Abstract
RNA molecules take advantage of prevalent structural motifs to fold and assemble into well-defined 3D architectures. The A-minor junction is a class of RNA motifs that specifically controls coaxial stacking of helices in natural RNAs. A sensitive self-assembling supra-molecular system was used as an assay to compare several natural and previously unidentified A-minor junctions by native polyacrylamide gel electrophoresis and atomic force microscopy. This class of modular motifs follows a topological rule that can accommodate a variety of interchangeable A-minor interactions with distinct local structural motifs. Overall, two different types of A-minor junctions can be distinguished based on their functional self-assembling behavior: one group makes use of triloops or GNRA and GNRA-like loops assembling with helices, while the other takes advantage of more complex tertiary receptors specific for the loop to gain higher stability. This study demonstrates how different structural motifs of RNA can contribute to the formation of topologically equivalent helical stacks. It also exemplifies the need of classifying RNA motifs based on their tertiary structural features rather than secondary structural features. The A-minor junction rule can be used to facilitate tertiary structure prediction of RNAs and rational design of RNA parts for nanobiotechnology and synthetic biology.
INTRODUCTION
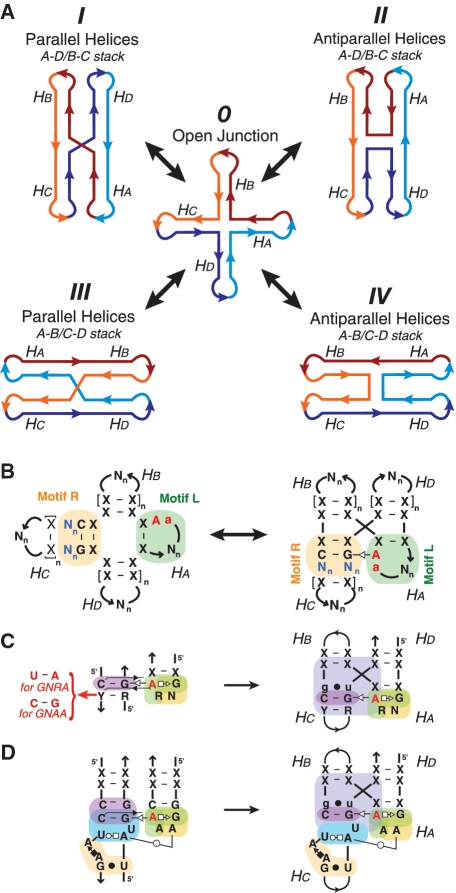
Recent studies strongly support the concept that the conformational search of RNA folding is highly context dependent, relying on hierarchical rules for guiding the compaction of RNAs into well-defined complex shapes in the presence of divalent ions (1). These context-dependent rules can be identified by looking for recurrent structural and sequence patterns (or motifs) in X-ray structures of RNA molecules. Among others, multi-helix junctions are important structural motifs that play key biological roles in DNA recombination as well as structural roles in numerous biological RNA molecules, including ribozymes, ribosomes and riboswitches (2,3). Like DNA helix junctions, RNA helix junctions formed of regular RNA helices are known to fluctuate between multiple different folded states during the conformational search for the most stable native structure (4). These states consist of different alternative helical stacking conformers with either parallel or anti-parallel arrangement of adjacent stacks (Figure 1A, conformers I, II, III and IV) that interchange through the open conformer (Figure 1A, conformer 0) (2,5). Pre-formed RNA helices have a tendency to stack coaxially on adjacent helices to maximize base stacking (6), stabilizing the different possible stacking conformers versus the open conformer 0 (Figures 1A and Supplementary Figure S1).
Figure 1.
A-minor topological rule and 4WJ motifs. (A) Alternative stacking conformers of a 4WJ motif. In this model, adjacent helices can be oriented either parallel (I and III) or antiparallel (II and IV). Two possible helix stacking patterns are possible, Ha over Hd and Hb over Hc (I and II), or Ha over Hb and Hd over Hc (III and IV). Four-way RNA junctions can rearrange to the different conformers through the open state 0 (4) (see also Supplementary Figure S1). However, the A-minor interaction can properly form between stems Ha and Hc only when the 4WJ is in conformation I. (B) Definition of the topological folding rule for RNA A-minor multi-helix junctions: a loop-receptor interaction between Motif L on stem Ha and Motif R on stem Hc stabilizes the parallel helix stacking conformer. (C) Diagrams of the generic GNRA/helix interaction with phylogenetic variations (left) and the A-minor junction of the region (H57-H59) from archaeal 23S rRNA (right). (D) Diagram of the GAAA/11 nt as a loop–receptor interaction and as embedded in the context of the A-minor junction. For the annotation of the motif sequence signatures, see legends of Figures S3 and 4.
The precise control of the folding of some RNA multi-helix junctions can be obtained through sequence-specific tertiary interactions between two constituent helical elements positioned adjacent to the strand-crossing junction, stabilizing one species in the conformational search over the three other possible arrangements. For example, the hairpin ribozyme is a self-cleaving RNA that consists of an anti-parallel four-way junction (4WJ) stabilized by two internal loops that interact to form the catalytic site (7,8) (see also Supplementary Figure S2A). In absence of the interacting loops the hairpin ribozyme junction fluctuates between parallel and anti-parallel helix orientations (4).
The isolated hairpin ribozyme junction (lacking catalytic site) has previously been adapted to construct tectoRNAs that assemble into fibers through loop-receptor interfaces under conditions of high magnesium salt (15 mM Mg2+) (9,10). It was found that the isolated hairpin junction adopts a single-dominant conformation (conformer II) at high-salt conditions (Figure 1A and Supplementary Figure S2). However, at low magnesium salt (1 mM Mg2+), the junction was found to be in equilibrium between multiple conformers.
In contrast to the fast dynamics observed for the hairpin ribozyme junction, numerous three- and four-way RNA junctions found in ribosomal and other RNA crystal structures are likely to be structurally stable (9,11,12). Many natural RNA junctions containing bulges of non-canonical base pairs (bp) are known to form stable coaxial stacking interactions (6), although they might be difficult to predict computationally in absence of structural data. Furthermore, a large number of natural three- and four-way junction motifs in RNA structures appear to be stabilized through tertiary interactions either present within the junction or adjacent to the junction (e.g. hairpin ribozyme, hammerhead ribozyme and ribosome). Interestingly, some of these RNA junctions are markedly different from the anti-parallel hairpin ribozyme junction as they can specify for parallel-helix stacks through stabilization by A-minor interactions (3,13,14) (Supplementary Table S1).
Herein, we describe a class of motifs called A-minor junctions (Figure 1B) (11). While three-way junction (3WJ) motifs belonging to this class were previously reported (3), we show that this class is defined by a more general topological rule that controls helical stacks in multi-helix junctions. This junction can be engineered such that numerous A-minor interacting motifs can be equivalently interchanged, with some permutations leading to novel and previously unidentified RNA folds (Figure 1C and D). Using a sensitive self-assembling molecular system, we have experimentally investigated the preferential helical topology induced by several natural and artificial A-minor junctions by site-directed mutagenesis, native polyacrylamide gel electrophoresis (PAGE) assays and atomic force microscopy (AFM). This work emphasizes the remarkable modularity of this class of topological motifs, exemplifying that different structural motif can contribute to the formation of topologically equivalent helical stacks. It also illustrates the importance of classifying RNA motifs based on their tertiary structural features rather than secondary structure elements.
MATERIALS AND METHODS
Identification of A-minor junction tertiary motifs in X-ray crystal structures
A-minor junction motifs were identified within published X-ray crystal structures by manually searching for anti-parallel multi-helix junctions with A-minor interaction signatures. In addition, an automated search for A-minor junction signatures was conducted using the computer program FR3D (15). The results of this exhaustive search are presented in Supplementary Table S1. Base-pair (bp) interactions and tertiary contacts used to annotate the various sequence signatures of motifs presented herein are indicated in the legend of Figures S3 and 4.
TectoRNA design and computer modeling
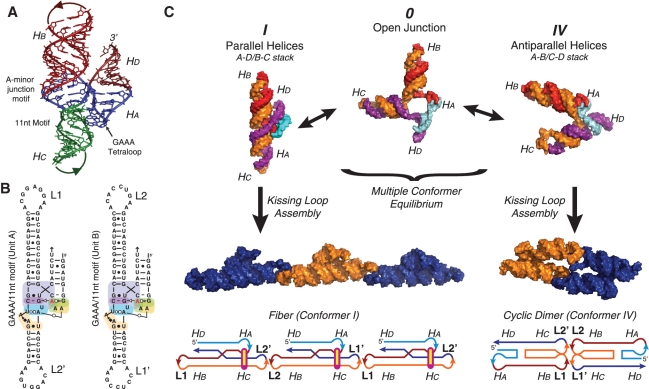
A-minor junction tectoRNAs were engineered by designing their 3D models with the computer program swissPDB viewer (16) following the RNA architectonics guidelines (17). The construction of their generic 3D template was created by compositing the X-ray structures of the A-minor junction (H57–H59) from Haloarcula marismortui (H. marismortui) 23S rRNA (PDB_ID: 1JJ2) and the DIS HIV kissing–loop interaction (PDB_ID: 1JJM; see also Supplementary Figure S4). The lengths of helices Hb and Hc were chosen such that the kissing–loops (KL) interactions are oriented in the same direction relative to the long axis of the structure. This ensures that both KL interactions are able to link up when the tectoRNAs fold into conformer IV to form a cyclic dimer (Figure 1A and 2C). The crystal structure of the hairpin ribozyme junction (PDB_ID: 1M5K) was used to guide the modeling of conformer IV (Figure 2C). The generic 2D structures corresponding to variants of the general A-minor junction tectoRNAs were all designed to keep the length of Hc and Hb constant. Additionally, computer models corresponding to the conformer I fibers and conformer IV cyclic dimers were built to estimate their scale. In the model of the conformer I fibers, the 4WJ motif creates a bulky density with a periodic spacing between units of ∼9 nm. In comparison, the computer model of conformer IV cyclic dimers is a rod ∼13-nm long with the bulky junction motifs located at the ends. The sequences of the RNAs were manually designed to fold into the desired 2D scaffold structure and were optimized using the computer software mFold to select the best folding of numerous manually submitted sequences (18). The list of sequences from all A-minor tectoRNAs used in this study is provided in the Supplementary Data (Supplementary Table S2).
Figure 2.
Assembly scheme for tectoRNAs. (A) Computer model of the A-minor junction showing the shape of the 4WJ in the context of the tectoRNA. (B) 2D sequence of the two molecules (A+B) used to build GAAA/11-nt-motif fibers. Each motif is tested in a tectoRNA system composed of two molecules that self-assemble end to end (via KL interactions between loops L1 and L1′, and loops L2 and L2′, see also Supplementary Figure S4). This is the same 2D template used to construct all other variants of the A-minor junction. (C) TectoRNAs undergo a folding equilibrium between stacking conformers I and IV. Depending the flexibility of the fiber, the tectoRNAs might assemble into intermediate structures that are closed rings of even numbers of RNA units (an even, rather than odd, number of units results from the A+B KL programming). Structural models were visualized in PyMOL (pymol.sourceforge.net).
TectoRNA synthesis and assembly
TectoRNAs were synthesized by in vitro T7 run-off transcription from PCR generate templates, purified by denaturing PAGE and 3′-[32P]pCp labeled as previously described (19). Self-assembly samples are prepared by mixing equimolar concentrations of two tectoRNAs in water. After denaturation (3 min, 90°C; 3 min, 4°C; 3 min, 30°C), samples were renatured in 10 mM Tris–borate pH 8.2 (TB), 50 mM KCl and 1.0 mM Mg(OAc)2 at 30°C for 30 min. For AFM and native PAGE, Mg(OAc)2 concentration was typically raised to 15 mM and heated at 50°C for 10 min before cooling to 4°C. For visual monitoring on PAGE gels, 1 nM of molecule A from each set has been radiolabeled with [32P]pCp.
Native PAGE
Native 7% (29:1) PAGE experiments were performed as previously described (20). RNA samples were prepared at variable concentrations in water, subjected to denaturing/renaturing protocol of heating to 90°C for 2 min, and cooling on ice (4°C) for 3 min prior to adjustment to final volume in 1 mM Mg(OAc)2, 50 mM KCl and TB 1× buffer. RNAs were incubated at 30°C for 30 min and cooled on ice before addition of blue loading buffer and PAGE migration. All gels contained 1 mM Mg(OAc)2 and TB 1×, and were migrated at a maximum temperature of 10°C for 4 h.
AFM
Samples were prepared as for PAGE studies in 1 mM Mg(OAc)2, 50 mM KCl and TB 1× buffer. Twenty microliters of tectosquare samples (100 nM RNA) stabilized in 8 mM Mg(OAc)2 buffer were directly deposited on freshly cleaved mica surface for 1 min, rinsed with a 2 mM Mg(OAc)2 solution and water, then dried with nitrogen. Imaging in air at ∼20°C was performed in tapping mode on a Multimode AFM equipped with a Nanoscope IIIa controller (Veeco, Santa Barbara). Silicon probes (model NSC12 from MikroMesch) with resonance frequency ∼150–250 kHz and spring constant ∼4–8 N/m (Nanodevices, Santa Barbara, CA, USA) were used. Images were processed by NanoScope® (DI) and leveled by a first order plane fit in order to correct the sample tilt. Samples of the 11-nt GAAA fibers prepared and incubated at higher magnesium concentration (15 mM Mg2+) and visualized by AFM assembled primarily into dimers (data not shown).
RESULTS
A topological rule for specifying parallel helical stacks in RNA multi-helix junctions
A tertiary topological rule is defined by a recurrent set of conserved tertiary contacts that promotes a recurrent structural topology of RNA helices. Careful analysis of all known RNA X-ray structures recently revealed such a rule in several natural RNA molecules, including the 16S and 23S ribosomal RNAs (rRNAs) and RNase P RNAs. For instance, several structural motifs present at the level of multi-helix junctions, such as 3WJ and 4WJs (Supplementary Table S1), share a conserved A-minor interaction (13,21–23) that promotes parallel-oriented coaxial stacking of helices (Figure 1A, conformer I). This topological rule is best described by the schematic displayed in Figure 1B. The A-minor interaction occurs between helix Ha comprising an adenine bearing motif that is typically a terminal or internal loop and helix Hc comprising a receptor motif containing a conserved G:C Watson-Crick (WC) bp, and favors the parallel coaxial stack of Ha over Hd and Hb over Hc in the junction (Figure 1B). In each of the alternative stacking conformers, the formation of the stabilizing A-minor interaction is not possible either because the loop motif in Ha and the receptor in Hc are not adjacent (Figure 1A, conformers II and IV) or because the interacting side of the loop motif in Ha does not face the minor groove of Hc (Figure 1A, conformer III) (see also Supplementary Figure S1 for stereo-views). Multi-helix junction motifs that follow this topological rule are called A-minor multi-helix junctions. Interestingly, they can have tertiary structures significantly different from one another, with a root mean square deviation (RMSD) of their ribose-phosphate backbone of up to 5 Å. The conserved A-minor interaction can indeed result from interacting loop/receptor motifs with remarkably different local structures (Supplementary Figure S3). Moreover, the A-minor topological ‘rule’ remains conserved irrespective of the number of helices involved and the connectivity of strands between them. Some motifs can lack stems Hb or/and Hd, leading to kink turns or 3WJ motifs (Supplementary Table S1A).
4WJ-tectoRNA design and self-assembly assay
Based on the recurrence of the generic A-minor junction fold in several natural RNA molecules, we hypothesized that A-minor junction motifs could promote parallel coaxial stacking in engineered self-assembling RNAs (4WJ-tectoRNAs), irrespective of the global structural context of their molecules of origin. Following the RNA architectonics design approach (17), the 3D structure of the A-minor junction (H57-H59) of the H. marismortui 23S rRNA was used as the generic 3D template to engineer a self-assembling bi-molecular (A+B) system. It consists in various pairs of 4WJ-tectoRNAs able to self-assemble through two different HIV-like KL interactions localized at the end of helical stems Hb and Hc (Figure 2 and Supplementary Figure S3, ‘Materials and Methods’ section) (24). The length of Hb and Hc were designed such that these tectoRNAs (Figure 2B) may either assemble end-to-end into linear or circular multimers, when Hb is stacked on Hc as in conformer I, or into cyclic dimers, when Ha is stacked on Hc as in conformer IV (Figure 2C). As all sets of 4WJ-tectoRNAs that we tested assemble through identical KL interactions, the self-assembly properties of the tectoRNAs can be used to assess and compare the contribution of different A-minor junctions. Indeed, based on the sequence, tertiary structure and energetic stability of the A-minor junctions present within these tectoRNAs, the conformational equilibrium between conformers I and IV can be modulated by varying RNA concentration to lead to three possible trends. When the A-minor junction locks the structure into a rigid conformer I, tectoRNA assembly through KL should favor linear multimers or fibers (Figure 2C, left panel). In contrast, unstable, highly dynamic A-minor junctions that can easily adopt conformer IV should lead to the entropically driven formation of cyclic dimers (Figure 2C, right panel). Stable but more dynamic A-minor junctions should lead to a somewhat intermediate situation, with formation of circular multimers or rings of an even number of units.
The 4WJ-tectoRNA self-assembly equilibrium is dependent on RNA concentrations as well as salt concentrations, especially magnesium, which contributes to the stability of most if not all RNA tertiary motifs. For comparing the self-assembly properties of various 4WJ-tectoRNAs, the magnesium concentration, therefore, needs to be carefully adjusted as it should promote stabilization of the junction motifs as well as induce supra-molecular assembly through KL interactions at low RNA concentrations.
Exploring 4WJ-tectoRNA self-assembly by native PAGE
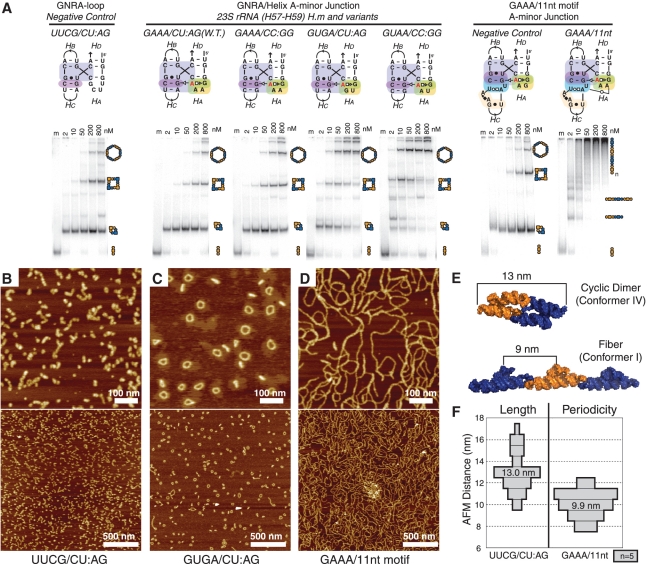
Non-denaturing PAGE (native PAGE) was used as the primary technique for comparing the assembly properties of various bi-molecular A+B 4WJ-tectoRNAs differing from one another at the level of their A-minor junction. After folding and assembly of corresponding pairs of 4WJ-tectoRNAs in ‘one pot’ at various RNA concentrations, the resulting self-assembly products were separated by native PAGE analysis (see ‘Materials and Methods’ section). Experiments were mostly done at 1 mM Mg2+. Previous results indicated that KL1 and KL2, the two KLs used for the construction of the A+B bimolecular system, assemble at 0.2 mM Mg2+ and 10°C with apparent Kds of 5.6 nM and 15.8 nM, respectively (25). In contrast, at 1 mM Mg2+, their Kds are in the high picomolar range. These conditions were shown to favor tectoRNA assembly into a variety of self-assembly products within the range of RNA concentrations tested (2–800 nM) (Figure 3).
Figure 3.
Analysis of different A-minor 4WJ motifs. (A) Native PAGE at 1 mM Mg(OAc)2 and TB 1× showing the end-to-end assembly of tectoRNAs containing different GNRA/helix and GNRA/receptor A-minor junction sequence signatures. Each experiment includes one lane (lane m) loaded with only one molecule (A) of the two (A+B), as a monomer standard. The other lanes in each set contain equal concentrations (nanomolar) of the two molecules A+B, assembled at 30°C in presence of 1 mM Mg(OAc)2, 50 mM KCl and TB 1×buffer. Higher molecular weight products are indicative of increased stability of conformer I, while low molecular weight products likely represent assemblies forced into conformer IV by the entropic closure of KL interactions. (B–D) A 600-nm scale AFM images were acquired in air under tapping mode. RNA samples were prepared at 200 nM, in 1 mM Mg(OAc)2, 50 mM KCl, TB 1× buffer and were adjusted to 100 nM RNA and 8 mM Mg(OAc)2 immediately before deposition on mica and imaging. (B) Complexes formed from UUCG/CU:AG A-minor junction motif. (C) Complexes formed from GUGA/CU:AG A-minor junction motifs. (D) Complexes formed from the GAAA/11-nt A-minor junction hybrid motif. (E) 3D computer model showing the expected size of the cyclic dimer and the expected periodicity of bulky features on the fiber (see also Supplementary Figure S5). (F) Histogram showing the average measured size of cyclic dimers observed in the UUCG 4WJ sample and average measured periodicity measured for fibers in the GAAA/11-nt-motif sample. W.T., wild type; H.m, H. marismortui.
The A+B 4WJ-tectoRNA system was first used to investigate the behavior of the hairpin ribozyme junction (Supplementary Figure S2), previously characterized to be in dynamic equilibrium between various conformers at 1 mM Mg2+ (10). As expected, the hairpin ribozyme tectoRNAs primarily assemble into cyclic dimers regardless of the orientation of the junction within the tectoRNA (Supplementary Figure S2C). However, increasing the magnesium concentration to 15 mM, a condition previously shown to stabilize the hairpin junction (10), failed to induce multimerization of the tectoRNAs designed to fold into the conformer II direction (data not shown). The KL interactions apparently become stabilized by magnesium ions to a greater extent than the junction motif, trapping the 4WJ-tectoRNAs in cyclic dimers. All subsequent 4WJ-tectoRNAs self-assembly experiments were therefore performed at 1 mM Mg2+.
Characterization of A-minor junctions involving GNRA tetraloops
The H57–H59 A-minor junction from the 23S rRNA from H. marismortui and several variants were then compared with the hairpin ribozyme junction to determine if energetic differences in the folding of A-minor junctions can have appreciable effects on the 4WJ-tectoRNA assemblies. While this particular A-minor junction is not universally conserved in all rRNAs, it comprises a GNRA tetraloop/helix interaction that is one of the most prevalent and well-characterized A-minor interactions found in stable RNAs (20,26,27) (GNRA loop sequences can have any nucleotide (N) at the second loop position and any purine (R) at the third loop position). In the context of H. marismortui H57-H59 junction, the GNRA/helix interaction involves a GAAA tetraloop recognizing a CA:UG bp duplex within a regular A-form helix. Interestingly, this sequence combination, while properly folded within the context of the 23S rRNA X-ray structure (13,23), deviates from most phylogenetically conserved GNRA/helix interactions found in stable RNAs, like group I introns (26,28) and RNase P RNAs (29) (Figure 1C). Typically, CA:UG bp duplexes are recognized by GYRA tetraloops, with a preference for GYGA versus GYAA. In contrast, CC:GG duplexes are essentially recognized by GNAA tetraloops, with a strong preference for GYAA versus GRAA.
Besides the H. marismortui H57–H59 natural sequence, other A-minor 4WJ-tectoRNAs employing different combinations of tetraloops with helix duplexes (either CA:UG or CC:GG) were created to determine if the phylogenetic trend for GNRA/helix interactions applies within the context of multi-helix junctions. Additionally, a set of 4WJ-tectoRNAs replacing the GNRA tetraloop by a UUCG tetraloop, with a completely different structure unable to form the A-minor interaction, was built as a negative control (UUCG/CU:AG in Figure 3A). As shown in Figure 3A, H. marismortui H57-H59 4WJ-tectoRNAs (GAAA/CU:AG) form larger self-assembly products than the hairpin constructs at RNA concentrations >200 nM. However, in the conditions tested (1 mM Mg2+), there is almost no difference with the negative control. In contrast, all 4WJ-tectoRNA variants with phylogenetically conserved GNRA/helix interactions show a markedly improved ability to generate large molecular species in comparison with H. marismortui H57–H59 and negative control 4WJ-tectoRNAs. For instance, as predicted by the GNRA/helix covariations, the CU:AG helical duplex is recognized best by GUGA with very few 4WJ-tectoRNAs remaining trapped into cyclic dimers at 800 nM. The CC:GG helical duplex clearly improves the ability of the GAAA tetraloop to promote multimerization of 4WJ-tectoRNAs. However, as predicted by the phylogeny rule, the CC:GG duplex assembles best when combined to GUAA. Above 200 nM, GUAA/CC:GG 4WJ-tectoRNAs form essentially large, discreet self-assembly species similar to those obtained with GUGA/CU:AG 4WJ-tectoRNAs. These results are also in good agreement with the trend in thermodynamic stability observed for these various GNRA/helix interactions within the context of self-assembling RNA dimers as determined by Surface Plasmon Resonance (S. Baudrey, J. Hoebeke and L. Jaeger, unpublished data). Our data demonstrate that the natural H. marismortui H57–H59 A-minor junction is suboptimal for promoting efficient tectoRNA multimerization. Therefore, this motif is likely to be meta-stable in absence of other stabilizing interactions within the 23S rRNA context.
In order to characterize the structure of the self-assembly products identified by native PAGE gels at 200 nM of RNAs and 1 mM Mg2+, AFM analysis were performed on UUCG and GUGA/CU:AG 4WJ-tectoRNAs assembled in similar conditions (25). As expected from native PAGE, UUCG 4WJ-tectoRNAs essentially assemble in small RNA particles (Figure 3B) that are ∼13 nm in length. According to computer 3D modeling, this corresponds to the size expected for cyclic dimers formed of two 4WJ-tectoRNAs in conformer IV (Figure 3E and F). In contrast, GUGA/CU:AG tectoRNAs essentially form discreet nano-rings containing an even number of end-to-end assembled units (Figure 3C). The approximate circumference for these discrete objects corresponds mostly to the size expected for hexamers and octamers, and to a lesser extent tetramers and dodecamers (see ‘Materials and Methods’ section and Supplementary Figure S5). Interestingly, the relative proportion of nano-objects of different sizes estimated by AFM is in good agreement with the relative proportion of self-assembled species visualized by native PAGE.
Fiber formation through A-minor 4WJ-tectoRNAs stabilized by complex GNRA/receptor interactions
In contrast to GYRA tetraloops that frequently interact with regular A-form helices, GAAA tetraloops are often involved in a much more selective and stable interaction with the 11-nt receptor, a prevalent motif found in stable natural RNAs (20,30) (Figure 1D). In addition to the A-minor interaction, some nucleotides within the 11-nt receptor form additional specific contacts with the second position of the GAAA tetraloop, leading to a tertiary interaction highly selective for GAAA (20,31). The GAAA/11-nt receptor interaction is one of the most stable tertiary interactions mediated by GNRA tetraloops, and its sequence signature is known to tolerate nucleotide variations in some of its structural components. For instance, one of the usually conserved C:G bp adjacent to the bp directly involved in the A-minor interaction is found substituted by a G:U bp in some natural RNA sequences [Figure 11 in reference (27)]. Within the context of tectoRNA dimers (19,20,32), this variation accounts for a 0.55 kcal/mol decrease in stability with respect to the canonical 11-nt receptor at 15 mM Mg2+ (E. Calkins and L. Jaeger, unpublished data).
To our knowledge there is no instance of natural sequence where a GAAA/11-nt receptor interaction is found within the context of the A-minor junction topological rule. Nevertheless, based on the respective sequence signatures and 3D structures of the GAAA/11nt and GNRA/A-minor junction motifs, this new composite A-minor interaction can easily be engineered (Figures 1D and 2A and B). Considering that the GAAA/11-nt receptor is thermodynamically more stable than the GYRA/helical receptor interaction, we anticipated that its incorporation within the context of 4WJ-tectoRNAs would promote the formation of longer multimers. As observed by native PAGE at 1 mM Mg2+, GAAA/11-nt 4WJ-tectoRNAs assemble with a markedly different assembly pattern than the GNRA/helix constructs (Figure 3A). At RNA concentrations >50 nM, rather than forming several discreet bands corresponding to well-defined circular multimers, these 4WJ-tectoRNA form high molecular weight self-assembly products that remain essentially trapped on the top of the native gels suggesting the formation of long RNA nano-fibers. AFM structural characterizations of GAAA/11-nt 4WJ-tectoRNAs assembled at 200 nM in similar conditions reveal spaghetti-like fibers of several hundred nanometer in length, some of them involving more than 50 tectoRNAs units (Figure 3D). These fibers have a string-of-pearls appearance (Figure 3D) with a periodic spacing between peaks roughly corresponding to the expected unit length of the 4WJ-tectoRNA unit of ∼10 nm (Figure 3E and F). While most fibers appear to be linear, the relatively few observed circular structures are clearly larger than those observed for GYRA/helix 4WJ-tectoRNAs, demonstrating that the composite GAAA/11-nt A-minor junction is significantly more rigid than those based on GYRA/helix interactions.
In agreement with previous data that demonstrated the remarkable selectivity of the 11-nt receptor for GAAA tetraloop (20), a single point mutation replacing the GAAA loop by GGAA within the 11-nt-based 4WJ-tectoRNAs essentially abolishes nanofiber formation and mostly leads to cyclic dimer formation like for the UNCG negative control (Figure 3A, right panel). Interestingly, A-minor 4WJ-tectoRNAs based on the receptor R1, an in vitro selected receptor selectively recognizing GGAA tetraloop (20), were shown to behave like the 11-nt constructs, assembling either into nanofibers when recognizing their cognate loop (GGAA), or into cyclic dimers when combined with a non-cognate loop (GAAA) (Supplementary Figure S6A).
Characterization of other A-minor junction motifs
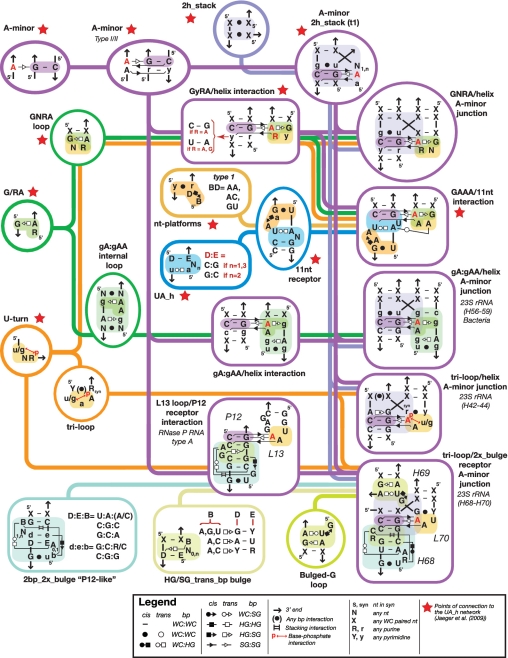
Besides GNRA/A-minor junctions, several other A-minor junctions have been identified in X-ray structures of natural RNA molecules (Supplementary Table S1). As emphasized by the results described above and the sequence motif network corresponding to several A-minor junctions (Figure 4 and Supplementary Figure S3), the remarkable aspect of the A-minor junction topological rule is that it is highly modular and can take advantage of different structural motifs as long as these motifs contribute to the formation of A-minor interactions. Small prevalent motifs enter into the makeup of various motifs of greater complexity, and can be interchanged with one another to promote the same topology of helices in a multi-helix junction. For instance, besides GNRA tetraloops, other loops such as triloops Y(u/gAA)R capped with a Y:R bp, or GA:NAA and sarcin internal loops can also interact with helices or receptors such as the ‘double-locked bulge motif’ (2bp_2x_bulge), to form A-minor interactions within the context of the A-minor junction (Figure 4 and Supplementary Figure S3).
Figure 4.
Structural motif network associated to A-minor junctions described herein. Submotifs combine in multiple ways to generate a variety of functionally equivalent A-minor junction motifs. The lines on the network indicate which motifs can be combined. Interactions are indicated according to the Leontis Westhof nomenclature (55) (see Legend). The Watson-Crick (WC), Hoogsteen (HG) and Shallow Groove (SG) edges of two nucleotides can interact with one another in cis or trans configuration to form the 12 major types of base pairings.
The 4WJ-tectoRNA system was used as an assay to compare several of these A-minor junctions to the previously characterized GNRA/A-minor junctions by assessing their ability to promote efficient parallel helical orientation. Furthermore, we also tested various natural long-range A-minor interactions within the short-range A-minor junction 4WJ-tectoRNA context to compare their stability with respect to other prevalent A-minor interactions.
A-minor junctions with GNRA-like motifs (GA:NAA internal and GN1-2RAN terminal loops)
Within the ribosome, some peripheral regions of the 23S rRNA can significantly vary between different branches of life. For example, the region H57–H59 of archaeal and bacterial 23S rRNAs adopt different global structures in domain 3. In contrast to the archaeal 23S rRNA and eukaryotic 28S rRNA, the H57–H59 interaction from the 23S rRNA of most bacteria features internal loop/helix interactions that replace the GNRA loop–helix interaction observed in H. marismortui H57–H59. These internal loops are GNRA-like structures that precisely mimic the molecular shape of the A-minor interacting face of GNRA tetraloops as they are built from the same small G/RA submotif known to participate in A-minor interactions (Figure 5); they are grouped into a class of motifs called GA:NAA internal loops that also include UAA:GAN (or GAN:UAA) interacting loops (11,33). Note that these GNRA-like internal loop motifs can also be seen as a subset of a broader class of motifs initially described as GNn:RA loop motifs (34), which includes all GNRA and GNRA-like terminal loop motifs (Supplementary Figure S7A). GNRA and GA:NAA internal loops are interesting examples of two classes of motifs with 3D structural similarities but different strand topologies (Supplementary Figure S8A and D).
Figure 5.
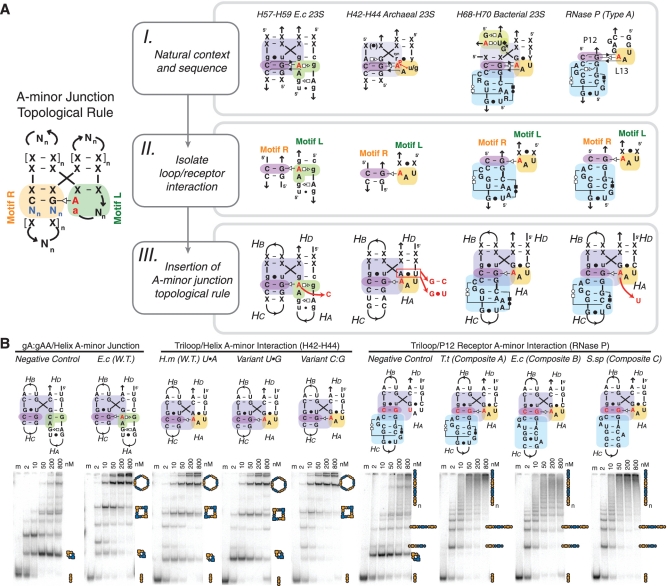
A-minor junction motif structural variants. (A) Scheme describing the engineering of new motifs fitting the A-minor-junction topological rule. (I) Examples of A-minor motifs as they appear in their natural context. Consensus sequences are shown for region H42–H44 of archaea and region H68–H70 of bacteria. (II) Isolation of the loop/receptor interaction from the natural motif. The interaction (left to right) are gA:gAA/helix interaction, triloop/helix interaction, triloop/H68–receptor interaction and L13-loop/P12–receptor interaction. (III) Insertion of the loop/receptor interaction in the context of the A-minor-junction topological rule. Red arrows indicate different mutants that are experimentally tested in (B). (B) Native PAGE at 1 mM Mg(OAc)2 and TB 1×buffer showing the end-to-end assembly of tectoRNAs containing the different 4WJ sequence signatures shown in (A). Negative controls expected to form cyclic dimers are indicated. PAGE gels were conducted under the same conditions as those presented in Figure 3A. W.T., wild type; 23S, 23S rRNA; E.c, E. coli; H.m, H. marismortui; T.t, T. thermophilus; S.sp, Synechococcus sp. PCC6301.
The H57–H59 gA:gAA/A-minor junction (Supplementary Figure S8D) from the 23S rRNA of Escherichia coli (E. coli) was incorporated within the 4WJ-tectoRNAs system to assess its contribution to self-assembly. As shown in Figure 5B, the natural gA:gAA/A-minor junction from E. coli is significantly more stable than the natural H57–H59 GAAA/CC:GG junction from the 23S rRNA of H. marismortui. In fact, it behaves remarkably similarly to the GUAA/CC:GG junction, one of the best variants of the GNRA/A-minor junction of H. marismortui H57–H59 (Figures 3A and 5B). As demonstrated by the negative control gA:gAC, which has a key interacting adenine from the gA:gAA loop changed into a C, the disruption of the A-minor interaction leads to destabilization of the parallel helical orientation (Figure 5B).
The prevalent A-minor interaction mediated by GAN:UAA loops was similarly tested in the 4WJ-tectoRNA system (Supplementary Figures S7B and S8E). In comparison with the previous GA:NAA interaction, the GAN:UAA/A-minor interaction proved to be comparatively less stable. It behaves like the natural H. marismortui H57–H59 GAAA/CC:GG junction. Despite lower stability, the A-minor contact in the GAN:UAA/A-minor interaction is, however, still important for favoring formation of higher order self-assembly products. Our results indicate that this particular GAN:UAA loop might likely be less thermodynamically stable than the gA:gAA internal loops. Nevertheless, the sequence that we tested might not be the most optimal GAN:UAA internal loop as other sequence variants are possible (33).
We investigated the assembly of an additional set of 4WJ-tectoRNAs to estimate the possible stabilizing contribution of another conserved natural GNRA-like/receptor from the 23S rRNA, which contributes to the docking of domain 2 with domain 5. This A-minor interaction occurs between the terminal loop L39 (pentaloop in Archaea, hexaloop in Bacteria) of domain 2 and a receptor in the stem H89 of domain 5 (Supplementary Figures S7AB and S8I). The receptor belongs to a class of prevalent motifs called ‘double-locked bulge motifs’ (2bp_2x_bulges) (see Supplementary Table S1B for a list). The nucleotide sequence corresponding to the loop/receptor interaction from H. marismortui 23S rRNA, however, does not promote multimer assembly of the corresponding 4WJ-tectoRNAs, suggesting that the proper assembly of this natural interacting motif is likely to be promoted by additional factors within the context of the ribosome. For instance, some of the nucleotide positions of the receptor in H89 are known to be post-transcriptionally modified in the E. coli 23S rRNA and are therefore likely to contribute to the folding and stability of this natural interaction.
A-minor junctions involving triloops
Triloop motifs Y(u/gNR)R are other small recurrent terminal loops characterized by a 3-nt loop with a Y:R non-canonical closing bp (Figure 5) (35,36). When the 3-nt loop sequence is (u/gNR), it typically folds as a U-turn submotif (37,38). The closing bp can adopt either an unusual Y:R cis WC:WC bp with the purine R in syn conformation, or a trans WC:HG bp. These types of triloops have been identified at the level of several A-minor junctions in five different locations within X-ray structures of RNA (Supplementary Table S1A). They contribute to the formation of a type I/II A-minor interaction with 2-bp duplexes much like in the GNRA/A-minor junctions (Supplementary Figure S8C). For instance, at several locations (e.g. H42–H44 and H90–H92 regions of the 23S rRNA and P10-P10.1 region of RNase P RNA of type B), UAA or GAA triloops are found interacting with CC:GG duplexes.
The sequences of Y(UAA)R/CC:GG A-minor junctions found in nature typically present a few additional nucleotide insertions/variations within their structures. Therefore, in order to facilitate comparison with our previous A-minor constructs involving GNRA/helix interactions, we generated various sets of 4WJ-tectoRNAs to test the contribution of the Y(UAA)R/CC:GG A-minor interaction only (Figure 5A). Several variants differing at the level of the closing bp of the triloop [variants U(UAA)G, U(UAA)A and C(UAA)G] or at the level of the triloop sequence [variant U(UAU)G] were generated to assess the contribution of these different structural features to the stability (and rigidity) of the A-minor junction. While disruption of the A-minor interaction [variant U(UAU)G) leads to 4WJ-tectoRNAs assemblies similar to the previous suboptimal GAAA/CU:AG 4WJ-tectoRNAs, all the closing bp variants promoted similar or better multimerization than GUGA/CU:AG constructs (Figures 3A and 5B). The observed trend is that the closing bp of U(UAA)G, U(UAA)A and C(UAA)G triloops lead to more stable A-minor junctions in the following order U:G < U:A < C:G (Figure 5B).
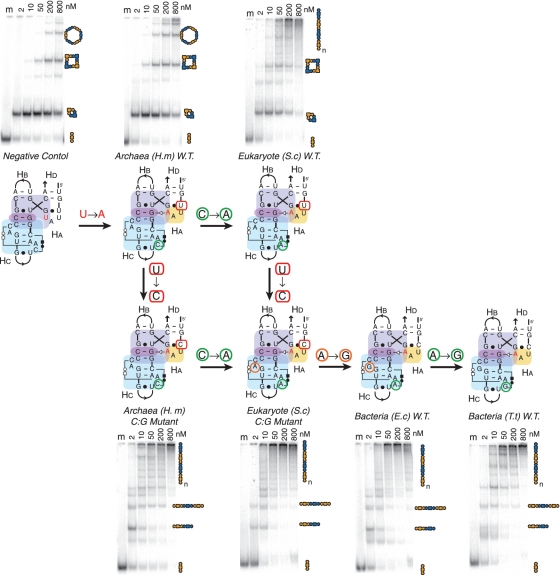
Like GNRA tetraloops, triloops can also recognize more elaborate receptors such as 2bp_2x_bulge motifs. For instance, the conserved H68–L70 triloop/A-minor junction from the 23S rRNA involves a 2bp_2x_bulge localized in H68. This type of motif is found at different independent locations in known RNA molecules (Supplementary Table S1B). More precisely, this triloop receptor 2bp_2x_bulge motif forms two specific base triple interactions, with one of them contributing to the formation of a stabilizing platform for the second nucleotide position of the L70 triloop (Figures 4 and Supplementary Figure S8H). The triloop/receptor interaction was transposed within the 4WJ-tectoRNA contexts (Figure 5A). A series of different versions of the H68–H70 interaction were tested, covering the highly conserved sequences from archaea (H. marismortui), bacteria [E. coli and Thermus thermophilus (T. thermophilus)] and eukaryotes [Saccharomyces cerevisiae (S. cerevisiae)] large rRNAs.
In comparison with the previous triloop/helix constructs, the H68–H70 junctions from bacteria (E. coli and T. thermophilus) behave more like the GAAA/11-nt receptor junction (Figure 6). The bacterial H68–H70 4WJ-tectoRNAs lead to the formation of multimeric self-assembly products that likely correspond to 1D fibers (Figure 6). Interestingly, the sequence of the triloop closing bp has a greater effect on the behavior of triloop/receptor A-minor junctions than on the previous triloop/helix constructs (Figures 5B and 6). In contrast to the bacterial (E. coli and T. thermophilus) H68–H70 A-minor junctions, which present C:G closing bps, the archaeal H68–H70 and eukaryotic H68–H70 A-minor junctions have both triloops with U:G closing bps. While a significant fraction of the wild-type archaeal and eukaryotic constructs assemble into cyclic dimers, the single point mutation changing the triloop closing bp from U:G to C:G dramatically enhances the formation of fibers in both cases (Figure 6), leading to self-assembly patterns similar to those observed for bacteria constructs. Additionally, the sequence of the base triples that lock the tertiary structure of the triloop receptor into a 2bp_2x_bulge motif can significantly contribute to the stabilization of the A-minor junctions. For instance, the wild-type archaea H. marismortui H68–H70 junction is far from being optimal for fiber self-assembly. Rather than promoting assembly into 1D fibers, it behaves like the U(UAA)G/CC:GG junction, forming instead well-defined multimeric nano-rings. In contrast, the wild-type eukaryote S. cerevisiae H68–H70 constructs, which differ from the H. marismortui junction by a single-point mutation (C to A) in one of the base triples stabilizing the 2bp_2x_bulge motif, assemble much more efficiently into longer 1D fiber products than the wild-type archaeal constructs (Figure 6). Apparently, based on our self-assembly assay, the cis WC:WC:HG base triples favors G:C:A and G:C:G combinations versus G:C:C. In contrast, the other single-point mutation distinguishing the eukaryote receptor from the bacterial receptor motif has no significant effect on self-assembly (Figure 6). It is, however, worth noticing that receptor positions not directly involved in the A-minor contacts can significantly affect its formation.
Figure 6.
Comparison of A-minor junctions from bacteria, archaea and eukaryotes. Variants of the H68-H70 A-minor junctions based on the natural sequences from H. marismortui (H.m), S. cerevisiae (S.c), E. coli (E.c) and T. Thermophilus (T.t) 23S rRNAs are compared by PAGE analysis. The receptors were designed by the method illustrated in Figure 5A. Additionally, two mutant H68–H70 junctions were prepared to allow all the receptors to be bridged by a series of single-point mutations. PAGE gels were run under the same conditions as those shown in the previous figures.
Another loop/receptor interaction occurring between the structural elements P12 and L13 from type A RNase P RNAs presents remarkable structural similarities with the H68–H70 triloop/receptor interaction (Figures 4 and 5A). For instance, L13 is a 7-nt loop with a U-turn sub-motif that forms an A-minor interaction with a 2x_bulge receptor located in P12. The 3D structures of the P12 and H68 receptors are similar (Supplementary Table S1B and Figure S8G and H). The sequences of the 2x_bulge receptors from various bacterial RNase P RNA species are, however, quite different from the H68 2x_bulge. Several composite A-minor junction 4WJ-tectoRNAs were generated using the L13 loop/P12 receptor interaction from RNase P RNAs of E. coli, T. thermophilus and Synechococcus sp. PCC6301 (S.sp PCC6301) (Figure 5B). Remarkably, they all assemble very effectively into 1D fibers. Likewise, the disruption of the A-minor interaction of the T. thermophilus P12 constructs leads primarily to cyclic dimers, verifying that these new composite A-minor junctions are truly A-minor interaction dependent (Figure 5B).
Modulation of RNA nanostructure assembly
We have demonstrated the effect of magnesium concentration and RNA unit concentration on the length of nanostructures formed, but the extent of formation of a particular set of self-assembling tectoRNA units into nanostructures can also be modulated by other means. By mixing and matching units containing comparatively stronger and weaker 4WJ-folds, we can further change the distribution of products. For example, when combining 1 unit of an A-minor junction tectoRNA that forms predominantly cyclic dimers with another that forms mostly hexamers, we obtain a product that is mostly tetramers (Supplementary Figure S6B). The assembly of several A-minor-junction motifs can be modulated to favor the formation of non-cyclic structures by altering the unit stoichiometry. For instance, at a fixed RNA concentration shifting the ratio of the A+B units moves the equilibrium toward formation of B-A-B trimeric species. A band corresponding to linear trimers was produced in all examples studied (Supplementary Figure S9). The T. thermophilus P12 and the E. coli H68-L70 constructs assembled into many linear species corresponding to odd numbers of units with the strongest band at the expected trimer level (Supplementary Figure S9). In contrast, the GAAA/CC:GG motif still assembled most dominantly into cyclic dimers despite a 23:1 stoichiometric ratio. Motifs that tend to circularize in the A+B assembly scheme become trapped as cyclic dimers even in presence of a large excess of 1 unit, while motifs that are rigid and lead to A+B fibers can have their size modulated to form trimers, pentamers and other odd-numbered length linear species when 1 unit is in excess.
DISCUSSION
The A-minor junction is a widespread class of structural motifs found in numerous different contexts and natural RNA molecules. We have experimentally characterized a variety of different A-minor junctions using supramolecular self-assembly of tectoRNAs as an assay. Our results exemplify the remarkable modularity of natural RNA molecules and demonstrate several structural principles useful for comprehensively describing RNA structural motifs (see detailed discussion below). The A-minor junction topological rule can also be used to generate stable new motifs not yet found in nature. Additionally, our results are also indicative of some of the constraints that pertain to the structural evolution of RNA.
Modularity and principles of topological equivalence and scaffolding
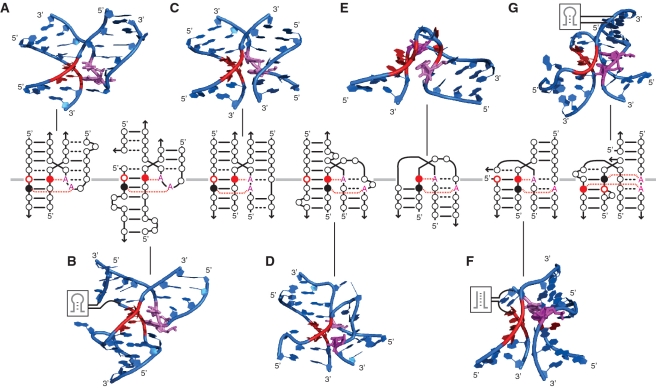
The A-minor junction appears in several locations within the 16S and 23S rRNAs, in ribozymes such as RNase P RNAs, Groups I and II introns, and in at least two different classes of riboswitches (Supplementary Table S1 and Figure 7). This class of motifs follows a topological rule that directs the parallel coaxial stacking of adjacent helices (conformer I) through a conserved A-minor interaction (between Ha and Hc) that can be mediated by a remarkably wide variety of different local structural motifs (Supplementary Table S1, Figures S3 and S10). For instance, we have demonstrated that GNRA, GNRA-like internal loops and triloops in interaction with a helix or specific receptor can all substitute for one another within the topological context of the A-minor junction (Figure 5 and Supplementary Figure S7B). Moreover, by taking advantage of stable natural and unnatural A-minor interactions, new A-minor junction motifs can be engineered that are significantly more stable than their natural counterparts. As such, the A minor junction is a good example of a class of topologically equivalent motifs that can accommodate various structurally distinct classes of smaller motifs (11).
Figure 7.
Motifs following the parallel helix A-minor junction topological rule. In the diagrams, nucleotides are indicated by circles. WC bps are shown as solid lines, and non-WC bps are shown as black dashed lines. The Type I A-minor interactions are indicated by the red A interacting with solid red circle. Type II A-minor interactions are indicated by the red A interacting with solid black circle. (A) GNRA/helix A-minor junction: region H57–H59 from H. marismortui 23S rRNA (PDB ID:1JJ2). (B) Triloop/2x_bulge receptor A-minor junction: region H68–H70 from H. marismortui 23S rRNA (PDB ID:1JJ2). (C) gA:gAA/helix A-minor junction: region H57–H59 from E. coli 23S rRNA (PDB ID: 2AW4). (D) Expanded kink turn (A-minor 3WJ_ABC junction): eukaryotic TPP riboswitch (PDB ID:2CKY). (E) Kink turn: H. marismortui 23S rRNA (PDB ID:1JJ2). (F) Bulged-G/helix A-minor junction (A-minor 3WJ_ACD junction): Oceanobacillus iheyensis Group II intron (PDB ID: 3BWP). (G) Internal loop/helix A-minor junction (A-minor 3WJ_ACD junction): region H14-15-16 from H. marismortui 23S rRNA (PDB ID:1JJ2). Structural models were visualized in PyMOL (pymol.sourceforge.net).
The diversity of structures capable of expressing the topological rule of the A-minor junction is clearly greater than we have thus far explored. Our extensive study suggests that the A-minor-junction class of motifs encompasses a wide range of natural two-, three- and four-helix branched structures (Figures 4, 7, Supplementary Figures S8 and S10) as the same parallel-helix topology can be achieved with various multi-helix junctions. For example, multiple 3WJs fold according to the A-minor topological rule (Figure 7D, F and G and Supplementary Figure S10), with either Hb or Hd removed compared with the original diagram displayed in Figure 1B. Even the common kink-turn motif can be considered a special case of A-minor junction, as it is a parallel helix motif forming an A-minor interaction (Figure 7E). Furthermore, the topological equivalence of triloop/helix and GNRA-like internal_loop/helix interactions illustrates a clear shortcoming in classifying RNA motifs simply based on their secondary structures, as the former can be seen as a 3WJ (Figure 7B), while the latter is a 4WJ (Figure 7C). Thus, rather than categorizing multi-helix junctions by their secondary structure (39), it is more appropriate to organize them according to the tertiary interactions that nucleate the folding of the junction (11). Therefore, like the UA_handle motif, the A-minor junction motif can be viewed as a scaffold for structural expansion (11).
Functional equivalence
Two predominant behaviors for A-minor-junction motifs were identified with the supramolecular self-assembly assay, one leading to circular structures and the other to extended 1D fibers. Interestingly, the difference in self-assembly observed between A-minor junction with helix and A-minor junction with receptor matches the observed structural differences previously reported between A-minor interactions of Type I/IIp and Type I/IIt (20). The latter interaction is typically stabilized by additional interactions from a receptor motif that can contribute stacking via a nucleotide platform and/or additional hydrogen bonds with specific nucleotides of the interacting loop. These two subclasses of A-minor interactions are illustrated in more depth in Supplementary Figure S8, where loop/helix interactions are predominantly Type I/IIp and naturally occurring loop/receptor interactions are predominantly in the category of Type I/IIt A-minor interaction. Therefore, based on self-assembly properties, A-minor junctions can be classified into two distinct classes of functional equivalence that are characterized by slightly different modalities of tertiary interactions.
Structural evolution
The self-assembly assay is very sensitive to small energetic changes within the A-minor interaction and thus suitable for ranking subtle biophysical differences between similar motifs. This is best exemplified by the series of variants tested for the H68–H70 junction from the 23S rRNA. Point mutations bridging natural sequences from archaea, eukaryotes and bacteria reveal that the stability/rigidity of this junction varies throughout the three domains of life. The natural motifs from archaea and eukaryotes are less stable than the sequences from bacteria (and especially T. thermophilus). Interestingly, the sequences of the H68–H70 interaction from archaea and eukaryotes can be made to fold more rigidly by making point mutations following logical structural rules (Figure 6). The mutational study supports our findings that Type I/IIt A-minor interactions involving loop/receptor interactions add considerable stability to the formation of the A-minor junction. Mutations physically distant from the location of the A-minor-junction strand-crossing have an appreciable effect on the stability of the resulting receptors, presumably because mutations at positions involved in the nucleotide triplex formation can contribute to stabilizing the A-minor interacting loop through base stacking on a nucleotide platform.
This study shows that motifs in the ribosome are not necessarily optimized for thermodynamic folding strength. The variation in folding behavior observed for natural sequences of H68–H70 demonstrates that the context of the motif and other functional constraints likely come into play. Therefore, the ribosome structure might require some degrees of structural flexibility to function, counter-selecting against some of the most optimal A-minor junction configurations over its evolution. Our finding that a single-point mutation to the conserved H68–H70 junction of H. marismortui results in a significant stabilization of the fold suggests that optimal self-assembly and stability is not the selective pressure for evolution of RNA within the ribosome. Additionally, some of our data also suggest that several of the conserved long-range RNA motifs from the ribosome are metastable folds in isolated context. For example, several motifs that take advantage of GNRA or GNRA-like internal or terminal loops in modern ribosomes such as the archaeal ribosome of H. marismortui do not assemble well in the 4WJ-tectoRNA context, suggesting that they require post-transcriptional modifications and/or protein contacts to fold stably (Supplementary Figure S7B). We suggest that this apparent sub-optimality might be a consequence of evolutionary constraints that shaped the maturation processes of the ribosomal machinery to be dependent on post-transcriptional modifications or/and proteins for regulatory purposes (40,41).
CONCLUSION
The use of natural RNA junctions to build up programmable self-assembling architectures for potential nanotechnology and synthetic biology applications is still relatively new (17,42). However, the few examples of RNA nanostructures in the literature take advantage of a wide assortment of structured multi-helix junctions consisting of two (43), three (25), four (10) and five (25,44) helices as building blocks for nanoconstruction (17). In contrast, with DNA as a medium for nanoconstruction, Holliday junctions have been the primary motif implemented to design self-assembling architectures (45,46) including folding the DNA up like an origami (47,48) or building up algorithmic structures (49,50). Particularly, many DNA self-assembly designs take advantage of the double crossover (DX or PX) motif to lock-in a particular conformation of DNA (51). The level of refined structural control of junctions is well-exemplified by a DNA machine that can transition between three different geometries of junction (52). Similar parallel-strand paranemic double crossover junctions made of RNA have recently been engineered (53). While the possibility exists to further apply the DNA engineering paradigm to RNA, single-crossover RNA junctions based on the A-minor junction rule offer an advantage as they are able to specify a dominant conformation without the need for a second crossover.
The A-minor junction rule greatly simplifies the identification of parallel-helix motifs, and can be applied to identify potential tertiary structures in non-coding RNA as well as aid in the solving of new RNA structures. For example, based on the triloop/A-minor junction topological folding rule (Figure 4), we can predict the presence of an A-minor junction interaction in the AdoCbl aptamer (11) [see Figure 7C in reference (11)]. Automated computer-aided search has led to the compilation of large databases of RNA junctions such as the RNA_junction database (54). It is our hope that a future combination of tertiary topological rules identified through the architectonics approach with computer-generated RNA databases will lead to great improvements in both the automated design of RNAs for nanotechnology and the automated identification of new RNA structures from genes. We have used our controlled supramolecular system to compare a number of natural and engineered A-minor junctions, but this approach could clearly be extended to the study of other motifs. The successful implementation of RNA structural rules to create new tertiary structures suggests that computational approaches might soon lead to the ability to rationally design large domains of RNAs for bionanotechnology and synthetic biology.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
This work was funded by the National Institutes of Health (R01-GM079604 to L.J.). Funding for open access charge: National Institutes of Health (R01-GM079604 to L.J.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ann Woodard for her technical assistance synthesizing RNA for this study, and Prof. Helen Hansma for the use of her AFM facilities. L.J. wishes to dedicate this paper to Professor Jérôme Lejeune. Author contributions: C.G. and L.J. designed research; CG performed biochemical and biophysical experiments; C.G. and A.C. performed AFM; C.G. and L.J. analyzed data; and C.G. and L.J. wrote the article.
REFERENCES
- 1.Woodson SA. Compact intermediates in RNA folding. Annu. Rev. Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lilley DM. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 3.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DMJ, Ha T. Conformational flexibility of four-way junctions in RNA. J. Mol. Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 5.McKinney SA, Tan E, Wilson TJ, Nahas MK, Declais AC, Clegg RM, Lilley DMJ, Ha T. Single-molecule studies of DNA and RNA four-way junctions. Biochem. Soc. Trans. 2004;32:41–45. doi: 10.1042/bst0320041. [DOI] [PubMed] [Google Scholar]
- 6.Tyagi R, Mathews DH. Predicting helical coaxial stacking in RNA multibranch loops. RNA. 2007;13:939–951. doi: 10.1261/rna.305307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 8.Walter NG, Burke JM, Millar DP. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- 9.Jaeger L, Leontis NB. Tecto-RNA: one-dimensional self-assembly through tertiary interactions. Angew. Chemie. Int. Ed. Engl. 2000;14:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 10.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 2006;34:1381–1392. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 14.Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 15.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 17.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 23.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 24.Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Structure and dimerization of HIV-1 kissing loop aptamers. J. Mol. Biol. 2001;311:475–490. doi: 10.1006/jmbi.2001.4879. [DOI] [PubMed] [Google Scholar]
- 25.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 27.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 29.Massire C, Jaeger L, Westhof E. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA. 1997;3:553–556. [PMC free article] [PubMed] [Google Scholar]
- 30.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 33.Lee JC, Gutell RR, Russell R. The UAA/GAN internal loop motif: a new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions. J. Mol. Biol. 2006;360:978–988. doi: 10.1016/j.jmb.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 34.Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J. Mol. Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- 35.Lisi V, Major F. A comparative analysis of the triloops in all high-resolution RNA structures reveals sequence structure relationships. RNA. 2007;13:1537–1545. doi: 10.1261/rna.597507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JC, Cannone JJ, Gutell RR. The lonepair triloop: a new motif in RNA structure. J. Mol. Biol. 2003;325:65–83. doi: 10.1016/s0022-2836(02)01106-3. [DOI] [PubMed] [Google Scholar]
- 37.Jucker FM, Pardi A. GNRA tetraloops make a U-turn. RNA. 1995;1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 38.Gutell RR, Cannone JJ, Konings D, Gautheret D. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- 39.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl Acad. Sci. USA. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen-Chalamish S, Hasson A, Weinberg D, Namer LS, Banai Y, Osman F, Kaempfer R. Dynamic refolding of IFN-gamma mRNA enables it to function as PKR activator and translation template. Nat. Chem. Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. [DOI] [PubMed] [Google Scholar]
- 42.Saito H, Inoue T. RNA and RNP as new molecular parts in synthetic biology. J. Biotechnol. 2007;132:1–7. doi: 10.1016/j.jbiotec.2007.07.952. [DOI] [PubMed] [Google Scholar]
- 43.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 44.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. A polyhedron made of tRNAs. Nat. Chem. 2010;2:772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeman NC. An overview of structural DNA nanotechnology. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeman NC. DNA Components for Molecular Architecture. Acc. Chem. Res. 1997;30:357–363. [Google Scholar]
- 47.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 48.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothemund PW, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2004;2:e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujibayashi K, Hariadi R, Park SH, Winfree E, Murata S. Toward reliable algorithmic self-assembly of DNA tiles: a fixed-width cellular automaton pattern. Nano Lett. 2008;8:1791–1797. doi: 10.1021/nl0722830. [DOI] [PubMed] [Google Scholar]
- 51.Zhang S, Fu TJ, Seeman NC. Symmetric immobile DNA branched junctions. Biochemistry. 1993;32:8062–8067. doi: 10.1021/bi00083a002. [DOI] [PubMed] [Google Scholar]
- 52.Gu H, Chao J, Xiao S-J, Seeman NC. Dynamic patterning programmed by DNA tiles captured on a DNA origami substrate. Nat. Nanotechnol. 2009;4:245–248. doi: 10.1038/nnano.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc. 2008;130:93–102. doi: 10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA. RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res. 2008;36:D392–D397. doi: 10.1093/nar/gkm842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.