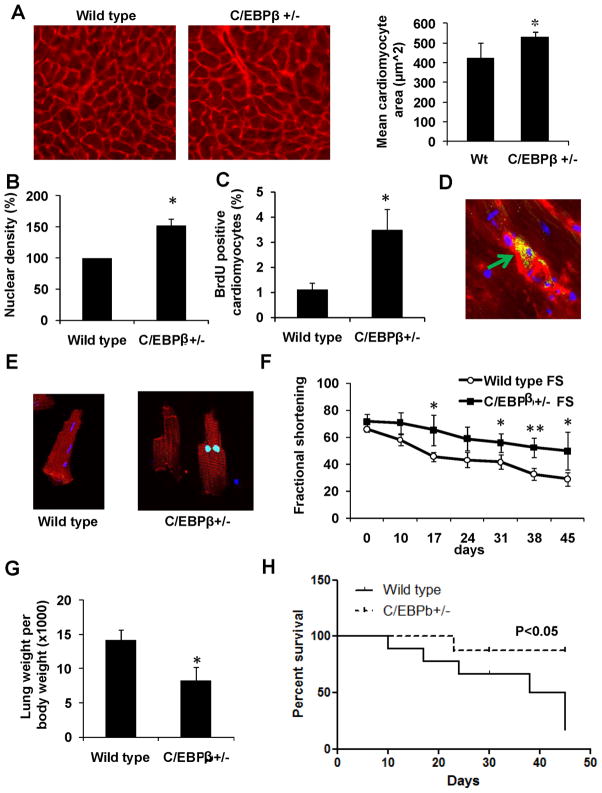
Mice with a complete loss of C/EBPβ show pre- and perinatal lethality, but heterozygotes are relatively normal. Mice heterozygous for C/EBPβ have cardiac C/EBPβ mRNA levels reduced to a similar level as observed in exercised mice (50% reduction; Supplemental Figure VI). These heterozygous animals were therefore used to study the role of this factor in mammalian cardiac function. The exercise-induced gene set defined above was analyzed in hearts of the C/EBPβ+/− compared to wild-type littermates, and a pattern of expression changes similar to that seen with exercise was seen for multiple genes (8/19 genes–Supplemental Figure VI). Furthermore, measurements of cell size (done in a blinded manner) revealed cardiomyocyte hypertrophy (Figure 7A), and an increase in heart weight (Supplemental Figure VI). Echocardiographic studies revealed a small but significant increase in fractional shortening, an important parameter of improved heart function (Supplemental Table III). No differences in heart wall thickness were observed.
Figure 7. C/EBPβ+/− mice have cardiac hypertrophy, increased proliferation markers and are resistant to pressure overload.
Cardiomyocyte size was measured in cardiac tissue from wild type (n=5) and C/EBPβ+/− (n=7) mice using WGA membrane staining (A). B-C. Quantification of nuclear density assayed (B) and BrdU incorporation (C) from indicated mice. D. Typical image of Aurora B kinase (green) positive cardiomyocyte from C/EBPβ+/− mouse heart, counterstained with anti-α-actinin (red) and dapi (blue). E. Immunohistochemistry against phosphohistone H3 (green) in primary cardiomyocyte from an adult C/EBPβ+/− mouse heart counterstained with α-actinin (red) and dapi (blue). F. Wild type and C/EBPβ+/− (n=6 and 7) mice subjected to TAC and assayed for fractional shortening at indicated times. G. Lung weight 52 days after TAC intervention. H. Kaplan Meyer plot of cardiac failure free fraction at indicated times in wild type and C/EBPβ+/− mice following TAC procedure. * indicates p<0.05 vs respective control using students t-test. Se also Supplemental Figure S7.
Cardiomyocyte proliferation was also assayed at baseline in the C/EBPβ+/− mice. Notably, there was a clear increase in cardiomyocyte nuclear density (Figure 7B), consistent with increased expression of proliferation markers in the cardiomyocytes that was also observed (Supplemental Figure VI). BrdU injections resulted in an increased number of positive cardiomyocytes in the C/EBPβ+/− mice (Figure 7C and Supplemental Figure VI). We also detected Aurora B kinase-positive cardiomyocytes in C/EBPβ+/− cardiac tissue (Figure 7D). To study adult cardiomyocyte proliferation directly, we also isolated adult cardiomyocytes from C/EBPβ+/− mice and littermate controls, incubated them in culture with BrdU for 48 hours, and stained for proliferation markers. Fewer than 0.05±-0.1% of cardiomyocytes from control mice were BrdU positive. However, cells from the C/EBPβ+/− mice displayed a 10-fold increase in positive nuclear staining (0.6±0.2% of cells, p<0.05 compared to control; Figure 7E).
We next performed the exercise protocol on the C/EBPβ+/− mice, together with littermate controls. As shown in Supplemental Figure VII, there was no further reduction in C/EBPβ levels with exercise in the C/EBPβ+/− mice compared to wild types. In addition, we could only detect a minor increase in heart mass in the exercised C/EBPβ+/− mice (Supplemental Figure VII). More interestingly, however, the C/EBPβ+/− mice displayed a clear increase in exercise capacity at baseline (Supplemental Figure VI) suggestive of a functional consequence of C/EBPβ reduction in vivo.
To critically assess the possible protective role of the cardiac changes seen in the C/EBPβ+/− mice, we performed transverse aortic constriction (TAC) to induce pressure overload on the heart leading to pathological cardiac hypertrophy and dysfunction in mice. As expected, TAC resulted in progressive reduction in cardiac function, as judged by fractional shortening (FS) in the wild type mice. However, the C/EBPβ heterozygous mice displayed only a minor reduction in contractile function after TAC (Figure 7F). Pulmonary edema is a very important characteristic of cardiac failure in both mice and humans, and the differences in heart function between the two groups was also supported by a significant difference in lung weight (Figure 7G). Ventricular dilatation (as a marker of cardiac failure) also increased more rapidly in the wild-type mice than in the C/EBPβ+/− animals (Supplemental Figure VII).
According to our IACUC rules, mice subjected to TAC had to be sacrificed when cardiac function (fractional shortening) decreased by at least four standard deviations from the mean, as this represents a very substantial deterioration likely to be associated with heart failure. Using such criteria, Kaplan-Meier analysis demonstrated that wild-type littermates were significantly more likely to develop this degree of cardiac dysfunction after TAC than C/EBPβ+/− littermates (Figure 7H). Thus, the reduction in C/EBPβ expression in the hearts of heterozygous mice, quantitatively comparable to that seen after exercise training, results in increased improved cardiac function and resistance to a heart-specific pathological stress.
DISCUSSION
Despite the well-documented beneficial cardiovascular effects of exercise, our understanding of the mechanisms underlying these benefits, particularly in the heart, remains very limited. In particular, little is known about the transcriptional pathways underlying physiological hypertrophy, as compared to the wide variety of transcription factors implicated in pathological hypertrophy. Here we provide evidence for a cardiac pathway induced by exercise and mediated by the reduction of C/EBPβ, resulting in physiological hypertrophy. Interestingly, exercise or experimental decreases in C/EBPβ expression induce increases in both cardiomyocyte size and cell division, which are likely to contribute to some of the benefits observed. Moreover, these effects were tightly coupled to a beneficial metabolic gene set, including an induction of the PGC1α pathway. Lastly, we observe that reduction in C/EBPβ expression was itself sufficient to result in improved cardiac function and resistance to pathological changes/lethality induced in the heart by pressure overload.
Considerable attention has recently been focused on the potential for cardiomyocytes to proliferate, illustrated in several recent papers (Bergmann et al., 2009; Bersell et al., 2009; Kajstura et al., 2010). In fact, the proliferative behavior observed here in cardiomyocytes caused by the endurance exercise was quantitatively similar as to that seen with NRG1 treatment previously (Bersell et al., 2009). Data presented here also suggests that C/EBPβ is an important functional target of ErbB4-related signaling pathways, particulary AKT1, in cardiomyocytes.
Exercise and genetic reduction of C/EBPβ both resulted in the induction of a specific gene set in vitro and in vivo that is known to influence cardiac function. Tbx5, Nkx2.5, Gata4 and α-MHC have all been demonstrated to induce hypertrophy in adult cardiomyocytes (Akazawa and Komuro, 2003). It is thus likely that C/EBPβ exerts part of its hypertrophic actions via increases in this important gene set. It is also worth noting that these effects of reduced C/EBPβ expression do not require massive changes in mRNA or protein levels; 50%–60% changes through exercise or genetic alterations are sufficient to bring about rather profound changes in cardiomyocyte biology and the in vivo response to biomechanical stress. This strongly suggests a rheostat-like role for C/EBPβ, whereby small but highly regulated changes in this factor are reflected in a broad downstream response in the heart.
Among affected genes was also PGC1α, a transcriptional co-activator important for mitochondrial biogenesis and protection against pathological heart stress (Arany et al., 2006; Finck and Kelly, 2007). As gain of function studies in vitro did not suggest that PGC1α induce cardiomyocyte hypertrophy, it is more likely that PGC1α act via increased mitochondrial gene expression and function.
A key mechanism by which C/EBPβ controls Gata4 expression is through binding to SRF and sequestering this factor from direct binding to the Gata4 (and α-MHC) promoters. This mechanism of action may have rather broad implications for cardiac biology considering the available data on the crucial role of SRF in cardiomyocyte differentiation and the regulation of expression of Tbx5, Gata and Nkx2.5 (Akazawa and Komuro, 2003; Chai and Tarnawski, 2002). In fact, ablation of SRF is the only known genetic knockout model which completely abolishes cardiomyocyte differentiation (Fukushige et al., 2006). Possible cardioprotective effects of C/EBPβ reduction are also consistent with recent reports that have described increased C/EBPβ DNA binding in pathological models for negative cardiac remodeling (Oh et al., 2010).
CITED4 was originally cloned 8 years ago (Braganca et al., 2002; Yahata et al., 2002), but very little is known about its function. We demonstrate here that CITED4 is expressed in the heart and its expression is increased with exercise. CITED4 levels are negatively regulated by C/EBPβ expression, while experimental alterations of CITED4 levels in primary cardiomyocytes regulate proliferation. Forced expression of CITED4 also increased levels of cyclinD1, which is known to drive cardiomyocyte proliferation (Campa et al., 2008). Thus, it is highly plausible that C/EBPβ exerts its anti-proliferative actions via inhibition of CITED4 expression. However, we cannot exclude the possibility that other pathways might also contribute.
C/EBPβ reduction in the zebrafish embryo induced significant proliferation which, together with data provided from mice, suggests an evolutionarily conserved pathway in cardiomyocytes. There were no effect on cardiomyocyte sizes in the fish embryos; this could reflect a species difference or could be explained by the particular developmental stage at which the oligonucleoties were utilized.
The C/EBPβ heterozygous mouse closely phenocopied the endurance exercise model, as judged from cardiac hypertrophy, cardiomyocyte proliferation and expression of key genes of cardiac function. In fact, 42% of the gene set analyzed was regulated in the same manner by both of these perturbations in vivo. The magnitude of the effect on hypertrophy at baseline, however, was smaller than in the exercised mice, suggesting that additional pathways are likely to contribute to the changes induced in physiological hypertrophy.
Although exercise has well known positive health effects, possible benefits of physiological cardiac hypertrophy remain unclear (such as improved outcome following pathological provocations) in either mice or man. However, we were able to demonstrate that hearts in C/EBPβ heterozygous mice had improved tolerance to pressure overload. This included resistance to deterioration in cardiac function (fractional shortening) after trans-aortic constriction and improved survival. These data are also important because trans-aortic constriction is a very selective stress to the heart, and thus would not be confounded by systemic effects of reduced C/EBPβ in the total body heterozygotes. Mechanistically, the increase in cardiomyocyte proliferation could possibly contribute to the functional resistance to cardiac dysfunction; such an effect would be in accordance with resistance to injury connected to proliferation observed in other systems (Bersell et al., 2009). However, improved cellular energetics (via increased PGC1α expression), physiological, cellular hypertrophy or other functional alterations could also be important contributing factors. Better understanding of the pathways affecting C/EBPβ protein expression, or drugs that suppress C/EBPβ expression in the heart, could be of significant clinical value in cardiac pathological states.
EXPERIMENTAL PROCEDURES
Material
A list of utilized primers is presented in supplemental table II. Antibodies against PCNA, tubulin, C/EBPβ, actin, CITED4, ki67, phosphor-Histone 3, Aurora B, BrDU, β-tubulin, α-actinin and non-immune IgG were from Abcam. Antibodies to SRF and C/EBPβ were from Santa Cruz Biotechnology. HRP conjugated secondary antibodies were from Jackson Immunolaboratories research. Alexa Flour (488 and 633) conjugated antibodies were from Invitrogen. Pre-coated plates were from BD bioscience. Chamber slides for microscopy were from Nalge Nunc International. BioPix iQ 2.0 were obtained from BioPix AB (Gothenburg, Sweden). 35S-Methionine was purchased from Perkin Elmer (Boston, MA). The Chromatin Immunoprecipitation kit was from Cell signal.
Mice cohorts
For the swimming experiments, 12 week old B6 mice were used, purchased from the Jackson Laboratory, Bar Harbor, Maine. C/EBPβ heterozygous mice were purchased from the Jackson Laboratory and all wild type controls used were age matched littermates. The following IACUC approved protocols controlled the experiments; 005-2009, 110-2008, 056-2008, #04650.
Swimming exercise protocol
Endurance exercise was carried out as described (Taniike et al., 2008). Briefly, mice swam in a ramp protocol starting with 10 minutes 2 times daily with 10 minutes increase each day until 90 minutes, 2 times per day was reached. The protocol was ended after totally 14 days. The mice were closely observed at all times to avoid relative hypoxia. With this protocol and using these mice strains, there were no events of mice submerging under the water surface.
Transverse aortic constriction (TAC)
TAC was carried out as described in (Arany et al., 2006) using a 27 gauge needle. Subsequently, mice were followed weekly using echocardiography and sacrificed at fractional shortening <30 (4 S.D. Below baseline) and/or cachexia.
Echocardiographic studies
Cardiac sonograms were carried out as described (Arany et al., 2006). Briefly, the heart was first visualized in long/short axis views followed by m-mode analysis of short axis. Care was used to obtain exactly symmetrical short-axis images at the level of papillary muscles. At least three measurements were obtained and averaged for every data point from each mouse.
RT-PCR
QPCR was carried out following standard procedure using SYBR green. All data was normalized to 18S or indicted in-house gene and quantitative measures obtained using the delta-delta-CT method. A list of primers can be found in supplemental Figure II.
Western blot and quantification and co-precipitations
Protein amounts from all samples were assessed using the BCA-kit (Thermo Scientific) followed by protein concentration normalization prior to all western blot experiments. Western blot was carried out following standard procedure and final band intensity (QL-BG) was quantified using BioPix iQ(Bostrom et al., 2010). All data was normalized to background and loading controls. Precipitation of SRF was carried out as described in (Bostrom et al., 2007).
Adenoviral construction and preparation
Clones for CITED4 and C/EBPβ were purchased (Imagenes) as shuttle vectors, and moved to the pAD/CMV/V5-DEST vector using Gateway recombination (Invitrogen) in accordance with manufacturers’ instructions. Adenovirus were then produced using the ViraPower Adenoviral Expression System (Invitrogen) and viral stocks were purified using Vivapure Adenopack 100 (Sartorius Stedim Biotech). An MOI of 20 was used for all over expression experiments in primary cardiomyocytes. Characterization of over expression is shown in Supplemental Figure IV and VI. Adenovirus expressing AKT and the dnAKT was used as described in (Matsui et al., 2003; Nagoshi et al., 2005).
Immunohistochemistry from frozen sections
Tissues were snap frozen in OCT in liquid nitrogen and sectioned at 10μm. For staining, sections were fixed in 50% and 100% Acetone followed by washing in PBS. Next, samples were incubated in 5% BSA, 0,1% Triton x-100, PBS (buffert A) for 1 hour. After additional washing, slides were incubated over night with primary antibody in buffer A, and thereafter in secondary antibody for 1 hour. Finally, slides were washed extensively in PBS for 30 minutes and mounted in Prolong Gold with DAPI (Invitrogen).
Confocal miscroscopy and image quantification
Samples were visualized using a Leica SP5 confocal microscope at 40x magnification using standard procedure. All imaging was performed with group identity blinded where at least 20 random images were obtained from each slide or group. Images were then quantified in BioPix iQ 2.0 in accordance with manufacturer’s instructions (Bostrom et al., 2007; Hulten et al., 2010).
BrdU injections and subsequent staining procedure
BrdU was injected intraperitonally three days prior to the end of the swim protocol at 50 mg/kg. Cardiac sections were obtained as described above, and prior to staining, the slides were incubated in 1M HCl for 10 minutes on ice, followed by 2M Hcl for 10 minutes room temperature and 20 minutes in 37 degrees. Slides were then washed with 0,1M Borate buffer 10 minutes and rinsed in PBS. Thereafter, the previously described protocol for immunohistochemistry was utilized.
Rat neonatal primary cardiomyocyte culture and transfections
Primary rat neonatal cardiomyocytes were prepared as described in (Aoyama et al., 2005) with gradient centrifugation purification of cardiomyocytes. siRNA transfections was carried out using Lipofectamine 2000 (Invitrogen) as recommended by manufacturer and as described in (Bostrom et al., 2007). Two validated siRNA constructs against C/EBPβ and CITED4 with respective controls were purchased from Sigma Aldrich. Following examination of knockdown and lack of off-target effects, the most effective construct was used for subsequent studies.
Primary adult cardiomyocyte preparation
Primary mouse cardiomyocytes were isolated following Langendorf perfusion as described (Liao and Jain, 2007).
Nuclear density quantification
Nuclear density was assessed as described for “image quantification” above using confocal images. The number of α-actinin positive DAPI-nuclei per area of α-actinin positive cells was used.
Treadmill exercise
The acute exercise protocol included 40 minutes of forced exercise on a 0% incline treadmill (Columbus Instruments). The mice were trained twice briefly prior to experiment. The long term running was carried out 5 days per week for 4 weeks at a warm up speed of 5 m/min for 5 min. Every subsequent 5 min, the speed was increased by 5 m/min until a maximal speed of 20 m/min was reached to a maximum of 1 hour running. Estimate of maximal exercise capacity was performed using a V02-max treadmill with a ramp protocol maximizing running within 15 minutes at 20 degrees incline. Watts were calculated from using the incline per minute and mouse weight.
Cell size measurements and quantification in vitro and in vivo
Cell size measurements from cardiac tissue were performed from WGA (Invitrogen) stained tissues, where only exactly cross sectional cells (hexagonal) were used for BioPix iQ area measurements.
Protein biosynthesis assay
Protein biosynthesis was assessed in vitro after incubation with methionine-free medium followed by a 1h pulse of 500μCi/well 35S-Met. Cells were then scraped, and protein was extracted using TCA (Bostrom et al., 2007).
Chromatin immunoprecipitation
Chromatin Immunoprecipitation was performed using the Simple ChIP kit (Cell Signaling) in exact accordance with manufacture’s instruction. Briefly, ten 10cm dishes were used in triplicates for all groups, and chromatin complexes were fixed using formaldehyde. Next, nuclear fractions were isolated, and DNA was digested enzymatically. The quality and length of digested fragments were verified on 1,5% gels. Finally, SRF was immunoprecipitated in parallel with non-immune IgG control and DNA was extracted from complexes. Finallay, DNA was analyzed for presence of Gata4 or myh6 promotor fragments containing SRE sites (primers used are presented in supplemental table II)
Zebrafish experiments
An antisense morpholino directed against C/EBPβ or a control morpholino was injected as described(Milan et al., 2006) at the one cell stage. Embryos were then analyzed at 24 or 36 hours as described. Morpholinos (Genetools) used were CCTCGTAAAAACCGGCCACTTCCAT for C/EBPβ and CCTCTTACCTCAGTTACAATTTATA for control.
Statistical analysis
Data is presented as mean ± SEM unless otherwise indicated. Unpaired, two-sided students t-test was used when indicated with p<0,05 considered statistical significant. When assessing multiple groups, One-Way ANOVA was utilized with turkeys Post Hoc test. Kaplan Meyer statistics was performed using Gehan-Breslow-Wilcoxon test. The statistical software used was SPSS 17.1.
Supplementary Material
Acknowledgments
We thank Shingo Kajimura for valuable discussion during the progression of the manuscript. This project was supported by NIH grant R01 HL085593. PB was supported by the Swedish Heart and Lung foundation, SSMF and the Wenner-Gren Foundation. JW was supported by a postdoctoral fellowship from the American Heart Association (Founders Affiliate #09POST2010078). NM is a trainee the Harvard/MIT Health Sciences and Technology Program and is supported by a Howard Hughes Postdoctoral Fellowship. PQ is supported by a T32 training grant. This research was supported in part by a Leducq Foundation Network of Research Excellence (BS, AR) and grants from the NIH (AR, BS). AR also gratefully acknowledges support from Judith and David Ganz, and the Maxwell Hurston Charitable Foundation. AR is a principal faculty member of the Harvard Stem Cell Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Matsui T, Novikov M, Park J, Hemmings B, Rosenzweig A. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation. 2005;111:1652–1659. doi: 10.1161/01.CIR.0000160352.58142.06. [DOI] [PubMed] [Google Scholar]
- Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson BR, Fernandez-Rodriguez J, Ericson J, Nilsson T, Boren J, et al. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Andersson L, Vind B, Haversen L, Rutberg M, Wickstrom Y, Larsson E, Jansson PA, Svensson MK, Branemark R, et al. The SNARE protein SNAP23 and the SNARE-interacting protein Munc18c in human skeletal muscle are implicated in insulin resistance/type 2 diabetes. Diabetes. 2010 doi: 10.2337/db09-1503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Braganca J, Swingler T, Marques FI, Jones T, Eloranta JJ, Hurst HC, Shioda T, Bhattacharya S. Human CREB-binding protein/p300-interacting transactivator with ED-rich tail (CITED) 4, a new member of the CITED family, functions as a co-activator for transcription factor AP-2. J Biol Chem. 2002;277:8559–8565. doi: 10.1074/jbc.M110850200. [DOI] [PubMed] [Google Scholar]
- Campa VM, Gutierrez-Lanza R, Cerignoli F, Diaz-Trelles R, Nelson B, Tsuji T, Barcova M, Jiang W, Mercola M. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J Cell Biol. 2008;183:129–141. doi: 10.1083/jcb.200806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol. 2002;53:147–157. [PubMed] [Google Scholar]
- DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, Muslin AJ. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. doi: 10.1161/CIRCULATIONAHA.105.595231. [DOI] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) regulatory cascade in cardiac physiology and disease. Circulation. 2007;115:2540–2548. doi: 10.1161/CIRCULATIONAHA.107.670588. [DOI] [PubMed] [Google Scholar]
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon M, Sealy L. Ras regulates the association of serum response factor and CCAAT/enhancer-binding protein beta. J Biol Chem. 1999;274:14224–14228. doi: 10.1074/jbc.274.20.14224. [DOI] [PubMed] [Google Scholar]
- Hulten LM, Olson FJ, Aberg H, Carlsson J, Karlstrom L, Boren J, Fagerberg B, Wiklund O. 15-Lipoxygenase-2 is expressed in macrophages in human carotid plaques and regulated by hypoxia-inducible factor-1alpha. Eur J Clin Invest. 2010;40:11–17. doi: 10.1111/j.1365-2362.2009.02223.x. [DOI] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, Ferreira-Martins J, Goichberg P, Rondon C, D'Amario D, et al. Cardiomyogenesis in the Adult Human Heart. Circ Res. 2010 doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao R, Jain M. Isolation, culture, and functional analysis of adult mouse cardiomyocytes. Methods Mol Med. 2007;139:251–262. doi: 10.1007/978-1-59745-571-8_16. [DOI] [PubMed] [Google Scholar]
- Matsui T, Nagoshi T, Rosenzweig A. Akt and PI 3-kinase signaling in cardiomyocyte hypertrophy and survival. Cell Cycle. 2003;2:220–223. [PubMed] [Google Scholar]
- Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- Nagoshi T, Matsui T, Aoyama T, Leri A, Anversa P, Li L, Ogawa W, del Monte F, Gwathmey JK, Grazette L, et al. PI3K rescues the detrimental effects of chronic Akt activation in the heart during ischemia/reperfusion injury. J Clin Invest. 2005;115:2128–2138. doi: 10.1172/JCI23073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M, Dey A, Gerard RD, Hill JA, Rothermel BA. The CCAAT/enhancer binding protein beta (C/EBPbeta) cooperates with NFAT to control expression of the calcineurin regulatory protein RCAN1-4. J Biol Chem. 2010;285:16623–16631. doi: 10.1074/jbc.M109.098236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian T, Johnson PF. Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle. 2006;5:953–957. doi: 10.4161/cc.5.9.2733. [DOI] [PubMed] [Google Scholar]
- Taniike M, Yamaguchi O, Tsujimoto I, Hikoso S, Takeda T, Nakai A, Omiya S, Mizote I, Nakano Y, Higuchi Y, et al. Apoptosis signal-regulating kinase 1/p38 signaling pathway negatively regulates physiological hypertrophy. Circulation. 2008;117:545–552. doi: 10.1161/CIRCULATIONAHA.107.710434. [DOI] [PubMed] [Google Scholar]
- Yahata T, Takedatsu H, Dunwoodie SL, Braganca J, Swingler T, Withington SL, Hur J, Coser KR, Isselbacher KJ, Bhattacharya S, et al. Cloning of mouse Cited4, a member of the CITED family p300/CBP-binding transcriptional coactivators: induced expression in mammary epithelial cells. Genomics. 2002;80:601–613. doi: 10.1006/geno.2002.7005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.