Abstract
O-Phosphoseryl-tRNA kinase (PSTK) is the key enzyme in recruiting selenocysteine (Sec) to the genetic code of archaea and eukaryotes. The enzyme phosphorylates Ser-tRNASec to produce O-phosphoseryl-tRNASec (Sep-tRNASec) that is then converted to Sec-tRNASec by Sep-tRNA:Sec-tRNA synthase. Earlier we reported the structure of the Methanocaldococcus jannaschii PSTK (MjPSTK) complexed with AMPPNP. This study presents the crystal structure (at 2.4-Å resolution) of MjPSTK complexed with an anticodon-stem/loop truncated tRNASec (Mj*tRNASec), a good enzyme substrate. Mj*tRNASec is bound between the enzyme’s C-terminal domain (CTD) and N-terminal kinase domain (NTD) that are connected by a flexible 11 amino acid linker. Upon Mj*tRNASec recognition the CTD undergoes a 62-Å movement to allow proper binding of the 7-bp D-stem. This large reorganization of the PSTK quaternary structure likely provides a means by which the unique tRNASec species can be accurately recognized with high affinity by the translation machinery. However, while the NTD recognizes the tRNA acceptor helix, shortened versions of MjPSTK (representing only 60% of the original size, in which the entire CTD, linker loop and an adjacent NTD helix are missing) are still active in vivo and in vitro, albeit with reduced activity compared to the full-length enzyme.
INTRODUCTION
Selenium is essential for human health. In proteins, it is found as selenocysteine (Sec), a natural amino acid (aa) encoded by UGA. Sec is formed in a tRNA-dependent transformation of serine that was attached to tRNASec by seryl-tRNA synthetase. Archaea and eukaryotes then employ a two-step route: the essential enzyme PSTK phosphorylates Ser-tRNASec to Sep-tRNASec which is then converted to Sec-tRNASec by Sep-tRNA:Sec-tRNA synthase (SepSecS) (1–3). PSTK is the first kinase described that acts on a tRNA substrate (4). Biochemical analyses have characterized the kinase domain active site (5), and a phylogenetic history shows that PSTK and SepSecS co-evolved since the archaeal-eukaryotic divide (6). Recently, a crystal structure of MjPSTK complexed with adenosine diphosphate (ADP) or adenylyl-imidodiphosphate (AMPPNP) but in the absence of the tRNA substrate was reported (7). The active site was on the surface of the enzyme, which has a deep groove with positive electrostatic potential. Based on the shape of the groove, a docking model suggested this groove could accommodate the long acceptor helix of Methanocaldococcus jannaschii tRNASec.
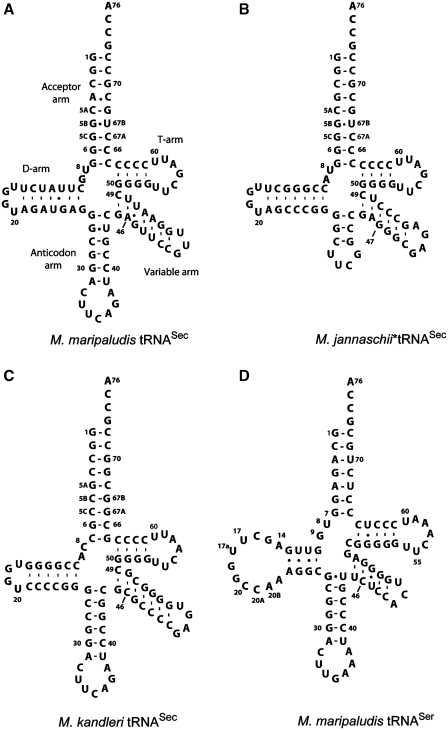
Selenocysteine tRNA (tRNASec) has an unusual structure (Figure 1). The eukaryotic and archaeal tRNASec species have a 9-bp acceptor helix and a 4-bp T-stem (8,9) assuming the so-called 9/4 structure (10). The D-stem/loop lengths are also variable (from 16–19 nt); the archaeal species have a 7-bp D-stem, whereas the eukaryotic ones have only 6 bp. Methanopyrus kandleri tRNASec is the only exception among 13 archaeal species [see legend to Figure 1 and ref. (11)]; it is a eukaryotic-like tRNA with only 6 bp like human tRNASec (9).
Figure 1.
The secondary structures of archaeal tRNASec and tRNASer species. The cloverleaf models are shown with the standard tRNA numbering as modified for tRNASec (10). (A) Methanococcus maripaludis tRNASec; (B) the anticodon-stem/loop truncated M. jannaschii tRNASec (Mj*tRNASec); (C) M. kandleri tRNASec; (D) M. maripaludis tRNASer. The M. maripaludis tRNASec has the 7-bp D-stem, which is found in all archaeal tRNASec species: Methanocaldococcus fervens AG86, Methanocaldococcus infernus, Methanocaldococcus jannaschii, Methanocaldococcus vulcanius M7, Methanococcus aeolicus, Methanococcus maripaludis S2, Methanococcus maripaludis C5, Methanococcus maripaludis C7, Methanococcus vannielii SB, Methanococcus voltae A3, Methanothermobacter thermolithotrophicus, Methanothermococcus okinawensis. The only exception is Methanopyrus kandleri tRNASec.
Here, we present the crystal structure of MjPSTK in complex with the anticodon-stem/loop truncated M. jannaschii tRNASec (Mj*tRNASec). The structure reveals that tRNA binding is accompanied by a large domain rearrangement in the enzyme, which could not be predicted from the tRNA-free structure. The orientation of the acceptor helix, therefore, does not reflect the earlier model. A similar structure of M. jannaschii PSTK complexed to M. kandleri tRNASec has just been reported (11). Comparison of both structures shows that MjPSTK is flexible in accommodating the differently sized D-stem for specific recognition of the unique tRNASec.
MATERIALS AND METHODS
Protein purification
In order to overproduce MjPSTK with a C-terminal His6-tag the Escherichia coli strain C41(DE3) (12) was transformed with the plasmid pET20b, containing the pstk gene. The cells were grown to A600 =0.6 and gene expression was induced with 0.4 mM isopropyl β-d-thiogalactopyranoside (IPTG), followed by cultivation at 20°C for 18 h. After centrifugation at 8000g for 5 min, the harvested cells were suspended in 50 mM Tris–HCl buffer (pH 7.0) containing 500 mM NaCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol, 10% (v/v) glycerol and 0.1 mM phenylmethanesulfonylfluoride (PMSF), and then were disrupted by sonication. The supernatant after centrifugation was purified by sequential chromatography on Ni-NTA (QIAGEN), Resource S (GE Healthcare), HiTrap Heparin HP (GE Healthcare) and Superdex200 10/300 GL (GE Healthcare) columns. The purified PSTK in the crystallization buffer was concentrated to 16 mg/ml and mixed with adenosine triphosphate (ATP) to a final concentration of 2 mM.
Mj*tRNASec purification
Mj*tRNASec was transcribed using T7 RNA polymerase, and subjected to denaturing polyacrylamide gel electrophoresis (PAGE). The RNA was purified by anion-exchange chromatography on Resource Q (GE Healthcare). The purified Mj*tRNASec was dissolved in the crystallization buffer, containing 10 mM Tris–HCl (pH 8.0), 30 mM NaCl, 10 mM MgCl2 and 1 mM DTT, and heated at 70°C for 10 min for refolding.
Crystallization and data collection
To crystallize the MjPSTK•Mj*tRNASec complex, MjPSTK was mixed with Mj*tRNASec in a molar ratio of 1:1.2, respectively, at a final protein concentration of 7 mg/ml. The 0.1-μl aliquot of the complex solution was mixed with 0.1 μl of a crystallization solution and was equilibrated by sitting-drop vapor diffusion against a 50-μl reservoir solution of 100 mM phosphate buffer (pH 6.2) containing 0.2 M NaCl and 41% (v/v) PEG 200. The crystals of MjPSTK•Mj*tRNASec complex were grown within 3 weeks at 20°C.
For data collection, the drop solution was slowly equilibrated against the same reservoir solution as for crystallization. The crystals obtained were briefly transferred to 1.2 × reservoir solution and were flash-cooled in a cryo-stream of nitrogen gas at 100 K. The data sets of the crystals were collected at station BL41XU of Spring-8 (Harima, Japan). The data set of the MjPSTK•Mj*tRNASec complex was collected at 100 K in a cryo-stream of nitrogen gas. The collected data sets were processed with HKL2000 (13).
Structure determination and refinement
The structure was determined by molecular replacement, using the NTD of the tRNA-free MjPSTK structure as a search model. Molecular replacement was done by MOLREP (14); tRNA models were built with program COOT (15). The solution was refined by REFMAC5 (16) and PHENIX (17). In the resultant 2m|Fo|–D|Fc| and m|Fo|–D|Fc| maps, we clearly found tRNA electron densities. The crystallographic, data collection and refinement statistics are presented in Supplementary Table S1. Molecular graphics were illustrated with CueMol (http://www.cuemol.org/).
RESULTS
Crystallization
Attempts to crystallize MjPSTK with full-length M. jannaschii tRNASec failed. Because the anticodon is not required for tRNASec recognition (5), and tRNAs with a shortened anticodon stem/loop replaced by a UUCG tetraloop have proved to be useful in tRNA crystallography (18), we designed three such tRNAs. One tRNA (Figure 1), designated Mj*tRNASec, provided well diffracting crystals and is an excellent substrate for PSTK (Supplementary Figure S1).
Overall structure
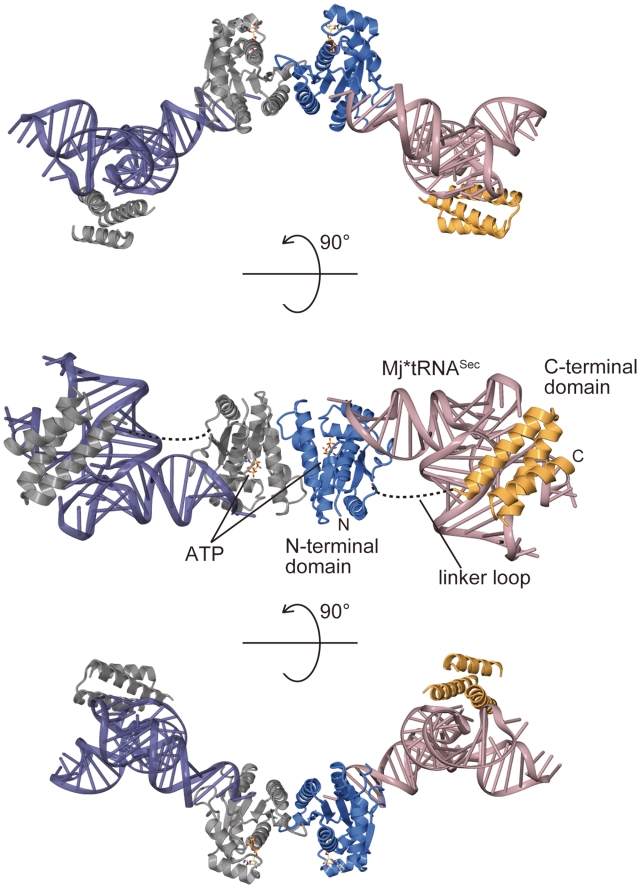
Methanocaldococcus jannaschii PSTK is a 252-aa-long protein with an NTD separated by an 11-aa linker from the CTD (7). The crystal structure of MjPSTK complexed with ATP and Mj*tRNASec was determined by molecular replacement, using the NTD of tRNA-free MjPSTK as a search model. The final model was refined at 2.4-Å resolution to free R-factor of 29.2%. The detailed data collection and refinement statistics are summarized in Table S1. The asymmetric unit contains one MjPSTK monomer and one Mj*tRNASec. By crystallographic symmetrical operation, the complex forms a dimer (Figure 2). Each monomer can be divided into an N-terminal kinase domain (NTD) (residues 1–177) and a C-terminal domain (CTD) (residues 189–252), connected by an 11-aa loop. In the MjPSTK•Mj*tRNASec complex, the NTD establishes several base-specific contacts with the acceptor stem, and the CTD binds to the tRNA D-loop (Figure 2, middle). Whereas the electron densities of the NTD, the C-terminal tRNA-binding domain and of tRNASec are well defined, the structure of the linker loop is completely disordered, reflecting the intrinsic flexibility of the CTD.
Figure 2.
Overall structure of MjPSTK•Mj*tRNASec complex. Crystal structure of MjPSTK is depicted by ribbon representation, consisting of the NTD (3–177, blue) and the CTD (188–252, orange) in one monomer. The other monomer is colored gray. Two bound tRNA molecules are colored pink and indigo blue. The bound ATP molecule is shown as a ball stick model. The complex structure is viewed from three angles for clarity.
A concurrent report of an MjPSTK•M. kandleri tRNASec structure (11) reveals basically the same features; the amino acid numbering differs in the two structures, as different reference sequences (accession: Q58933 versus NP_248546) were used for cloning and protein preparation.
Acceptor stem recognition
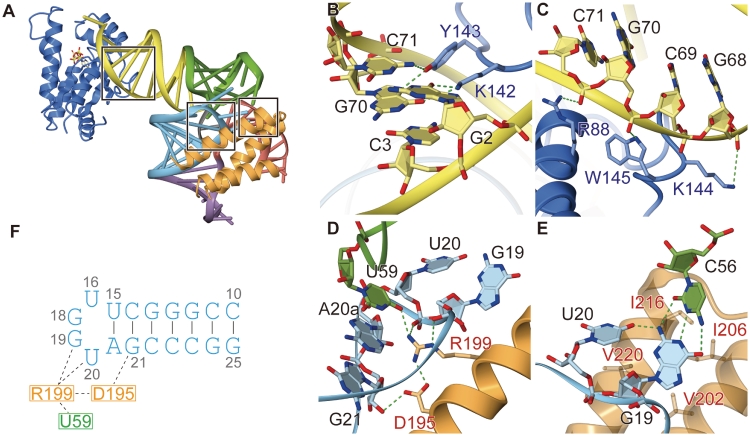
We reported previously (5) that PSTK recognizes the base pairs 2•71 and 3•70 in the acceptor helix of tRNASec. In the complex structure, the ε-amino group of Lys142 hydrogen-bonds to the O6 and N7 positions of G2 in tRNASec, while the side-chain hydroxyl group of Tyr143 hydrogen-bonds to the N7 group of G70 and the N4 group of C71 (Figure 3B). Thus, Lys 142 and Tyr143 from the NTD contribute to the base-specific recognition of the tRNASec acceptor stem. Furthermore, the side-chain nitrogen groups of Arg88, Lys144 and Trp145 hydrogen-bond or electrostatically interact with the phosphate backbone of G68, G70 and C72 in the 3′-strand of the tRNASec acceptor helix (Figure 3C).
Figure 3.
tRNA recognition by MjPSTK. (A) The overall structure highlighting the tRNA recognition sites as rectangles. (B) The zoomed view of the base-specific recognition of the acceptor stem. tRNA is colored yellow, while the amino acid residues from the NTD are colored blue. (C) The zoomed view of the phosphate backbone recognition in the acceptor stem. (D) The zoomed view of the D-loop recognition by MjPSTK. (E) The zoomed view of the recognition of the base triplet at the TΨC-loop•D-loop interaction site. (F) The key interactions by D195 and R199 are summarized in a pattern diagram.
D-arm recognition
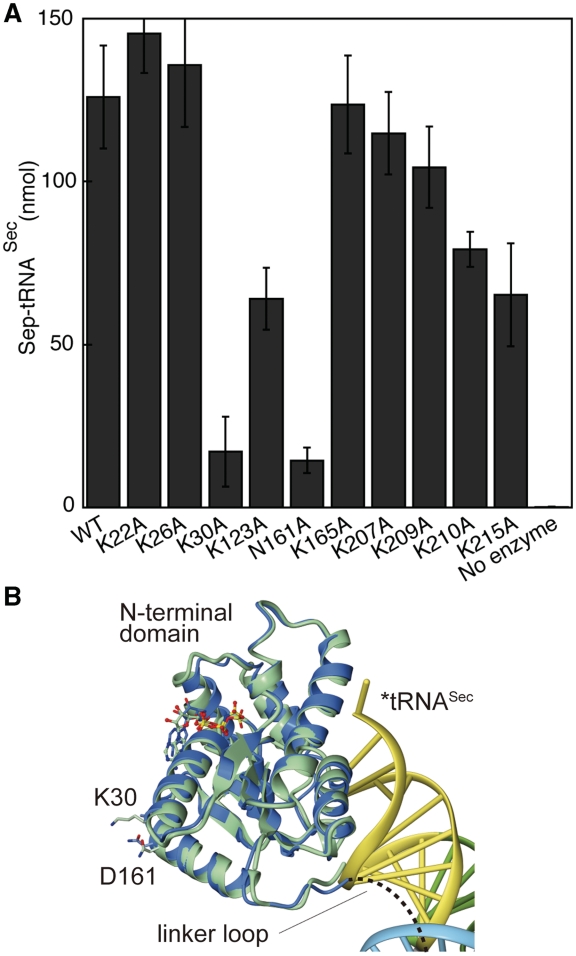
The C-terminal domain of MjPSTK consists of a three-helix bundle. The first helix of the bundle approaches the tRNA D-loop and recognizes the characteristically compact tRNASec D-loop, which is a tetraloop smaller than typical tRNA D-loops (Figure 3F). Arg199 penetrates into the D-loop to interact electrostatically with the phosphate backbone of U20 (Figure 3D and F). To stabilize the L-shape of the tRNA, the D-loop interacts with the TΨC-loop. In the present complex structure, the O4 group of U59 in the TΨC-loop is specifically recognized by Arg199. Additionally, Asp195 interacts electrostatically with the side-chain amino group of Arg199 to fix its position (Figure 3D and F), while at the same time, the side-chain of Asp195 hydrogen-bonds to the O2′ atom of G21. In archaeal tRNASec, C56 forms a Watson–Crick-type tertiary base pair with G19, forming a base triplet between C56, G19 and U20, which stabilizes the TΨC-loop•D-loop interaction in the tRNA core (Figure 3E). In the MjPSTK•M. kandleri tRNASec complex (11) the same base triplet (C56, G19 and U20) is recognized by Lys207. This Lys207 residue is structurally disordered in our MjPSTK•Mj*tRNASec complex, and the activity of a Lys207Ala mutant was only slightly reduced (Figure 4). Therefore, the D-loop recognition might be flexibly adapted to tRNASec species from different organisms.
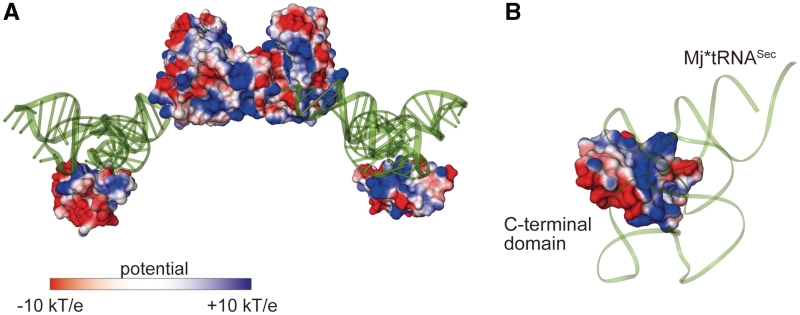
Figure 4.
The electrostatic surface potential model. (A) The protein electrostatic surface potential model of the MjPSTK•Mj*tRNASec complex, calculated by the program APBS (24). The Mj*tRNASec molecules are shown by tube model and colored green. (B) Close-up view of the interaction surface between the C-terminal domain and D-arm.
Additionally, Val202, Ile206, Ile216 and Val220 in the second and third helices of the C-terminal helix bundle form a platform, providing hydrophobic interactions with the base triplet (Figure 3E).
Biochemical and genetic analysis of mutant MjPSTK enzymes
These analyses employed established tests, which are very sensitive (5,6). The in vitro assay accurately monitors conversion of Ser-tRNASec to Sep-tRNASec; the assay is based on nuclease P1 digestion of [32P]-end-labeled aminoacyl-tRNA and separation of the resulting aminoacyl- [32P]AMP derivatives by thin-layer chromatography (5). Formate dehydrogenase H is the only selenoprotein in E. coli, and the presence of selenocysteine is essential for having an active enzyme. The sensitive in vivo assay measures Sep-tRNASec formation by the ability of wild-type or mutant PSTK genes to restore the benzyl viologen reducing activity (observed as violet color) in our reporter protein formate dehydrogenase H in the E. coli selA deletion strain JS1 (19).
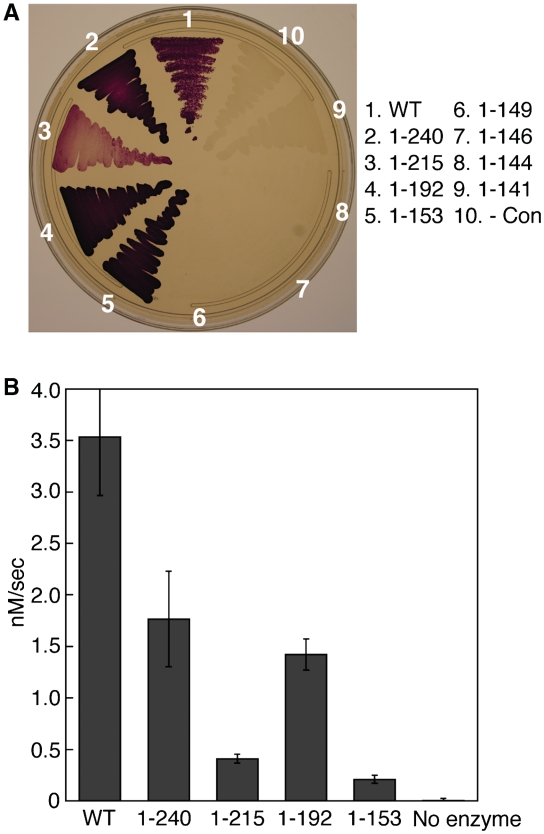
First, we investigated the role of the CTD of MjJPSTK for Sep-tRNASec formation. Sequential truncations were made from the C-terminus of the 252-aa full-length protein. As shown in Figure 5B, deletions of up to 98 aa (MjPSTK1–153) still show activity, albeit at a much reduced level (6% of the initial velocity of the intact enzyme). However, in vivo MjPSTK1–153 is still able to maintain the level of activity that compensates for the E. coli selA deletion (Figure 5A). MjPSTK1–192 lacks the entire CTD domain, while the MjPSTK1–215 and MjPSTK1–240 mutants lack the terminal two and one helices of the helix bundle, respectively. In the in vitro assay, MjPSTK1–192 exhibits 40% activity as compared to the full-length enzyme (WT) (Figure 5B), which can be ascribed to the specific D-loop recognition by the C-terminal domain. MjPSTK1–215 mutant showed remarkably reduced activity (Figure 6B), presumably due to the instability of the remaining first helix.
Figure 5.
Activity of progressive MjPSTK deletions. (A) Formation of Sep-tRNASec in vivo is assayed by the ability of the wild-type PSTK and C-terminal deletion mutants (1–240, 1–215, 1–192, 1–153, 1–149, 1–146, 1–144 and 1–141) to restore the benzyl viologen reducing activity (seen as a violet color) of the selenoprotein FDHH in the E. coli selA deletion strain JS1. (B) Initial velocities were calculated for the phosphotransferase activity of PSTK deletion mutants. Wild-type PSTK (10 nM) was incubated with 600 nM 32P-labeled Ser-tRNASec for 80 s at 37°C; 10-s time points were taken. Truncation mutants (1–240, 1–215, 1–192 and 1–153) were incubated with 600-nM 32P-labeled Ser-tRNASec for 8 min at 37°C; 1-min time points were taken. Those reaction aliquots were quenched with 100 mM sodium citrate, pH 5.0 and digested with 0.66 mg/ml nuclease P1 for 35 min at room temperature. Samples were then spotted onto a PEI-cellulose TLC plate and developed in 100 mM ammonium acetate, 5% acetic acid for 75 min. The plates were exposed to an imaging plate (FujiFilms), scanned on a Molecular Dynamics Storm 860 PhosphorImager, and quantified using ImageQuant software. Error bars represent the standard deviation of three experiments.
Figure 6.
(A) In vitro assays of MjPSTK mutants. The catalytic activity of the mutants K22A, K26A, K30A, K123A, N161A, K165A, K207A, K209A, K210A and K215A was tested using the phosphotransferase assay. Ten nanomolar enzyme was incubated with 1.5 µM 32P-labeled Ser-tRNASec for 30 s at 37°C. Error bars represent the standard deviation of three experiments. Reactions were analyzed as described in Figure 5. (B). The vicinity of Lys30 and Asn161. The N-terminal domain of the tRNA-free form and complexed form are depicted with the same color code as in Figure 7A. Mj*tRNASec is colored according to the domains, in the same color code as in Figure 3A. The amino acid residues (Lys30 and Asp161) and the bound ATP molecules are shown as a ball stick model.
Next, we determined whether single amino acid changes affected Methanococcus maripaludis tRNASec (with 7-bp D-stem) recognition in terms of Sep-tRNASec formation or tRNASec binding. Ala mutations of several amino acids involved in specific tRNA binding of the NTD or CTD (see above) did not abolish enzyme activity, and thus did not abolish tRNA recognition (Figure 6). Defective phosphorylation activity was observed in MjPSTKK30A and MjPSTKN161A mutants. In the present structure, Lys30 and Asn161 are suggested to form a salt bridge, which may fix the orientation of the linker between the N-terminal and C-terminal domains (Figure 6C). This linker orientation is likely important to discriminate the long acceptor stem of tRNASec, as described subsequently. In contrast, other active site mutations (e.g. MjPSTKK17A or MjPSTKS18A) did abolish enzyme activity and inhibited tRNASec recognition (6). Benzyl viologen assays reveal that the PSTK mutants K142A, W145A and D146A are active in vivo (data not shown). However, the MjPSTK double mutant K142A/Y143A showed severely reduced activity with the M. kandleri tRNASec substrate (11).
When M. maripaludis tRNASec binding was measured, single amino acid mutations of PSTK showed some reduction of substrate affinity (Supplementary Table S2). However, the truncated enzymes (Figure 5) did not show any binding under these conditions. Thus, their KD is estimated to be >40 μM which is 50–2000-fold higher than the experimentally determined KD values of the single mutant enzymes.
We conclude that the NTD of PSTK has all the elements required for Sep-tRNASec formation but has a much lower affinity for tRNASec than the intact protein, suggesting that CTD of PSTK solely contributes to the binding and orientation of tRNASec.
DISCUSSION
tRNA recognition with domain movement
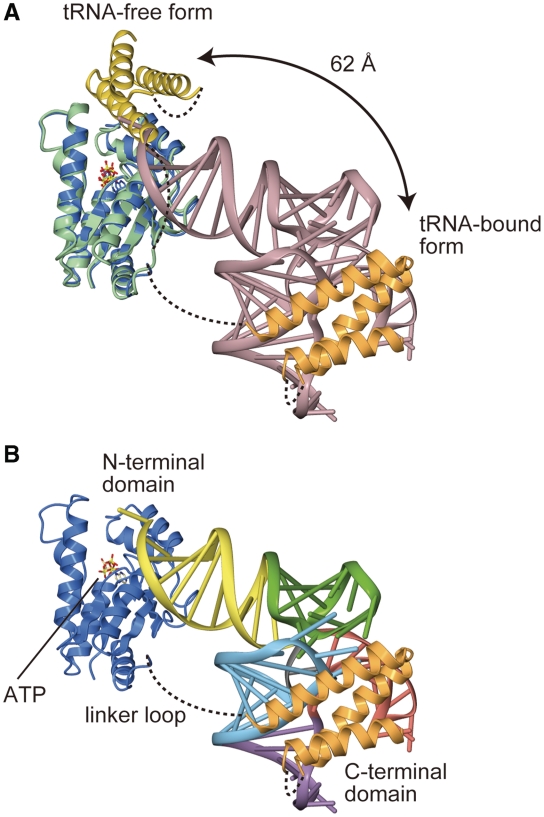
In contrast to the canonical tRNA species, tRNASec has an unusually long acceptor-TΨC stem composed of 13 bp and an unusual D-arm composed of 7 bp and tetraloop (Figure 1). As the tRNA-free MjPSTK structure previously suggested (7) the high flexibility of the C-terminal domain, the complex structure revealed that the C-terminal domain moves away from the NTD to bind tRNASec (Figure 7A). Although the NTD recognizes the acceptor stem and the CCA-terminus, the C-terminal domain directly interacts with the characteristically compact D-loop of tRNASec (Figure 7A). As the CTD appears to move far away from the NTD upon tRNA binding, the linker loop must be long enough for the CTD to reach and bind the D-loop of the tRNA. The linker is disordered, so its precise conformation is likely unimportant for tRNA binding, and the linker does not establish specific contacts to the tRNA. The linker varies in length from 10 aa (M. kandleri) to 41 aa (Trypanosoma) among the PSTK sequences.
Figure 7.
Structure of an MjPSTK•Mj*tRNASec complex. (A) Superposition of the tRNA-free form (green and yellow) and complexed form (blue and orange) of MjPSTK. The large movement of C-terminal domain upon binding with tRNA is highlighted. (B) Ribbon representation of the crystal structure of the complex, with the same color code as in Figure 2. Mj*tRNASec is differently colored according to the domains, in the same color code as in Figure 3. The bound ATP molecule is shown as a ball stick model.
The electrostatic potential on the solvent-accessible surface model of tRNA-bound MjPSTK showed that the positively charged patch is spread over the CTD. The compact D-loop of tRNASec is snugly accommodated to this patch (Figure 4A and B). Such large domain movements upon substrate binding are frequently observed in tRNA-modifying enzymes. For instance, in the archaeal Trm5 enzyme, which methylates tRNAs bearing G37 to m1G37, the C-terminal domain moves upon tRNA binding to recognize the molecule’s core region comprising the TΨC- and D-loops; this verifies the canonical mature L-shape tRNA structure (20). In the tRNA-free form of PSTK and Trm5, the C-terminal domain is intrinsically flexible, and captures the tRNA at the initial recognition step. In contrast, in bacterial TilS that forms lysidine at C34 of tRNAIle2, the C-terminal domain is fixed by hydrophobic interactions with the central domain, and moves considerably upon complex formation to recognize the tRNA acceptor end (21). This mechanism of sequential RNA recognition coupled with domain movements enables the enzyme to strictly recognize tRNAIle2 embedded in the precursor, because TilS starts modification during pre-tRNA processing. The large domain movement of PSTK as well as of Trm5 somewhat resembles the ‘fly-casting’ mechanism of an intrinsic disordered protein (22).
Recognition of tRNASec by PSTK
Our earlier biochemical work (5) showed that the tRNASec acceptor helix contains crucial tRNA identity elements for PSTK recognition. This was borne out (as seen in both structures) by the specific interaction of NTD residues with these specific bases in the acceptor helix (see above), and also by the enzyme inactivation in the MjPSTKK142A/Y143A double mutant (11). tRNA identity experiments with a chimera of tRNASec and tRNASer showed that the transplantation of only 3 bp from the acceptor helix of M. maripaludis tRNASec into the body of M. maripaludis tRNASer led to a tRNA that is recognized by PSTK almost as efficiently as wild-type tRNASec (tRNA chimera in Table 1 in ref. 6). In contrast to tRNASec (Figure 1), which has a 7-bp D-arm and 4-base D-loop, the tRNASer body has a shorter D-arm (4 bp) and a longer D-loop (11 bases), so the specific architecture of the D-arm and D-loop are not a basis upon which PSTK discriminates tRNASec from tRNASer. Taken together, these data show that the NTD of MjPSTK contains the main features necessary for recognition of tRNASec. The CTD interaction with the tRNASec D-arm is, therefore, of secondary importance, but the CTD does provide significantly tighter binding to the tRNA.
Herein we produced artificially truncated PSTK variants (i.e. only the NTD) that are still weakly active with Mj*tRNASec, and a similar result was found with M. kandleri tRNASec (11). In fact, PSTK open reading frames exist in nature that consist of the NTD with dramatically shortened or non-existent CTDs, and such PSTK genes are present in the genomes of Methanothermococcus okinawensis (http://genome.jgi-psf.org/metok/metok.home.html) and Schistosoma japonicum (23). These interesting PSTK variants await further experimentation and are expected to be catalytically competent enzymes that may display weaker tRNA affinity. An alternative possibility is that these naturally truncated PSTKs establish compensatory interactions with their tRNA substrates, leading to equivalent enzymatic activity compared to full-length PSTKs.
FUNDING
The National Institute of General Medical Sciences GM22854 and the Department of Energy (to D.S.); the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology (to O.N.); Science Research of the Ministry of Education, Culture, Sports, Science and Technology (to R.I. and O.N.); Mitsubishi Foundation (to O.N.); Japan Society of the Promotion of Science (JSPS) research fellowship for young scientists (to Y.A.); National Institute of General Medical Sciences (NIGMS), Ruth L. Kirschstein National Research Service Award (NRSA) fellow (to R.L.S.); the Schweizerischer Nationalfonds, postdoctoral fellowship (to C.A.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACCESSION NUMBER
The crystallographic coordinates are deposited with PDB under ID: 3AM1.
ACKNOWLEDGEMENTS
We thank Patrick O'Donoghue, Jiqiang Ling, Abby Plummer and Lennart Randau for invaluable discussions, William Whitman and Ken Takai for questions with the M. okinawensis genome, and the beam-line staff (at SPring-8 and at KEK PF-AR) for assistance with data collection.
REFERENCES
- 1.Yuan J, O'Donoghue P, Ambrogelly A, Gundllapalli S, Sherrer RL, Palioura S, Simonovic M, Söll D. Distinct genetic code expansion strategies for selenocysteine and pyrrolysine are reflected in different aminoacyl-tRNA formation systems. FEBS Lett. 2010;584:342–349. doi: 10.1016/j.febslet.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palioura S, Herkel J, Simonovic M, Lohse AW, Söll D. Human SepSecS or SLA/LP: selenocysteine formation and autoimmune hepatitis. Biol. Chem. 2010;391:771–776. doi: 10.1515/BC.2010.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl Acad. Sci. USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherrer RL, Ho JM, Söll D. Divergence of selenocysteine tRNA recognition by archaeal and eukaryotic O-phosphoseryl-tRNASec kinase. Nucleic Acids Res. 2008;36:1871–1880. doi: 10.1093/nar/gkn036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherrer RL, O'Donoghue P, Söll D. Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 2008;36:1247–1259. doi: 10.1093/nar/gkm1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araiso Y, Sherrer RL, Ishitani R, Ho JM, Söll D, Nureki O. Structure of a tRNA-dependent kinase essential for selenocysteine decoding. Proc. Natl Acad. Sci. USA. 2009;106:16215–16220. doi: 10.1073/pnas.0908861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palioura S, Sherrer RL, Steitz TA, Söll D, Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh Y, Chiba S, Sekine S, Yokoyama S. Crystal structure of human selenocysteine tRNA. Nucleic Acids Res. 2009;37:6259–6268. doi: 10.1093/nar/gkp648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturchler C, Westhof E, Carbon P, Krol A. Unique secondary and tertiary structural features of the eucaryotic selenocysteine tRNASec. Nucleic Acids Res. 1993;21:1073–1079. doi: 10.1093/nar/21.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba S, Itoh Y, Sekine SI, Yokoyama S. Structural basis for the major role of O-phosphoseryl-tRNA kinase in the UGA-specific encoding of selenocysteine. Mol. Cell. 2010;39:410–420. doi: 10.1016/j.molcel.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 18.Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 19.Yuan J, Palioura S, Salazar JC, Su D, O'Donoghue P, Hohn MJ, Cardoso AM, Whitman WB, Söll D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl Acad. Sci. USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi K, Bonnefond L, Kimura S, Suzuki T, Ishitani R, Nureki O. Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase. Nature. 2009;461:1144–1148. doi: 10.1038/nature08474. [DOI] [PubMed] [Google Scholar]
- 22.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl Acad. Sci. USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.