Abstract
Flap endonuclease-1 (FEN1) is a member of the Rad2 structure-specific nuclease family. FEN1 possesses FEN, 5′-exonuclease and gap-endonuclease activities. The multiple nuclease activities of FEN1 allow it to participate in numerous DNA metabolic pathways, including Okazaki fragment maturation, stalled replication fork rescue, telomere maintenance, long-patch base excision repair and apoptotic DNA fragmentation. Here, we summarize the distinct roles of the different nuclease activities of FEN1 in these pathways. Recent biochemical and genetic studies indicate that FEN1 interacts with more than 30 proteins and undergoes post-translational modifications. We discuss how FEN1 is regulated via these mechanisms. Moreover, FEN1 interacts with five distinct groups of DNA metabolic proteins, allowing the nuclease to be recruited to a specific DNA metabolic complex, such as the DNA replication machinery for RNA primer removal or the DNA degradosome for apoptotic DNA fragmentation. Some FEN1 interaction partners also stimulate FEN1 nuclease activities to further ensure efficient action in processing of different DNA structures. Post-translational modifications, on the other hand, may be critical to regulate protein–protein interactions and cellular localizations of FEN1. Lastly, we also review the biological significance of FEN1 as a tumor suppressor, with an emphasis on studies of human mutations and mouse models.
INTRODUCTION
DNA replication, recombination and repair are essential for maintaining the integrity of genomes. These DNA metabolic pathways create various DNA intermediates that must also be efficiently processed, otherwise severe genomic instability will result. Many of the DNA intermediates formed are bifurcated nucleic acid structures that require nucleases to recognize the structure of the intermediate and to only cleave the appropriate phosphate diester, regardless of sequence. A nuclease that typifies these qualities is flap endonuclease-1 (FEN1), which is the archetypal member of the Rad2 nuclease family (1,2). FEN1 is a structure-specific metallonuclease best known for its essential roles in the penultimate steps of Okazaki fragment maturation and long-patch base excision repair (BER) (1,3,4). Furthermore, FEN1 has also been implicated in other major DNA metabolic pathways, including resolution of tri-nucleotide repeat sequence-derived secondary structures, rescue of stalled DNA replication forks, maintenance of telomere stability and apoptotic fragmentation of DNA (5–10).
How FEN1 is involved in multiple pathways that are seemingly contradictory (i.e. genome maintenance versus apoptotic DNA fragmentation) has been of interest to our lab and others. The multiple functions of FEN1 may be regulated via three interplaying mechanisms: (i) formation of complexes with different protein partners, (ii) cellular compartmentalization and (iii) post-translational modifications. First, FEN1 is known to rely on interactions with other DNA metabolic proteins for recruitment to different machineries for DNA replication, repair or degradation. Some of these interaction partners of FEN1 also stimulate its nuclease activities to facilitate efficient processing of various bifurcated DNA intermediates. To date, 34 proteins from various DNA metabolic pathways have been identified as interacting with FEN1. Second, FEN1 was shown to localize to the nucleus in a DNA damage and cell cycle-dependent manner (11), implicating the dynamic nuclear localization of FEN1 as playing an important role in mediating its functions in DNA replication and repair. More recently, FEN1 was shown to super-accumulate in the nucleolus, where it is thought to maintain the stability of tandem repeats of ribosomal DNA (12). Furthermore, FEN1 was also shown to localize to the mitochondrion, where it plays an important role in mitochondrial DNA (mtDNA) replication and repair (13). Last, FEN1 can be acetylated, phosphorylated and methylated, and these post-translational modifications may be important for the regulation of the nuclease activities, protein partner selection and/or subcellular compartmentalization (14–16).
FEN1 displays a severe mutator phenotype in yeast when deleted (17), and therefore, functional deficiency of FEN1 has been suggested to cause genomic instability and predisposition to cancer in mammals. Recently, a group of FEN1 mutations was identified in human cancer specimens (9). Most of these mutations abrogated the ability of FEN1 to cleave model exonuclease (EXO) and gap-endonuclease substrates in vitro, while retaining the ability to cleave FEN substrates (9). To demonstrate the etiological significance of these somatic mutations, a mouse line that harbors an E160D Fen1 point mutation was generated to represent the mutations identified in human cancers (9). Selective elimination of the EXO and GEN hydrolytic specificities led to frequent spontaneous mutations and accumulation of incompletely digested DNA fragments in apoptotic cells. In addition, the mutant mice were predisposed to autoimmunity, chronic inflammation and cancers. Thus, the mutator phenotype results in the initiation of cancer, whereas chronic inflammation promotes cancer progression (9).
MULTIPLE STRUCTURE-SPECIFIC NUCLEASE ACTIVITIES AND THEIR BIOLOGICAL FUNCTIONS
Mammalian FEN1 was first characterized as DNase IV, an obligate 5′–3′ dsDNA exonuclease, during a survey of nucleases from rabbit tissues (18). After bacterial 5′–3′ exonucleases in eubacteria were discovered to be flap structure-specific nucleases (19), a mammalian FEN1 homolog was reported and determined to be the same as DNase IV, discovered >20 years earlier (20). Since then, much work to determine how FEN1 recognizes its substrates has been conducted (21–31). The predominantly accepted model for mammalian FEN1 has been the track-down model (27,32). Based on studies in which access to the 5′-end of the flap was blocked by streptavidin, FEN1 was postulated to initially recognize the free 5′-end of the flap structure and then track down the 5′-flap until reaching the double-stranded DNA (dsDNA) region where it then cleaved the scissile phosphate (27,32). Despite the popularity of the track-down model, more recent biochemical evidence suggests that mammalian FEN1 first recognizes the dsDNA two-way junction, and then places the scissile phosphate in the active site by threading or clamping the single-stranded DNA (ssDNA) flap (22,33). The latter mechanism, first proposed by Joyce and co-workers for eubacterial FEN1 homologs (31), is more consistent with the ability of FEN1 to perform exonucleolytic cleavage and is likely the best description of the protein's substrate recognition mechanism in vitro. Furthermore, the latter model is also consistent with the notion that Rad2 nucleases are a family of DNA junction recognition enzymes (34).
In contrast to previous reports that the presence of dsDNA in the flap strand prevented FEN1 from cleaving (27,35), two separate studies determined that FEN1 has GEN activity, which cleaves single-stranded regions with gaps of four or more nucleotides (6,10). CRN-1, the FEN1 homolog in Caenorhabditis elegans, was shown to cleave the template strand of a gapped DNA duplex that resembled an intermediate DNA structure generated during apoptotic DNA fragmentation (6), whereas human FEN1 was found to cleave the template strand of gapped DNA fork and bubble substrates that mimic a stalled DNA replication fork. Furthermore, the presence of intrinsic FEN1 GEN activity was supported by mutational analysis. FEN1 mutations such as R70A, R326A/R327A, K244A/R245A at the dsDNA binding region abolished the EXO and/or GEN activities, but had little effect on the FEN activity (26). The E178A mutation, which occurs near the active center and the dsDNA binding region, was shown to eliminate 95% of the wild-type GEN activity, but retained most of its 5′-FEN activity (10). In contrast, another mutant, Q112R, identified in human cancer cells and located in the helical loop involved in ssDNA binding, was observed to increase GEN activity by 3-fold while retaining wild-type levels of the other two activities (9).
Characterization of the hydrolytic specificities of FEN1 has provided mechanistic insights into how a nuclease with a single active site can perform three separate activities. Kinetic analysis revealed that FEN1 displays its highest catalytic efficiency on the so-called double flap substrate, which is characterized by having a 5′-ssDNA flap of any length and a single nucleotide 3′-flap (26). Consistent with this, equilibrating flap substrates, which can adopt a double flap conformation, are thought to be the in vivo FEN1 substrate (21,25). In comparison to the catalytic efficiency on double flap substrates, the kcat/KM value for the EXO activity on a nick substrate (i.e. no 5′- or 3′-flap) was ∼15-fold less than the FEN activity, whereas the efficiencies of the GEN activity on gapped-fork and gapped-duplex substrates were 6- and 250-fold less than the FEN activity, respectively (26). Despite the wide range of cleavage efficiencies, these three manners of hydrolyzation (FEN, EXO and GEN) all share a commonality: predominant incision 1 nt into the downstream dsDNA with minor cleavages 5′ or 3′ of this site. Because structural studies of FEN1 revealed that FEN1 contains two dsDNA-binding regions and a helical arch associated with ssDNA-binding (36), the ability of FEN1 to hydrolyze a wide range of substrates using the same active center was explained by differences in the manner that FEN1 binds to various substrates (26). Moreover, cleavage patterns can be explained if the downstream dsDNA region (i.e. dsDNA that is incised) is bound by the H3TH motif with the active site in proximity to the scissile phosphate and subsequent placement of other substrate structural elements with the upstream dsDNA binding region and/or helical arch. In addition, the efficiency of cleavage could also be partially rationalized using this model (26). These findings support the concept that FEN1 employs distinct DNA substrate-binding modes to interact with different DNA substrates.
The ability of FEN1 to cleave nick and gapped-fork substrates is greatly enhanced by the addition of a 1 nt 3′-flap, similar to that observed with flap substrates (22). Recently, we and others have conducted more detailed kinetic studies with various FEN1 substrates with a 3′-flap. These studies suggest that the paramount structural feature of substrates recognized by FEN1 is a 3′-flap (22,30) rather than a 5′-flap (27). Although it was previously known that substrates bearing a 3′-flap were cleaved at a faster rate by FEN1 (37), the presence of the 3′-flap was further shown to increase single-turnover rates of catalysis in addition to its role in increasing enzyme-substrate affinity (22,30,37). Thus, occupation of 3′-flap binding pocket is not only critical for initial substrate capture, but for increasing subsequent first order rates of reaction after formation of the enzyme substrate complex by an unknown mechanism. With an increase in kcat and concomitant decrease in KM observed with double flap substrates, it is not surprising that the second order rate constants of reaction with double flap substrates approached levels suggesting that encounter of substrate and enzyme is rate-limiting.
FEN activity is critical for RNA primer removal and BER pathways
FEN activity, the most dominant activity of FEN1, is considered to play a major role in RNA primer removal during Okazaki fragment maturation of lagging strand DNA replication, as well as in removal of flap structures formed during long-patch BER. Details of the role of FEN1 in these two classical pathways were previously reviewed (1,4,38,39). It should be noted that the mechanisms of RNA primer removal in human and yeast are likely different. During lagging strand DNA synthesis in eukaryotic cells, Polα creates RNA/DNA primers containing ∼10 nt of RNA and 10–20 nt of DNA to initiate synthesis of Okazaki fragments. In human and other mammalian cells, Polδ displaces the RNA/DNA primer to create a flap for FEN1 to cleave; however, the size of the flap generated by strand-displacement synthesis in mammals is still unknown. The essential role of FEN1 in RNA/DNA primer removal in mammalian cells has been validated by several in vivo studies using mouse models that showed that FEN1 deficiency causes defects in DNA replication, failure of cell proliferation and embryonic lethality (40–42). In contrast, deletion of RAD27 (FEN1 yeast homolog) is tolerated in yeast, implicating the involvement of other nucleases in this process (43,44). Furthermore, recent studies suggest that long flap structures occur frequently during Okazaki fragment maturation in yeast. The displacement of flaps longer than 30 nt during Okazaki fragment maturation in yeast is envisioned to attract RPA, which inhibits FEN1, but recruits Dna2 endonuclease to cleave a large portion of the primer, as Dna2 can efficiently clamp onto the site. The cleavage of the long flap structure by the Dna2/RPA complex generates a short flap (∼5–7 nt) that resists binding and cleavage by RPA and Dna2, respectively (45). The FEN1 nuclease can then precisely remove the remaining flap to produce a substrate for ligation. However, the RPA-governed sequential actions of Dna2 and FEN1 for Okazaki fragment processing may only occur in yeast, and not higher eukaryotes. In mammalian cells, DNA2 predominantly migrates into mitochondria (46), and the residual nuclear DNA2 is not associated with DNA replication foci (46,47), suggesting that DNA2 is not a primary component in the DNA replication machinery. These findings support the hypothesis that FEN1 may be the primary nuclease in nuclear RNA primer removal in mammals, whereas coordinated actions of FEN1 and Dna2 are important in this process in yeast.
The FEN activity is also necessary for repairing DNA lesions that have an oxidatively-damaged sugar moiety, which cannot be removed by the dRP lyase of Polβ (13,48). In such a case, a few nucleotides are added by polymerase β or δ to produce a short flap, which is subsequently removed by FEN1 in complex with the processivity factor proliferating cell nuclear antigen (PCNA) (49). This alternative FEN1/PCNA-dependant BER pathway is termed long-patch BER (LP-BER) (49). A recent study by Wilson's group further suggested that the FEN and EXO activities of FEN1 might work together because the Polβ activity is only active on DNA duplexes that contain a gap, but not a nick. Therefore, the EXO activity of FEN1 may be important for cleaving the nick substrate and generating an optimal gap intermediate for Polβ during LP-BER (50). The short flap resulting from displacement DNA synthesis would then be removed by the FEN activity. In support of this model, the E160D FEN1 mutation, which specifically eliminates the EXO activity, causes mouse embryonic fibroblasts to be sensitive to DNA damaging agents such as methylmethane sulfonate (9).
Concerted action of EXO and GEN activities for resolution of DNA hairpin structures and apoptotic DNA fragmentation
The two other ‘minor’ activities of FEN1 (EXO and GEN) may be important for other DNA metabolic pathways, including resolution of tri-nucleotide repeat (TNR) sequence-derived secondary structures and apoptotic DNA fragmentation. Much evidence indicates that FEN1 efficiently cleaves ssDNA flaps, but inefficiently processes flaps that adopt secondary structures and lack a 3′-flap (27,51). However, absence of Fen1 in yeast results in a significant increase in TNR expansion (51). Furthermore, the expression level of FEN1 has recently been shown to correlate with TNR expansion in mice (52). Therefore, three possibilities exist: (i) TNRs do not always form stable secondary structures, (ii) FEN1 uses an alternative approach to resolve the secondary structures (5,8) or (iii) increased unequal sister chromatid exchange, a phenotype associated with FEN1 deletion (53), causes TNR contraction and expansion. We have tested the hypothesis that concerted action of the EXO and GEN activities of FEN1 plays a role in resolution of secondary structures formed by (CTG)n and (GAA)n repeats (8). Using a yeast FEN1 point mutant, E176A, which is deficient in EXO and GEN activities but retains almost all of its FEN activity, we showed severe defects in the ability of this mutant to cleave various TNR intermediate substrates (8). Precise knock-in of the E176A point mutation in yeast caused an increase in both the expansion and fragility of a (CTG)n tract in vivo (8). Taken together, the combined biochemical and genetic analyses suggested that although FEN activity is important for ssDNA-flap processing, the EXO and GEN activities may also contribute to the suppression of TNR contraction and expansion. Therefore, we proposed the following model: in the presence of TNRs, which can base pair and fold back to form hairpin structures or loops, FEN1 endonucleolytic cleavage is suppressed. This increases the half-life of the flap, which can form an internal loop and ligate into the genome, resulting in TNR expansion. However, as a back-up approach, the EXO activity of FEN1 can remove the hairpin nucleotides in a gap or ssDNA/dsDNA junction, on which GEN cleavage can occur (Figure 1).
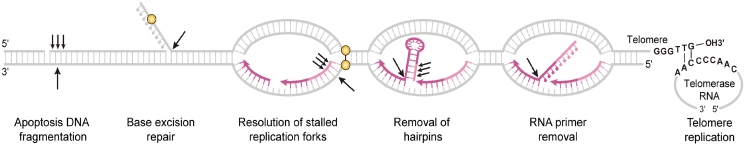
Figure 1.
DNA metabolic pathways and corresponding biochemical activities of FEN1. FEN1 is implicated in six DNA metabolic pathways. Its FEN activity is essential for cleaving the single-stranded flap resulting from displacement DNA synthesis during RNA primer removal and in long-patch base excision repair. The 5′-EXO activity of FEN1 removes a few nucleotides from 5′-blunt ends or nicks to generate a gap, which can be cut by the gap-endonuclease (GEN) activity of FEN1 at the opposite template strand. The concerted action of the EXO and GEN activities produces DNA double strand breaks, which are important for removal of hairpin structures, rescue of stalled replication forks and apoptotic DNA fragmentation. FEN1 activities may also be important for maintenance of the telomere, but how they act at the chromosome end is unknown. Arrows: FEN1 nuclease cleavage sites; Red: newly synthesized RNA and DNA; yellow circles: damaged DNA or cross-links.
The finding that FEN1 plays an important role in suppression of TNR expansion in yeast has stirred great interest in exploring whether FEN1 is critical in destabilization of the TNR hairpin structures and whether deficiency of FEN1 in humans contributes to TNR-related genetic diseases, including Huntington disease (HD) (51,54). However, whether FEN1 deficiency plays a role in development of HD or other TNR-related diseases remains controversial. A survey for FEN1 mutations in human HD patients did not identify any mutations in this subset of patients (55). More recently, another study showed that haploinsufficiency of FEN1 did not accelerate the expansion of (CTG)n (CAG)n (n: 105–110) repeats, which resemble the myotonic dystrophy type 1 disease locus, in mice (129Ola genetic background) (56). On the other hand, Yang and Freudenreich (57) showed that haploinsufficiency of FEN1 promotes expansion of (CAG)120 repeats that mimic the human HD locus in mice (CBAxC57BL/6 genetic background. We note that these two TNR mouse models have different genetic backgrounds, which may contribute to the discrepancies in TNR phenotypes. It is likely that FEN1 mutations on their own may not significantly promote TNR expansions or contractions due to redundant pathways in maintenance of TNR stability, but in combination with other genetic mutations, FEN1 deficiency may contribute to TNR instabilities and related diseases in humans. Supporting this view, a previous study showed that microsatellite instabilities were not promoted in mice heterozygous for FEN1 knockout, but haploinsufficiency of FEN1 in a genetic background with an adenomatous polyposis coli (APC) gene mutation did significantly increase microsatellite instabilities (40).
The concerted action of the EXO and GEN activities of FEN1 may also play a role in oligonucleosomal fragmentation of chromosomes in apoptotic cells (58). In collaboration with the Xue group, we have shown that crn-1, a C. elegans homolog of human FEN1, is important for apoptosis (6). Reduction of CRN-1 activity by RNA interference resulted in cell death phenotypes similar to those displayed by a mutant lacking the mitochondrial endonuclease CPS-6/endonuclease G (Endo G). We proposed that CRN-1 associates and cooperates with CPS-6 to promote stepwise DNA fragmentation, whereby CPS-6 nicks the dsDNA and the EXO activity of CRN-1 subsequently removes a few nucleotides from the 5′-end to generate a gap. The GEN activity of CRN-1 then cuts the template strand at the DNA gap, leading to DNA fragmentation (Figure 1). Supporting this hypothesis, the EXO- and GEN-deficient FEN1 mutation, E160D, retards the disposal of apoptotic DNA and results in anti-dsDNA circulating in the blood system of mutant mice (9).
FEN1 functions in other DNA metabolic pathways
FEN1 has been implicated in the rescue of stalled replication forks caused by exogenous physical and chemical insults. FEN1 forms a complex with WRN in response to DNA damage that arrests replication forks (10,59), suggesting a role of FEN1 in resolution of stalled replication forks. Results from the Brosh group suggest that the FEN activity may be important for cleaving regressed replication forks to resolve the chicken foot structure (59). Alternatively, stalled replication forks may be restarted via the break-induced recombination pathway, in which a DNA double-stranded break (DSB) is created on the stalled replication fork to initiate downstream recombination cascades (60). ssDNA gaps that accumulate on stalled replication forks are potential targets for endonucleases to generate DSBs. In such a case, the GEN activity, which is dramatically stimulated by WRN, cleaves the fork to initiate the break-induced recombination (10). Human FEN1, but not the GEN-deficient mutant, E178A, is capable of rescuing yeast fen1 null mutants from UV- and campothecin-induced DNA damage, indicating the importance of the GEN activity of FEN1 in restarting stalled replication forks. Furthermore, we have recently shown that deficiency in FEN1 nuclease activities in yeast also causes defects in replication of rDNA, in which DNA replication forks are frequently stalled by naturally occurring fork barriers such as DNA-binding proteins (12).
Several lines of biochemical and genetic evidence have suggested that FEN1 plays a role in maintenance of the integrity of GC-rich repeat DNA sequences, including CEB-1 minisatellites, rDNA regions and telomeres. Deletion of Rad27 in yeast causes instabilities of the human minisatellite locus CEB1 inserted into the yeast genome (61,62). Two models have been proposed to explain how FEN1 deficiency causes minisatellite expansions or contractions. In the first model, deficiency in the FEN activity leads to unprocessed 5′-flap structures. The GC-rich minisatellites, such as CEB1 sequences, that are prone to forming secondary structures, fold back and anneal to the template strand (61,62), although such secondary structure can be processed by the concerted action of EXO and GEN activities of FEN1 (8,26). Alternatively, the non-cleaved DNA flap may result in DNA DSBs and trigger improper DNA recombination events, leading to expansions or contractions (17,62).
Recently, FEN1 has also been suggested to contribute to telomere stability in yeast (63,64) and human cells (7,65,66). Abrogation of FEN1 function may influence the transformation of a cell into a cancer cell by compromising telomere stability and driving genomic instability. The Steward group analyzed the telomeres of human cancer cells after FEN1 depletion, and showed that FEN1 is required for telomere stability in cancer cells that rely on the alternative lengthening of telomere (ALT) mechanism (7,65). FEN1 depletion resulted in telomere dysfunction that was characterized by formation of telomere dysfunction-induced foci (TIFs) and end-to-end fusions in ALT-positive cells. In contrast, no telomere dysfunction was observed in telomerase-positive cells with FEN1 depletion, suggesting that ongoing telomerase activity protected telomeres. The FEN1 mutations affect telomere stability and genome fidelity by promoting telomere fusions and anaphase–bridge–breakage cycles, which further drive genome instability, thereby contributing to the transformation process (7,65). In addition, Chai and colleagues have shown complementary data indicating that FEN1 is in complex with telomerase in vivo (66), and have demonstrated that FEN1 deficiency in mouse embryonic fibroblasts leads to an increase in telomere end-to-end fusions. In cancer cells, FEN1 deficiency induces gradual shortening of telomeres, but does not alter the single-stranded G-overhangs (66). This is the first evidence that FEN1 and telomerase physically co-exist as a complex and that FEN1 can regulate telomerase activity at telomeres in mammalian cells (66). A recent study shows that FEN1, but not exonuclease 1, can cleave varying model substrates containing a telomeric G-quadruplex structure, suggesting that FEN1 may be important in resolution of G-quadruplex structures during replication of telomere sequences (67). Because FEN1 can cleave both DNA and RNA, we postulate that FEN1 may also function to remove the telomerase RNA template in the last cycle of telomere extension (Figure 1).
REGULATORY MECHANISMS
Localization as a major means of functional regulation of FEN1
Despite the wealth of information available on the biochemical functions of FEN1 and its roles in genome stability and cancer avoidance, cellular compartmentalization and dynamics that correspond to FEN1's involvement in various DNA metabolic pathways have not been fully elucidated. Previously, we demonstrated that FEN1 migrates into the nucleus in response to DNA damage and during S-phase of the cell cycle (Figure 2) (11). Furthermore, primary sequence analysis of eukaryotic FEN1 identified a bipartite C-terminal motif that is rich in positively-charged amino acid residues as a putative nuclear localization signal, which was validated by site-directed mutagenesis (11). More recently, we found that FEN1 super-accumulates in the nucleolus (Figure 2) and plays a role in the resolution of stalled DNA replication forks that form at sites of natural replication fork barriers (12).
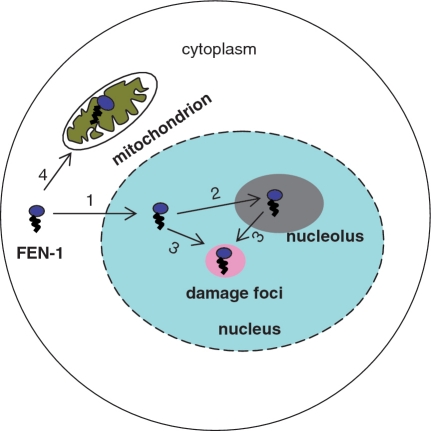
Figure 2.
Dynamic subcellular localization of FEN1. FEN1 localizes to four distinct subcellular compartments in mammalian cells: cytoplasm, mitochondria, nuclear plasm and nucleolus. In response to cell cycle transitions and DNA damage, FEN1 dynamically migrates via four different routes: (i) nuclear translocation from the cytoplasm, (ii) accumulation in the nucleolus from the nuclear plasm; (iii) formation of replication/DNA damage foci from the nuclear plasm and nucleolus and (iv) mitochondrial import from the cytoplasm. The nuclease domain of FEN1 is represented by an oval, and the C-terminus is indicated as a zig-zag tail.
High-resolution imaging of cells stained with anti-FEN1 antibodies revealed that FEN1 is also present in the mitochondria (Figure 2) (13,68). We conducted fluorescence microscopy on a C-myc-tagged FEN1 mutant protein (Y83H) to clearly show that this mutant protein localized to mitochondria and not to nuclei (13). These data were complemented by cell fractionation experiments, which identified FEN1 in the mitochondrial extract. Further support for FEN1 migration into the mitochondrion comes from the work of Kalifa and Sia, demonstrating the mitochondrial localization of yeast FEN1 using immunohistochemical and genetic approaches (69).
The replication and repair of mtDNA depends on many factors that are encoded in the nucleus, as the gene products of the mtDNA genome are limited to 22 tRNAs, 2 rRNA subunits and 13 proteins involved in energy production (70). All other genes required for mitochondrial function are encoded in the nuclear genome, and their gene products must be imported (70). Recently, other DNA replication and repair proteins encoded in the nuclear genome, in addition to FEN1, were shown to be localized in mitochondria in addition to the nucleus (13,71,72). Because reactive oxygen species are mainly generated in mitochondria, mtDNA is profoundly susceptible to oxidative damage, the accumulation of which is positively correlated with aging and age-related diseases like neurodegenerative disorders. Studies of mtDNA damage have shown that a significant portion of oxidative damage occurs on the deoxyribose moiety. Such damage results in the formation of deoxyribolactone, which if repaired by short-patch BER, gives rise to protein-DNA cross-links. Therefore, the majority of deoxyribolactone products are preferentially processed by LP-BER. More recently, we have shown that immunodepletion of FEN1 or DNA2 nuclease impairs the ability of mitochondrial extracts to process model substrates representing intermediate structures during RNA primer removal, suggesting that FEN1 plays a role in mtDNA replication as well. Because FEN1 is the nuclease responsible for flap cleavage in RNA primer removal and LP-BER, its presence in the mitochondria is not surprising. However, the mechanism by which it enters the mitochondrial matrix is unknown, as it is devoid of known canonical mitochondrial targeting sequences. We suggest that FEN1, like DNA2 nuclease, may be imported into mitochondria via an internal mitochondrial targeting motif.
Protein–protein interaction: FEN1's functional complexes
The types of proteins with which FEN1 interacts suggest its pivotal role in several DNA metabolic pathways. We therefore carried out an extensive investigation of the network of all possible physical protein–protein interactions (PPIs) of human FEN1 and its homologs in Saccharomyces cerevisiae (Rad27) and C. elegans (CRN-1). Manual curation of PPIs from the literature began by species-specific identification of PPIs through the PPI database STRING (http://string-db.org). This method identified 105, 137 and 42 PPIs from human, S. cerevisiae, and C. elegans, respectively, with a default confidence of 0.4. We ultimately identified 34 PPI partners of FEN1 that were previously evaluated by various methods, including co-immunoprecipitation and yeast two hybrid. Based on the known effects of these interactions on the biochemical activities and biological functions of FEN1, its interacting partners can be grouped into four functional categories (Figure 3).
Figure 3.
Protein–protein interactions mediate FEN1's actions in different DNA metabolic pathways. FEN1 interactive proteins were categorized into five functional groups: DNA replication, DNA repair, apoptotic DNA degradation, maintenance of telomere stabilities and post-translational modifications, based on the known biochemical activities and pathways of these proteins. Protein–protein interactions (PPIs) of FEN1 that occur in mammalian cells are colored in gray. The physical and functional interactions of FEN1 (Rad27) with Pol4 and FEN1 with Dnl4/Lif1, which are important for the processing of DNA ends in S. cerevisiae (91,117), are colored in yellow. Two nucleases, CPS-6 and CRN-1, the C. elegans homologs of EndoG and FEN1, respectively, have been implicated in mediating apoptotic DNA degradation in C. elegans (6,118) and are colored in blue. APEX1, AP endonuclease 1; BLM, bloom syndrome protein; CDK1, cyclin-dependent kinase 1; CDK2, cyclin-dependent kinase 2; CPS-6, ortholog of human mitochondrial endonuclease G (EndoG); CRN-1, cell-death-related nuclease family member (crn-1); CRN-3, cell-death-related nuclease family member (crn-3); CRN-4, cell-death-related nuclease family member (crn-4); CRN-5, cell-death-related nuclease family member (crn-5); Cyclin A, cyclin A2; CYP-13, cyclophilin homolog; DNA2L, DNA2-like helicase; Dnl4, DNA ligase IV homolog; EP300, E1A binding protein p300; FEN1, flap structure-specific endonuclease 1; hCHLR1, DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11; HMG1, high-mobility group box 1; HNRNPA1, heterogeneous nuclear ribonucleoprotein A1; Hus1, HUS1 checkpoint homolog; LIG1, DNA ligase 1; NEIL1, Nei endonuclease VIII-like 1; Nej1p, homologous to mammalian XRCC4 protein; PCNA, proliferating cell nuclear antigen; POL α, DNA polymerase α-catalytic subunit; POL β, DNA polymerase β; POL δ, DNA polymerase δ-catalytic subunit; POL ε, DNA polymerase ε, catalytic subunit A; Pol4, DNA polymerase IV; PRMT5, protein arginine methyltransferase 5; Rad1, DNA repair exonuclease rad1 homolog; Rad17, homolog of human and S. pombe Rad1 and Ustilago maydis Rec1 proteins; RAD27, 5′–3′ exonuclease, 5′-FEN; Rad9, DNA repair exonuclease rad9 homolog A; RFC, replication factor C; RPA, replication protein A; TERT, telomerase reverse transcriptase; TRF2, telomeric repeat binding factor 2; WAH-1, worm AIF homolog; WRN, Werner syndrome ATP-dependent helicase; Solid line, physical and functional interactions; dashed lines, genetic interactions; rightwards arrow: stimulatory to the enzyme activities; multimap, inhibitory to the enzyme activity; reverse assertion, post-translational modifications.
The first category includes protein partners that assist FEN1 in RNA primer removal during DNA replication. In eukaryotic cells, DNA synthesis is primed by the primase/polymerase-α (primosome), which synthesizes the RNA/DNA initiator primer (73). After primer synthesis, replication factor C (RFC) loads the homotrimeric ring-shaped protein known as PCNA onto dsDNA. PCNA acts as a sliding clamp molecular adaptor that localizes bound proteins to DNA. When the processive replisome encounters the downstream Okazaki fragment, a portion of the RNA/DNA primer is displaced to form a 5′-flap structure, which needs to be removed prior to ligating the remaining DNA segments. FEN1 interacts with PCNA, Pol α/ε, RPA, hnRNP A1 and DNA ligase I for efficient Okazaki fragment processing (74–78). RPA has been reported to inhibit FEN1, based on the ‘sequential nuclease cleavage’ model in yeast cells, in which DNA2 and FEN1 sequentially cleave the RNA primers (45).
FEN1 also interacts with members of the RecQ helicase family, including WRN, BLM and RecQ5. The possible physiological roles of these interactions with FEN1 in DNA replication and replication fork rescue have been discussed in detail elsewhere (59,79). In contrast to PCNA, Wrn protein stimulates the GEN activity, thereby cleaving the ssDNA region in the duplex DNA molecule (10). In addition, there is evidence that formation of a functional FEN1/Wrn complex is important for resolving stalled DNA replication forks. Moreover, phenotypes associated with yeast DNA2 mutants can be rescued by expression of BLM or WRN, which stimulate FEN1 activity (59,80). Therefore, it is interesting to speculate that the complex formed between FEN1 and either RecQ helicase is the functional homolog of DNA2. Furthermore, Imamura and Campbell have postulated that a helicase such as BLM may be necessary to facilitate strand displacement synthesis (80), which suggests that the RecQ helicases might coordinate formation and FEN1-mediated removal of 5′-flap structures during Okazaki fragment maturation. When dealing with the ends of chromosomes, FEN1 interacts with proteins involved in telomere stability, which include TRF2, Wrn and TERT (7,66). Furthermore, FEN1 has also been shown to interact with Chlr1, a member of the DEAD/DEAH helicases in the chromosome cohesion complex (81), which implies that FEN1 may play a role in communicating between the DNA replication and chromosome segregation machinery.
The second category of FEN1-interacting proteins is DNA repair proteins. In mammalian cells, single base lesions are repaired by single-nucleotide BER or LP-BER. Apurinic endonuclease 1 (APE1) recognizes the apurinic/apyrimidinic residues (AP) and cleaves 5′ to the AP sites after the actions of glycosylases such as Neil1, thereby generating a nick on which Pol β is loaded (82,83). If the lyase activity of Pol β cannot remove the deoxyribose moiety after it has added the complementary nucleotide to the free 3′-hydroxyl, strand displacement synthesis occurs, thereby creating a small 5′-flap (2–10 nt) that must be cleaved by FEN1 with subsequent ligation of the nick to complete the repair (48). The role of FEN1 in LP-BER is regulated and coordinated by physical interaction with important BER components such as Pol β, APE1, Lig 1 and PCNA (82,84,85). The LP-BER protein Neil1 has also been reported to interact and stimulate FEN1 nuclease activities in vitro (83). Furthermore, WRN has recently been shown in vitro to participate in LP-BER by facilitating Pol β strand-displacement synthesis via its helicase activity (86). Therefore, WRN's interaction with and stimulation of FEN1 may also be important for efficient 5′-flap removal in LP-BER. More recently, the FEN1/WRN complex was implicated in replication fork rescue through break-induced repair (10). FEN1 was also shown to interact with the human Rad9-Rad1-Hus1 checkpoint complex (9-1-1 complex) (87–89), a heterotrimeric protein that is similar to the PCNA toroidal sliding-clamp. Formation of this complex is stimulated by Rad17/RFC in yeast in a p53-dependent manner (90). Although the 9-1-1 complex is unable to stimulate Pol δ synthesis like PCNA, it can stimulate the FEN activity of FEN1 on flap substrates (89). The role of this interaction in vivo remains to be shown. Because 9-1-1 accumulates at sites of DNA damage, the interaction of FEN1 with 9-1-1 is likely important for stimulating FEN-1 activity for DNA repair. Recently, ScFEN1 (RAD27) was shown to interact with Pol4 and Dnl4/Lif1, which are components of the non-homologous end-joining dsDNA break repair pathway (91). The various functional interactions between FEN1 and proteins involved in different DNA repair pathways indicates its wide range of roles in DNA repair.
The third category consists of apoptotic proteins, and is exemplified by Endo G. Parrish et al. (6) demonstrated that CRN-1, the C. elegans FEN1 homolog, in association with CPS-6 (Endo G), mediates stepwise DNA degradation during apoptosis. This is the first example in a burgeoning field that is sure to lead to the identification of additional apoptotic FEN1-interacting proteins across many species. Furthermore, Parrish et al. (6) have identified seven novel nucleases that play a role in apoptotic DNA fragmentation in C. elegans. Most of these proteins form a complex called a ‘degradosome’ to efficiently break down the chromosomes and nucleic acids during development (6). Association of FEN1 with apoptotic proteins such as Endo G may be a key mechanism to switch FEN1's role from DNA replication and repair to apoptotic DNA fragmentation. FEN1 interaction with Endo G also greatly enhances its minor GEN and EXO activities, important for disposal of apoptotic DNA.
The final category of FEN1-interaction partners are those proteins that post-translationally modify FEN1, including p300, Cdk1-Cyclin A, Cdk2-Cyclin A and PRMT5 (14–16). These proteins and their involvement in FEN1 function are discussed in detail in the next section.
Despite the wide variety of proteins, many of these PPIs are predominantly mediated via FEN1's C-terminal extension (88). In the absence of a protein partner, the extended C-terminus is predicted to be intrinsically disordered (DisEMBL http://dis.embl.de and PONDR http://www.pondr.com); thus, FEN1 joins a growing list of proteins that are mostly folded, but have regions of local disorder that mediate PPIs (92,93). Locally disordered regions of proteins are known to undergo disorder-to-order transitions upon binding a target. This allows for the formation of complexes with high specificity and relatively low affinity, which is important for proteins that must initiate an action and then dissociate when finished with that action. The disordered region of a protein also confers the ability to interact with multiple protein partners and modifying enzymes thereof using the same motif (92,93). Thus, the nucleolytic action of FEN1 in the cell is likely directed by different protein partners by binding initially to the C-terminal region of FEN1 as a means to sequester the protein to specific sites in the nucleoplasm. Although high affinity interaction is mediated by the extended C-terminus, a crystal structure of FEN1 in complex with PCNA revealed that core nuclease domain interactions also exist (94), but are probably too weak to mediate interactions strong enough to be detected by non-equilibrium binding assays. Therefore, many of the proteins identified as FEN1 interaction partners probably utilize the extended C-terminus, but interactions are likely not exclusive to this region.
Recently, we have determined the residues critical for FEN1 interaction with its five different interactive partners (88). Consistent with the role of a disordered region, each interactive partner of FEN1 requires a distinct combination of extended C-terminal residues for binding. For instance, the Q337, L340, D342, F343, F344, V346, S353 and S352 residues within and next to the PIP box of FEN1 are essential for the FEN1/PCNA interaction, whereas the K366, K367, K375, R378 and K380 residues are required for the FEN1/WRN interaction (88). On the other hand, the lysine residues, including K375 and K380, of FEN1 have been shown to be acetylated by p300 (14). The distinct PPI signatures at the C-terminal region of FEN1, together with post-translational modifications, may provide a mechanism to allow FEN1 to interact with different proteins and be recruited to different DNA metabolic machineries in response to DNA damage.
Simple deletion of a gene is not a high-resolution approach to study the biological significance of a PPI, because a single protein may interact with multiple partners. In addition, the complete deletion of an essential gene is often embryonically lethal in animals. Therefore, to obtain high-resolution biological information for a specific interaction, we take an alternative approach of knocking-in rather than knocking-out identified mutations that specifically disrupt one interaction. Phe343 and Phe344 of FEN1 are key residues that mediate the interaction between the PIP-box of FEN1 and the interdomain connector loop motif of PCNA (49,95) (Figure 4A). Substitution of these two residues with two Ala residues (FFAA) abolishes the high affinity physical interaction between FEN1 and PCNA (49,95). Disruption of this interaction does not affect the in vitro nuclease activity of FEN1. Instead, it disrupts precise PCNA-mediated coordination among the polymerase, nuclease and ligase during the processes of Okazaki fragment maturation and LP-BER (Figure 4B). As a consequence, the FFAA mutation causes failure of recruitment of FEN1 to the replication foci and inhibits processing of Okazaki fragments, even when present only in the heterozygous form. Furthermore, heterozygous FFAA mutant mice develop aneuploidy-associated cancers, similar to what has been observed in human cancers (L. Zheng et al. unpublished data).
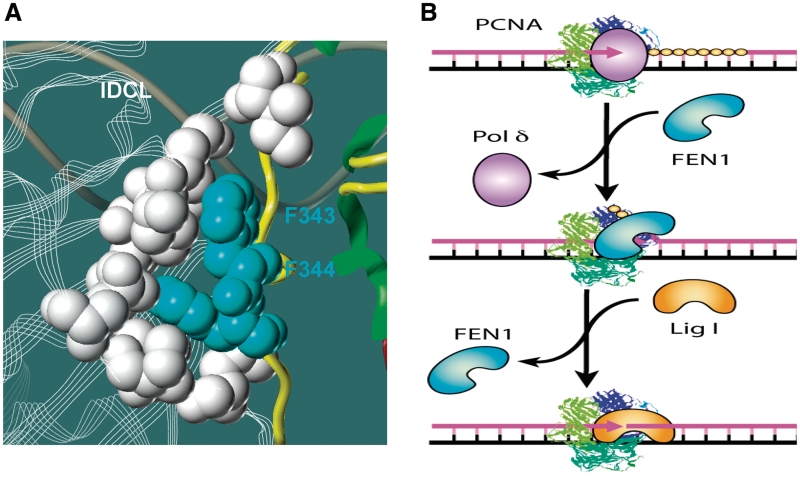
Figure 4.
Model of the PCNA-mediated sequential actions of Pol δ, FEN1 and Lig I. (A) Structural basis for FEN1/PCNA interactions. Three dimensional structure demonstrating that F343 and F344 of FEN1 are key residues that mediate the physical interaction between FEN1 and PCNA. The structural model was based on crystal structures of the FEN1/PCNA complex (36,94). IDCL: interdomain connector loop. (B) Sequential recruitment of Pol δ, FEN1 and Lig 1 via PCNA interaction. The PCNA trimmer encircles the DNA duplex and serves as a platform for sequential recruitment of Pol δ, FEN1 and Lig I, which are key enzymes in Okazaki fragment maturation.
Post-translational modifications: a primary event for functional regulation
FEN1 is a target for at least three post-translational modifications in vivo: phosphorylation (15), acetylation (14,96) and methylation (16). The Cdk1-cyclin A or Cdk2/cyclin E complex phosphorylates FEN1 at Ser187 in late S phase, and in vitro Cdk1-cyclin A phosphorylation of FEN1 reduces the endonuclease and exonuclease activities of FEN1 without affecting DNA binding (15). In addition, phosphorylation of FEN1 abolishes PCNA binding (15). Serine phosphorylation is hypothesized to be one of the cell cycle regulatory mechanisms of FEN1 activities, and Ser187 is the only FEN1 residue that has been shown to be phosphorylated (15). In late S phase, phosphorylation may be used to block FEN1's nuclease activities and its recruitment to the DNA replication site by PCNA. Therefore, this phosphorylation may contribute to halting the replication machinery, thereby ensuring exit from S phase (15). In addition, phosphorylation of FEN1 was also shown to regulate its nucleolar localization and to influence the dynamic roles of FEN1 in ribosomal DNA replication and damage repair (12). In response to UV irradiation and upon phosphorylation, FEN1 migrated to the nucleoplasm to participate in the resolution of UV crosslinks on DNA, most likely through the concerted action of its EXO and GEN activities (12). Based on yeast complementation experiments, the Ser187Asp mutation, which mimics constitutive phosphorylation, abolishes nucleolar accumulation of FEN1. On the other hand, replacement of Ser187 by Ala, which eliminates the only phosphorylation site, causes retention of FEN1 in the nucleoli. Both mutations cause UV sensitivity, impair cellular UV damage repair capacity and reduce overall cellular survival (12).
Furthermore, the methylation and phosphorylation of FEN1 offer a possible answer to one critical question of how FEN1 can efficiently bind to PCNA and DNA substrates, and then dissociate once the nuclease reaction is complete to avoid blockage of Lig1 reaction during DNA replication in the S phase of the cell cycle. Our recent discovery sheds light on this puzzle. We demonstrated that FEN1 is methylated at the R192 residue, and this methylation prevents FEN1 phosphorylation at the S187 residue (16). The methylated form of FEN1 interacts with PCNA, but the phosphorylated form of FEN1 causes prompt dissociation from PCNA, providing a mechanism for the nuclease to dynamically associate with and dissociate from PCNA, and thus, the DNA substrate (16). Based on our data, we postulate that in the early stages of Okazaki fragment maturation, FEN1 exists in a methylated form, which enables it to replace Pol δ and access PCNA and the flap structure. Upon cleavage of the RNA primer flap, FEN1 is demethylated, thereby allowing the nuclease to be phosphorylated by a cell cycle-dependent kinase, cdk1 or cdk2. Phosphorylation of FEN1 causes the nuclease to fall off PCNA and the DNA nicks, leading to Lig1 recruitment and DNA ligation. However, the mechanism by which FEN1 is demethylated remains to be investigated.
Another intriguing question is how FEN1 phosphorylation influences the FEN1/PCNA interaction. Previous studies have demonstrated that phosphorylation may induce conformational changes and modulates protein–protein interactions (97–99). We suggest that phosphorylation of the S187 residue adds a negative charge to the embedded S187 residue, leading to exposure of the phosphorylated S187 residue. This may in turn stabilize a conformation whereby the positively-charged C-terminal random coil folds back to interact with the negatively-charged nuclease core domain, masking the interface for PCNA binding. As a consequence, phosphorylated FEN1 fails to interact with PCNA. This hypothesis is consistent with a previous study showing that phosphorylation of PTEN induced conformational changes that led to the masking of the PZD binding domain and disruption of the interaction between PTEN and PZD domain-containing proteins (98). In addition, the theory of phosphorylation-induced conformational change can also explain why FEN1 phosphorylation at the S187 residue abrogates FEN1 nucleolar localization. FEN1 co-localizes with WRN and nucleolin (C23) in nucleoli (12), and the interactions between FEN1 and nucleolar proteins such as WRN and nucleolin are important to retain the nuclease within nucleoli. However, because the C-terminal region is also the key element for interaction with WRN, folding back of the C-terminal region in phosphorylated FEN1 may also disrupt FEN1 interaction with WRN or other proteins, leading to migration out of nucleoli.
FEN1 can also be post-translationally modified by acetylation (14,100). Hasan et al. (14) observed an acetylated form of FEN1 in HeLa cells, and the amount of this form was enhanced upon UV treatment. The p300 acetyltransferase interacts with FEN1 in vitro and in vivo and acetylates FEN1 in vitro (14). Mass spectrometry analysis of in vitro acetylated FEN1 revealed four lysine sites located the very C-terminus of FEN1 that could be acetylated (K354, K375, K377 and K380) (14). In vitro acetylated FEN1 had reduced PCNA-independent nuclease activity and DNA binding affinity, but intact PCNA-binding and stimulating capacity (14,96). The inhibitory effect of FEN1 acetylation on its enzymatic activity is puzzling, considering that the nuclease activities of FEN1 are important for processing different DNA intermediate structures in various DNA metabolic processes. Hubscher and colleagues suggested that FEN1 acetylation functions to avoid premature processing of Okazaki fragments (14). Inhibition of FEN1 would promote the formation of longer flaps that require the two-nuclease pathway (DNA2-FEN1) (14). In support of this, Bambara and colleagues showed that p300 acetylates Dna2, thereby stimulating its 5′–3′ endonuclease, helicase and DNA-dependent ATPase activities, and enhancing its binding with DNA substrates (35). More recently, three acetylation sites, K80, K267, K375, were identified in FEN1 purified from human cells (100). Among these three in vivo acetylation sites of FEN1, two are different from the in vitro identified sites. The functional roles of these new acetylation sites remain unknown. Because K267 is close to the downstream dsDNA binding region, it could regulate DNA binding. Like K267, K80 is solvent exposed, but on the opposite side of the DNA binding interface. Therefore, like acetylation of K375 in the extended C-terminus, these acetylations may regulate PPIs.
FEN1 MUTATIONS LINK THE MUTATOR GENE TO CANCER
Functional deficiency of FEN1 has been suggested to cause genomic instability and cancer predisposition, and the importance of FEN1 in the prevention of oncogenesis has recently been studied (40). In mice, homozygous knockout of FEN1 is embryonically lethal, consistent with the observation that Fen1 null mouse blastocysts are arrested in S phase (41). Fen1 heterozygous knockout mice are viable and appear to be free of disease. However, FEN1 heterozygous knockout mice that are also heterozygous for the APC gene develop adenocarcinomas that result in decreased survival (40). These results suggest that FEN1 is a tumor suppressor gene (101).
The initiation and development of cancer is considered a microcosm of the evolutionary process, a hallmark of which is the accumulation of numerous genetic abnormalities in multiple genes (102,103). Inherited mutations of specific genes, such as BRCA1 or BRCA2, and mismatch repair genes, which result in susceptibility to breast cancer and colorectal cancer, respectively, account for only 20% of cancers (104). In most cancers, genetic changes arise by somatic mutations (104). Based on our understanding of the FEN1 nuclease, we predict that there could be four classes of FEN1 mutations that may be identified in cancer cells, which might impair (i) the nuclease activities, (ii) compartmentalization, (iii) protein–protein interactions or (iv) post-translational modification sites. Thus far, we have identified FEN1 mutations that eliminate the EXO and GEN activities, but retain the FEN activity (9). To demonstrate the etiological significance of these somatic mutations, we established the first mouse line harboring the E160D mutation, which interferes with FEN1 binding to the magnesium ion responsible for substrate binding and specifically abrogates the EXO and GEN activities of FEN1 (9,105). This mouse line was used to model the FEN1 mutations identified in human cancers (9). Selective elimination of nuclease activities in this mouse line led to frequent spontaneous mutations and accumulation of incompletely digested DNA fragments in apoptotic cells. Two independent studies demonstrated that the E160D mutant mice are susceptible to the development of cancers (9,106). We further revealed that the mutator phenotype results in the initiation of cancer, whereas chronic inflammation promotes cancer progression (9). These findings support the view that FEN1 is required for genome stability and can therefore be considered a tumor suppressor gene (101).
The onset of tumorigenesis is rooted in the accumulation of genetic aberrations, such as point mutations, frame shifts, chromosomal defects and microsatellite expansions and contractions. The sequential progression from normal to malignant cells is accompanied by an increase in these alterations in cells, which continues within the heterogeneic tumor cell population during tumor growth. The mechanisms by which these errors accumulate are thought to be due in part to damage acquired by genes involved in genomic stability. This instability would result in further mutations to regulatory genes. The involvement of FEN1 in controlling genomic stability through multiple metabolic actions suggests that functional loss of FEN1 through mutations would facilitate further tumor mutagenesis.
Deficiency in the activity of FEN1 nuclease to degrade apoptotic DNA, on the other hand, can result in production of anti-DNA auto-antibodies and subsequently lead to autoimmune-related chronic inflammation, which may promote cancer progression, possibly via activation of NF-kB (9). The FEN1 E160D mutant mouse model will be useful for resolving additional issues in cancer biology, including the identification of essential mediators of chronic inflammation in lung tissue; the determination of whether suppression of inflammatory responses by altering cytokine profiling with NF-kB inhibitors, or by anti-inflammatory agents, reduces cancer incidence; and the establishment of whether E160D mutant mice, deficient in DNA repair and apoptotic DNA degradation, are more susceptible to tobacco-induced tumorigenesis. Answering these questions will be important to designing new cancer therapeutic regimens that act by the suppression of specific inflammatory responses.
Contrary to FEN1's role as a tumor suppressor, several studies from our and other groups suggest that FEN1 may also be required to support the growth and progression of cancers. FEN1 is detectable in all proliferative tissues, but is at very low levels in quiescent cells (107). Furthermore, FEN1 expression is elevated in cultured lung cancer cells as compared to normal cells (108), and FEN1 is over-expressed in many cancer types (109–112). In addition, the level of FEN1 expression in tumor tissues has been correlated with increased tumor grade and aggressiveness (109). Because FEN1 plays an essential role in DNA replication, it is thought that high levels of FEN1 are required to support hyperproliferation of cancer cells. Consistent with FEN1 over-expression in cancer tissues, several breast cancer cell lines have increased FEN1 expression at the mRNA and protein levels that correlate with decreased CpG2 methylation levels in comparison to matched normal tissues (111). Whether the epigenetic changes in the promoter region of FEN1, as mentioned above, occur in cancers other than breast cancer remains to be determined.
CONCLUDING REMARKS
Compelling evidence from in vitro and in vivo studies indicates that FEN1 is a multifunctional nuclease that participates in distinct DNA metabolic pathways. Interestingly, FEN1 can promote cancer in two very different ways: (i) mutation of the gene can result in genomic instability and initiate malignant transformation and (ii) overexpression of the gene confers a growth advantage to tumors. In response to the latter way to promote cancer, FEN1 has been suggested as cancer therapeutic target (113–116). However, like other current chemotherapeutics, inhibitors of FEN1 will likely be fraught with side effects and have the potential to propagate or even initiate new cancers. Nonetheless, such work should continue to increase our clinical cancer arsenal, with eventual hopes of novel drug delivery mechanisms.
Despite our substantial knowledge of FEN1 structure, biochemical activities and biological functions, many questions remain to be addressed. For instance, how does FEN1 dynamically translocate from one location to another; and in particular, how is FEN1 recruited to replication and repair foci to execute flap cleavage and subsequently dissociate from DNA substrates to avoid interference with Lig1-mediated DNA ligation? What is the mechanism for phosphate diester cleavage? How FEN1 is demethylated? How does FEN1 avoid constant phosphorylation and failure to interact with PCNA? How does FEN1 migrate into the mitochondrion for its function? Do FEN1 mutations that abolish FEN1 post-translational modifications, localization or a specific protein–protein interaction cause cancer and/or other diseases? Future studies will fill in the knowledge gaps regarding this structure-specific nuclease.
FUNDING
National Institutes of Health/National Cancer Institute (R01CA073764 and R01CA085344 to B.H.S. and F32CA117236 to L.D.F.). Funding for open access charge: National Institutes of Health/NCI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful for productive collaborations with R. A. Bambara (Rochester University), J. A. Tainer (Scripps), Jane A. Grasby (University of Sheffield, UK) and D. Xue (University of Colorado, Boulder), which resulted in several lines of evidence on novel structure and functions of FEN1 discussed in this review. We also thank Keely Walker for editorial assistance. The authors regret that this article could not cite all the pertinent articles due to space limitations.
REFERENCES
- 1.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson CG, Atack JM, Chapados B, Tainer JA, Grasby JA. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem. Soc. Trans. 2010;38:433–437. doi: 10.1042/BST0380433. [DOI] [PubMed] [Google Scholar]
- 3.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurray CT, Wilson SH. Coordination between Pol {beta} and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 2003;22:3451–3460. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh P, Zheng L, Chavez V, Qiu J, Shen B. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 2007;282:3465–3477. doi: 10.1074/jbc.M606582200. [DOI] [PubMed] [Google Scholar]
- 9.Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, Tsark W, Huang Q, Kernstine K, Zhang X, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 10.Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen B. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu J, Li X, Frank G, Shen B. Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J. Biol. Chem. 2001;276:4901–4908. doi: 10.1074/jbc.M007825200. [DOI] [PubMed] [Google Scholar]
- 12.Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L, Shen B. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol. Cell Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, III, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hubscher U, Hottiger MO. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell. 2001;7:1221–1231. doi: 10.1016/s1097-2765(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 15.Henneke G, Koundrioukoff S, Hubscher U. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene. 2003;22:4301–4313. doi: 10.1038/sj.onc.1206606. [DOI] [PubMed] [Google Scholar]
- 16.Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, Shen B. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 2010;6:766–773. doi: 10.1038/nchembio.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T, Gally JA, Edelman GM. Deoxyribonuclease IV: a new exonuclease from mammalian tissues. Proc. Natl Acad. Sci. USA. 1969;62:597–603. doi: 10.1073/pnas.62.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyamichev V, Brow MA, Dahlberg JE. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 20.Robins P, Pappin DJ, Wood RD, Lindahl T. Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J. Biol. Chem. 1994;269:28535–28538. [PubMed] [Google Scholar]
- 21.Bornarth CJ, Ranalli TA, Henricksen LA, Wahl AF, Bambara RA. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 1999;38:13347–13354. doi: 10.1021/bi991321u. [DOI] [PubMed] [Google Scholar]
- 22.Finger LD, Blanchard MS, Theimer CA, Sengerova B, Singh P, Chavez V, Liu F, Grasby JA, Shen B. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J. Biol. Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington JJ, Lieber MR. DNA structural elements required for FEN-1 binding. J. Biol. Chem. 1995;270:4503–4508. doi: 10.1074/jbc.270.9.4503. [DOI] [PubMed] [Google Scholar]
- 24.Hohl M, Thorel F, Clarkson SG, Scharer OD. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 2003;278:19500–19508. doi: 10.1074/jbc.M213155200. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser MW, Lyamicheva N, Ma W, Miller C, Neri B, Fors L, Lyamichev VI. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem. 1999;274:21387–21394. doi: 10.1074/jbc.274.30.21387. [DOI] [PubMed] [Google Scholar]
- 26.Liu R, Qiu J, Finger LD, Zheng L, Shen B. The DNA-protein interaction modes of FEN-1 with gap substrates and their implication in preventing duplication mutations. Nucleic Acids Res. 2006;34:1772–1784. doi: 10.1093/nar/gkl106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murante RS, Rust L, Bambara RA. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 28.Qiu J, Liu R, Chapados BR, Sherman M, Tainer JA, Shen B. Interaction interface of human flap endonuclease-1 with its DNA substrates. J. Biol. Chem. 2004;279:24394–24402. doi: 10.1074/jbc.M401464200. [DOI] [PubMed] [Google Scholar]
- 29.Storici F, Henneke G, Ferrari E, Gordenin DA, Hubscher U, Resnick MA. The flexible loop of human FEN1 endonuclease is required for flap cleavage during DNA replication and repair. EMBO J. 2002;21:5930–5942. doi: 10.1093/emboj/cdf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams R, Sengerová B, Osborne S, Syson K, Ault S, Kilgour A, Chapados BR, Tainer JA, Sayers JR, Grasby JA. Comparison of the Catalytic Parameters and Reaction Specificities of a Phage and an Archaeal Flap Endonuclease. J. Mol. Biol. 2007;371:34–48. doi: 10.1016/j.jmb.2007.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. Contacts between the 5′ nuclease of DNA polymerase I and its DNA substrate. J. Biol. Chem. 2001;276:30167–30177. doi: 10.1074/jbc.M100985200. [DOI] [PubMed] [Google Scholar]
- 32.Balakrishnan L, Gloor JW, Bambara RA. Reconstitution of eukaryotic lagging strand DNA replication. Methods. 2010;51:347–357. doi: 10.1016/j.ymeth.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart JA, Campbell JL, Bambara RA. Dna2 is a structure-specific nuclease, with affinity for 5′'-flap intermediates. Nucleic Acids Res. 2009;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Declais AC, Lilley DM. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opin. Struct. Biol. 2008;18:86–95. doi: 10.1016/j.sbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Balakrishnan L, Stewart J, Polaczek P, Campbell JL, Bambara RA. Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates. J. Biol. Chem. 2010;285:4398–4404. doi: 10.1074/jbc.M109.086397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 37.Friedrich-Heineken E, Hubscher U. The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 2004;32:2520–2528. doi: 10.1093/nar/gkh576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nazarkina ZK, Lavrik OI, Khodyreva SN. Flap endonuclease-1 and its role in the processes of DNA metabolism in eucaryotic cells. Mol. Biol. 2008;42:405–421. [PubMed] [Google Scholar]
- 39.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 40.Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, Kane MF, Fan K, Russell R, Brown AM, et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl Acad. Sci. USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng L, Dai H, Qiu J, Huang Q, Shen B. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol. Cell. Biol. 2007;27:3176–3186. doi: 10.1128/MCB.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 44.Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gary R, Park MS, Nolan JP, Cornelius HL, Kozyreva OG, Tran HT, Lobachev KS, Resnick MA, Gordenin DA. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell Biol. 1999;19:5373–5382. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 51.Spiro C, Pelletier R, Rolfsmeier ML, Dixon MJ, Lahue RS, Gupta G, Park MS, Chen X, Mariappan SV, McMurray CT. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 52.Goula AV, Berquist BR, Wilson DM, III, Wheeler VC, Trottier Y, Merienne K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro MS, Bi L, Bailis AM. A mutant allele of the transcription factor IIH helicase gene, RAD3, promotes loss of heterozygosity in response to a DNA replication defect in Saccharomyces cerevisiae. Genetics. 2007;176:1391–1402. doi: 10.1534/genetics.107.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 55.Otto CJ, Almqvist E, Hayden MR, Andrew SE. The "flap" endonuclease gene FEN1 is excluded as a candidate gene implicated in the CAG repeat expansion underlying Huntington disease. Clin. Genet. 2001;59:122–127. doi: 10.1034/j.1399-0004.2001.590210.x. [DOI] [PubMed] [Google Scholar]
- 56.van den Broek WJ, Nelen MR, van der Heijden GW, Wansink DG, Wieringa B. Fen1 does not control somatic hypermutability of the (CTG)(n)*(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett. 2006;580:5208–5214. doi: 10.1016/j.febslet.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 57.Yang J, Freudenreich CH. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene. 2007;393:110–115. doi: 10.1016/j.gene.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagata S. Apoptotic DNA fragmentation. Exp. Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 59.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 61.Lopes J, Debrauwere H, Buard J, Nicolas A. Instability of the human minisatellite CEB1 in rad27Delta and dna2-1 replication-deficient yeast cells. EMBO J. 2002;21:3201–3211. doi: 10.1093/emboj/cdf310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopes J, Ribeyre C, Nicolas A. Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol. Cell Biol. 2006;26:6675–6689. doi: 10.1128/MCB.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parenteau J, Wellinger RJ. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parenteau J, Wellinger RJ. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics. 2002;162:1583–1594. doi: 10.1093/genetics/162.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saharia A, Stewart SA. FEN1 contributes to telomere stability in ALT-positive tumor cells. Oncogene. 2009;28:1162–1167. doi: 10.1038/onc.2008.458. [DOI] [PubMed] [Google Scholar]
- 66.Sampathi S, Bhusari A, Shen B, Chai W. Human flap endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J. Biol. Chem. 2009;284:3682–3690. doi: 10.1074/jbc.M805362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vallur AC, Maizels N. Complementary roles for exonuclease 1 and flap endonuclease 1 in maintenance of triplet repeats. J. Biol. Chem. 2010;285:28514–28519. doi: 10.1074/jbc.M110.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair. 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Attardi G, Schatz G. Biogenesis of mitochondria. Annu. Rev. Cell. Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 71.Frossi B, Tell G, Spessotto P, Colombatti A, Vitale G, Pucillo C. H(2)O(2) induces translocation of APE/Ref-1 to mitochondria in the Raji B-cell line. J. Cell. Physiol. 2002;193:180–186. doi: 10.1002/jcp.10159. [DOI] [PubMed] [Google Scholar]
- 72.Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J. Biol. Chem. 1995;270:14659–14665. doi: 10.1074/jbc.270.24.14659. [DOI] [PubMed] [Google Scholar]
- 73.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 74.Biswas EE, Zhu FX, Biswas SB. Stimulation of RTH1 nuclease of the yeast Saccharomyces cerevisiae by replication protein A. Biochemistry. 1997;36:5955–5962. doi: 10.1021/bi962890u. [DOI] [PubMed] [Google Scholar]
- 75.Chai Q, Zheng L, Zhou M, Turchi JJ, Shen B. Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in alpha-segment processing during Okazaki fragment maturation. Biochemistry. 2003;42:15045–15052. doi: 10.1021/bi035364t. [DOI] [PubMed] [Google Scholar]
- 76.Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 77.Siegal G, Turchi JJ, Myers TW, Bambara RA. A 5′ to 3′ exonuclease functionally interacts with calf DNA polymerase epsilon. Proc. Natl Acad. Sci. USA. 1992;89:9377–9381. doi: 10.1073/pnas.89.20.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu FX, Biswas EE, Biswas SB. Purification and characterization of the DNA polymerase alpha associated exonuclease: the RTH1 gene product. Biochemistry. 1997;36:5947–5954. doi: 10.1021/bi962889v. [DOI] [PubMed] [Google Scholar]
- 79.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Imamura O, Campbell JL. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc. Natl Acad. Sci. USA. 2003;100:8193–8198. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, Hurwitz J. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J. Biol. Chem. 2008;283:20925–20936. doi: 10.1074/jbc.M802696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 83.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J. Biol. Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 85.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 86.Ahn B, Harrigan JA, Indig FE, Wilson DM, III, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J. Biol. Chem. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 87.Balakrishnan L, Brandt PD, Lindsey-Boltz LA, Sancar A, Bambara RA. Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J. Biol. Chem. 2009;284:15158–15172. doi: 10.1074/jbc.M109.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo Z, Chavez V, Singh P, Finger LD, Hang H, Hegde ML, Shen B. Comprehensive mapping of the C-terminus of flap endonuclease-1 reveals distinct interaction sites for five proteins that represent different DNA replication and repair pathways. J. Mol. Biol. 2008;377:679–690. doi: 10.1016/j.jmb.2007.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl Acad. Sci. USA. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 91.Tseng HM, Tomkinson AE. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J. Biol. Chem. 2004;279:47580–47588. doi: 10.1074/jbc.M404492200. [DOI] [PubMed] [Google Scholar]
- 92.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 93.Wright PE, Dyson HJ. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frank G, Qiu J, Zheng L, Shen B. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J. Biol. Chem. 2001;276:36295–36302. doi: 10.1074/jbc.M103397200. [DOI] [PubMed] [Google Scholar]
- 96.Friedrich-Heineken E, Henneke G, Ferrari E, Hubscher U. The acetylatable lysines of human Fen1 are important for endo- and exonuclease activities. J. Mol. Biol. 2003;328:73–84. doi: 10.1016/s0022-2836(03)00270-5. [DOI] [PubMed] [Google Scholar]
- 97.Johnson LN, Barford D. The effects of phosphorylation on the structure and function of proteins. Annu. Rev. Biophys. Biomol. Struct. 1993;22:199–232. doi: 10.1146/annurev.bb.22.060193.001215. [DOI] [PubMed] [Google Scholar]
- 98.Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 99.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 100.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 101.Henneke G, Friedrich-Heineken E, Hubscher U. Flap endonuclease 1: a novel tumour suppresser protein. Trends Biochem. Sci. 2003;28:384–390. doi: 10.1016/S0968-0004(03)00138-5. [DOI] [PubMed] [Google Scholar]
- 102.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc. Natl Acad. Sci. USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat. Rev. Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 104.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shen B, Nolan JP, Sklar LA, Park MS. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Larsen E, Kleppa L, Meza TJ, Meza-Zepeda LA, Rada C, Castellanos CG, Lien GF, Nesse GJ, Neuberger MS, Laerdahl JK, et al. Early-onset lymphoma and extensive embryonic apoptosis in two domain-specific Fen1 mice mutants. Cancer Res. 2008;68:4571–4579. doi: 10.1158/0008-5472.CAN-08-0168. [DOI] [PubMed] [Google Scholar]
- 107.Kim IS, Lee MY, Lee IH, Shin SL, Lee SY. Gene expression of flap endonuclease-1 during cell proliferation and differentiation. Biochim. Biophys. Acta. 2000;1496:333–340. doi: 10.1016/s0167-4889(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 108.Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, Minna JD. Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene. 2003;22:7243–7246. doi: 10.1038/sj.onc.1206977. [DOI] [PubMed] [Google Scholar]
- 109.Lam JS, Seligson DB, Yu H, Li A, Eeva M, Pantuck AJ, Zeng G, Horvath S, Belldegrun AS. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006;98:445–451. doi: 10.1111/j.1464-410X.2006.06224.x. [DOI] [PubMed] [Google Scholar]
- 110.Nikolova T, Christmann M, Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 2009;29:2453–2459. [PubMed] [Google Scholar]
- 111.Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, Kernstine KH, Lin D, Shen B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warbrick E, Coates PJ, Hall PA. Fen1 expression: a novel marker for cell proliferation. J. Pathol. 1998;186:319–324. doi: 10.1002/(SICI)1096-9896(1998110)186:3<319::AID-PATH184>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 113.McManus KJ, Barrett IJ, Nouhi Y, Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc. Natl Acad. Sci. USA. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Panda H, Jaiswal AS, Corsino PE, Armas ML, Law BK, Narayan S. Amino acid Asp181 of 5′-flap endonuclease 1 is a useful target for chemotherapeutic development. Biochemistry. 2009;48:9952–9958. doi: 10.1021/bi9010754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tumey LN, Bom D, Huck B, Gleason E, Wang J, Silver D, Brunden K, Boozer S, Rundlett S, Sherf B, et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 2005;15:277–281. doi: 10.1016/j.bmcl.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 116.Tumey LN, Huck B, Gleason E, Wang J, Silver D, Brunden K, Boozer S, Rundlett S, Sherf B, Murphy S, et al. The identification and optimization of 2,4-diketobutyric acids as flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 2004;14:4915–4918. doi: 10.1016/j.bmcl.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 117.Tseng HM, Tomkinson AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- 118.Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]