Abstract
Non-homologous end-joining (NHEJ) is a critical error-prone pathway of double strand break repair. We recently showed that tyrosyl DNA phosphodiesterase 1 (Tdp1) regulates the accuracy of NHEJ repair junction formation in yeast. We assessed the role of other enzymes in the accuracy of junction formation using a plasmid repair assay. We found that exonuclease 1 (Exo1) is important in assuring accurate junction formation during NHEJ. Like tdp1Δ mutants, exo1Δ yeast cells repairing plasmids with 5′-extensions can produce repair junctions with templated insertions. We also found that exo1Δ mutants have a reduced median size of deletions when joining DNA with blunt ends. Surprisingly, exo1Δ pol4Δ mutants repair blunt ends with a very low frequency of deletions. This result suggests that there are multiple pathways that process blunt ends prior to end-joining. We propose that Exo1 acts at a late stage in end-processing during NHEJ. Exo1 can reverse nucleotide additions occurring due to polymerization, and may also be important for processing ends to expose microhomologies needed for NHEJ. We propose that accurate joining is controlled at two steps, a first step that blocks modification of DNA ends, which requires Tdp1, and a second step that occurs after synapsis that requires Exo1.
INTRODUCTION
A critical type of DNA damage is DNA double strand break. Double strand breaks can be generated by environmental agents such as ionizing radiation, and also can occur when single strand lesions interfere with cellular processes such as replication. Two major pathways repair double strand breaks: homologous recombination and non-homologous end-joining. Homologous recombination pathways predominantly repair damage occurring in S and G2 when a sister chromatid is available to serve as a template for repair. In haploid yeast cells, non-homologous end-joining (NHEJ) is critical in G1 cells and stationary phase cells, when a homologous chromosome is unavailable to serve as a template for repair. Therefore, NHEJ has the advantage of being capable of repairing double strand breaks in many cellular contexts (1).
Most components of NHEJ are highly conserved throughout the eukaryotic kingdom. The key components of NHEJ include a DNA binding heterodimer composed of Ku70 and Ku80 (2–5), a specialized DNA ligase (DNA ligase IV) (6,7) and ligase accessory factors [XRCC4 in mammals, Lif1 in yeast (8,9)]. In yeast, the complex that includes Mre11, Rad50 and Xrs2 (the MRX complex, termed the MRN complex in mammalian cells) is also required, perhaps to bring broken ends in close proximity (10,11). The end-joining reaction includes recognition of broken ends by Ku70/Ku80, ultimately followed by ligation by ligase IV (12). A variety of proteins potentially participate in end-processing including a specialized nuclease Artemis (13,14) (apparently absent in yeast), DNA polymerases [polµ and polλ in mammals (15–17), in yeast POL4 (18,19)], and additional factors that may regulate ligase (Xlf/Cernunnos) (9,20,21). Mammalian cells also express a PI-kinase family member termed DNA-dependent protein kinase (DNA-PK) that plays an important role in some NHEJ reactions (22).
A key requirement in NHEJ is to alter DNA ends so that both strands can be ligated. This can include removal of damaged nucleotides, as well as the addition or removal of a small number of undamaged nucleotides. In general, the junctions formed during NHEJ will contain alterations that will include nucleotide additions or deletions. Since NHEJ does not use a homologous template to carry out repair, inaccuracy in the repair reaction is unavoidable. Nonetheless, the repair reactions at DNA ends are likely regulated to minimize mutational alterations. We recently demonstrated that yeast tyrosyl DNA phosphodiesterase I (Tdp1) prevents additions at repair junctions arising from broken DNA with 5′-extensions. We hypothesized that the 3′-nucleosidase activity of Tdp1 temporarily blocks the action of polymerases and other enzymes by generating ends with a 3′-phosphate (23). In this article, we demonstrate that Exo1 also plays a role in processing ends during NHEJ, and independently of Tdp1, limits addition reactions during NHEJ. We also show that Exo1 plays a role in processing blunt DNA ends during NHEJ.
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae strains used in this study were isogenic derivatives of the wild-type strain BY4741 (MATa hisΔ1 leu2Δ0 lys20 ura3Δ0). Individual yeast KANMX4 open reading frame (ORF) deletion mutants in BY4741 (exo1Δ, yku80Δ, tdp1Δ, and pol4Δ) were purchased from Open Biosystems (Huntsville, AL, USA). For generating double-disruption strains, URA3 marked deletions of TDP1 (23) or EXO1 [plasmid pHT133 (24)] were introduced into appropriate BY4741 mutant derivatives. Some experiments were also repeated in appropriate mutant derivatives of JN362a (23). The plasmid pHT133 was used to generate exo1Δ derivatives of JN362a.
Plasmid purification
The YCplac111 plasmid (a centromeric plasmid that carries the LEU2 gene) was digested with different restriction enzymes and used as an extrachromosomal NHEJ substrate (4). Products were separated on a 0.8% agarose gel and linearized DNA was extracted from gels (QIAquick Gel Extraction Kit, Qiagen, Valencia, CA, USA) and eluted in water in aliquots of 40 ng/µl and stored at 4°C.
Yeast transformation
For yeast transformation assays of NHEJ, yeast cells were grown overnight in yeast peptone dextrose adenine (YPDA) media. Cells were diluted the following morning to the OD of 0.4 and grown until the suspension reached an OD600 of ∼1. Typically, 100 ng of linearized or supercoiled DNA was transformed into yeast by electroporation (25). All experiments were performed with this carrier-free procedure to eliminate plasmid reactions with carrier DNA (19,26–28). Electroporation was performed at 0.75 V, 25 µF and 200 Ω, using cuvettes with a 0.1-cm gap width (Bio-Rad, Hercules, CA, USA).
In all cases, transformation with an undigested plasmid was performed in parallel to determine repair efficiency. Repair efficiency was expressed as the number of transformed colonies obtained using linearized plasmids to the number of colonies obtained with circular plasmids ×100.
Breakpoint junction analysis
By design, 100 independent colonies were analyzed per experiment. Plates containing less than 100 colonies were chosen, and all colonies on several plates were used for DNA isolation to minimize bias in selecting colonies. Plasmid DNA was extracted from colonies after transformation, as described earlier (23). The region containing the restriction enzyme site originally used to linearize the plasmid was amplified. For YCplac111, the primers were 5′-TAGCCGTAGTTAGGCCACCAC-3′ and 5′-ACCGCACAGATGCGTAAGGAG-3′. PCR products were then digested with the same restriction enzyme(s) initially used to linearize the plasmid. Digested products were scored as accurately repaired, and PCR products that were resistant to digestion were sequenced by using the primer 5′-CCAATACGCAAACCGCCTCTCC-3′.
For some substrates, experiments were repeated with independent DNA preparation. In those cases, results are shown for all colonies analyzed (a multiple of 100).
Statistical analyses
Significance of differences in the accuracy of repair (or the frequency of particular classes of events) was assessed using Fisher’s exact test, and values were calculated using Instat version 3.0.
RESULTS
Efficiency of NHEJ in exo1-deleted strains is not impaired
We recently reported that yeast tdp1Δ mutants exhibited reduced accuracy of NHEJ with substrates that carried 5′-extensions (23). The reduced accuracy was not accompanied by a change in repair efficiency. We were interested in testing whether other end-processing activities impaired the accuracy of NHEJ. In particular, we were interested in assessing various nucleases that might process unpaired nucleotides and thereby minimize insertions at repair junctions. One obvious candidate is Exo1, a nuclease important for both homologous recombination and mismatch repair (29,30). Earlier work of Wu et al. (31) did not detect a role for Exo1 at repair junctions during NHEJ; however, they did not assess the accuracy of repair using DNA with 5′-extensions.
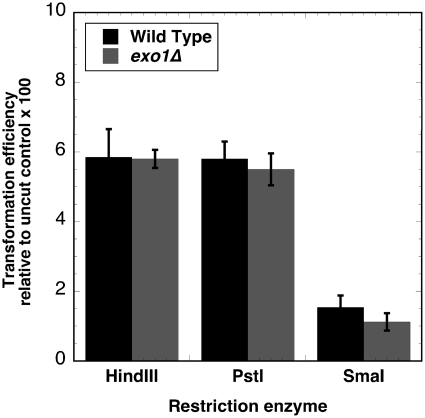
We tested potential roles for Exo1 in the accuracy of NHEJ. The plasmid YCplac111 was linearized with HindIII, PstI or SmaI to generate 5′-extensions, 3′-extensions or blunt ends. The digested plasmids were transformed into the haploid wild-type strain BY4741 or an isogenic exo1Δ strain. Uncut plasmid was transformed in parallel to normalize for transformation efficiency. The results, shown in Figure 1, express the repair efficiency as the ratio of transformants with linearized plasmid to those with uncut plasmid ×100. The efficiency of repair for HindIII, PstI or SmaI linearized plasmids was comparable between wild-type and exo1Δ strains, confirming that Exo1 does not play an essential role in end-joining with these types of DNA ends.
Figure 1.
Plasmid repair efficiency in wild-type and exo1Δ strains. Plasmid YCplac111 was linearized with HindIII, PstI or SmaI, and transfected into BY4741 (WT) or in an exo1Δ derivative by electroporation. Repair frequencies for each genotype are expressed as the ratio of colonies obtained with linear DNA divided by colonies obtained with circular DNA ×100. The results shown are the mean of at least three independent transfections. The error bars indicate SEM.
Exo1 plays a role in accurate joining of ends with 5′-extensions
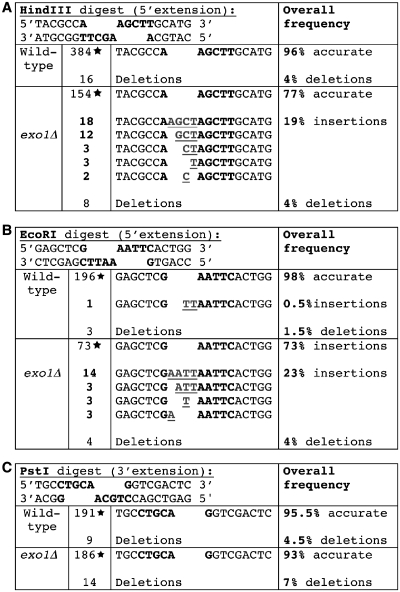
To assess the accuracy of the repair reaction, plasmids were isolated from the transformed cells, and the repair junctions were amplified by PCR. Precise repair of the plasmid maintains the original restriction site; therefore, PCR products from accurate repair reactions can be digested with the same restriction enzyme used to linearize the plasmid. PCR products that could not be cut with the original restriction enzyme were sequenced to confirm that the repair was inaccurate, and to determine the nature of the misrepair events. In the wild-type strain, repair of the HindIII linearized plasmids is largely accurate (96% of colonies analyzed showed accurate repair in four independent experiments), and the remaining 4% that were misrepair events consisted of 1–2 nt deletions (Figure 2A). As we observed earlier with tdp1Δ strains, deletion of exo1 also reduced the repair accuracy of linearized plasmids with 5′-extensions. In exo1Δ strains, 77% of colonies carried accurately repaired plasmids (Figure 2A). Of the 46 colonies carrying plasmids that were repaired inaccurately (23%), 38 isolates (19%) carried plasmids with insertions at the repair junction of 1–4 nt. In all cases, the insertions were templated and could be rationalized by partial or complete filling in of the 5′-extension. The eight other colonies carried small deletions, similar to what was observed with wild-type cells. The frequency of misrepair was significantly different between wild-type and exo1Δ strains (P < 0.0001 by Fisher’s exact test). We also examined the frequency of misrepair of HindIII linearized plasmid in JN362a exo1Δ, an independently derived exo1Δ mutant. We observed 22/100 colonies carrying insertions, compared with 0/100 for JN362a EXO1+ (data not shown). This experiment demonstrates that the observed effect does not depend on the BY4741 strain background.
Figure 2.
Spectra of junctions recovered from the wild-type and exo1Δ strains. The accuracy of repair of plasmids of linearized DNA with 5′-extensions (HindIII, A and EcoRI, B) or 3′-extensions (PstI, C) was determined in WT and exo1Δ deleted strains. Plasmid DNA was isolated, and the repair junction was amplified by PCR. PCR products were digested using the same restriction enzyme used to linearize the DNA, and samples that could be digested were scored as accurately repaired. Samples that failed to digest were analyzed by DNA sequencing. The enzyme recognition sites are highlighted in bold. The junctions with insertions are underlined in gray. Stars denote accurately repaired junctions.
Similar results to those obtained with HindIII were obtained with plasmids linearized with EcoRI, which also generates 5′-extensions (Figure 2B). As observed in the HindIII-linearized plasmid, the misrepair events were largely templated insertions. These results demonstrate that exo1Δ deletion mutants carry out inaccurate NHEJ with substrates carrying 5′-extensions, a property also exhibited by tdp1Δ strains.
In tdp1Δ strains, misrepair during NHEJ is specific for substrates with 5′-extensions. To determine the accuracy of repair of DSBs with 3′-extensions, we examined transformation of wild-type and exo1Δ strains with PstI linearized YCplac111. In the wild-type strain, repair of PstI linearized plasmids was accurate in 98% of colonies examined. In the first experiment in the exo1Δ strain, repair was accurate in 91/100 colonies, and the remaining 9/100 colonies showed inaccurate repair. One colony carried a deletion of 27 nt, and the other 8 inaccurate events were deletions of 1–3 nt. The difference between wild-type and exo1Δ strains is not quite statistically significant (P = 0.0582). Since the deletion frequency was slightly elevated, we examined a second set of 100 colonies, and the pooled data from the two experiments are shown in Figure 2C. The pooled data from the two experiments showed 95.5% accurate repair events in wild-type cells, and 93% accurate repair events in exo1Δ strains. For the pooled data, the difference between wild-type and exo1Δ strains is not significant (P = 0.1). These results strongly suggest that exo1Δ strains do not show an appreciable increase in misrepair of substrates with 3′-extensions. Therefore, both exo1Δ and tdp1Δ strains show misrepair during NHEJ that is specific for 5′-extensions.
Additions in the absence of EXO1 require NHEJ factors and POL4
In our earlier work with misrepair that occurs in tdp1Δ strains, we found that the generation of templated additions requires NHEJ components including Ku70, Ku80 and DNA ligase IV. These results showed that the additions occurred through the NHEJ pathway. We carried out a similar analysis with exo1Δ strains. We generated yku80Δ exo1Δ strains, and examined efficiency of plasmid and accuracy of plasmid repair.
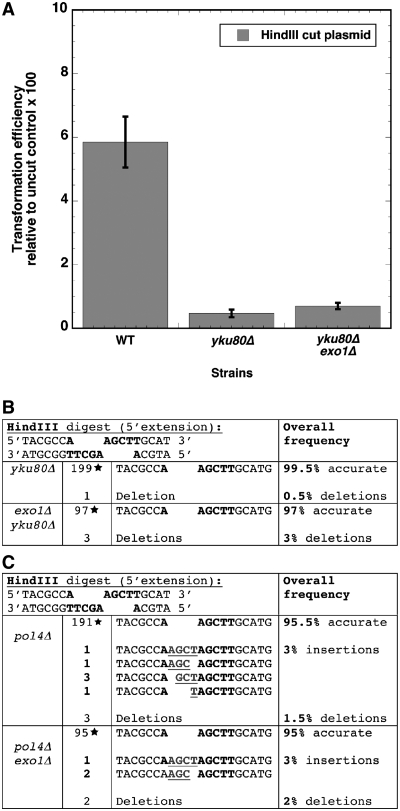
BY4741 derivatives lacking YKU80 have a >10-fold decrease in repair of plasmids with HindIII ends compared with isogenic wild-type strains (Figure 3A). There was a similar decrease in the yku80Δ exo1Δ strain, with no significant difference in repair between the yku80Δ strain and the yku80Δ exo1Δ strain (P = 0.24).
Figure 3.
Insertions generated in exo1Δ strains occur through the NHEJ pathway and require POL4. (A) HindIII linearized YCplac111 was transfected into strains carrying a deletion of yku80Δ or of both exo1Δ and yku80Δ. As in Figure 1, transformation efficiency was normalized to undigested YCplac111. The efficiency of transformation was substantially reduced in the yku80 strain compared with the wild-type strain and the exo1Δ yku80Δ strain showed a similar reduction compared with wild-type cells. (B) Accuracy of the repair of HindIII-linearized YCplac111 DNA was assessed in plasmids recovered from yku80Δ and exo1Δ yku80Δ strains. The data from the yku80Δ strain includes earlier published samples from our laboratory, as well as 100 additional isolates (23). The deletions in the exo1Δ yku80Δ strain ranged from 1 to 23 nt in length. (C) Accuracy of the repair of HindIII-linearized YCplac111 DNA was assessed in pol4Δ and pol4Δ exo1Δ strains. The data from the pol4Δ strain includes earlier published samples from our laboratory, as well as 100 additional isolates (23).
We next examined the accuracy of repair of HindIII linearized plasmid in the NHEJ deficient strains. In yku80Δ single mutants, the HindIII linearized plasmid was accurately repaired in 199/200 colonies examined. As we and others noted earlier (23,32), the accuracy of repair of cohesive ends in NHEJ deficient mutants (such YKU80) has been controversial, with some reports indicating highly accurate repair, and other indicating substantial reductions in accuracy. In our experiments, we observed accurate repair in NHEJ deficient mutants. In the yku80Δ exo1Δ strain, 97/100 isolates showed accurate repair, and importantly, no insertions were recovered (Figure 3B). We conclude that the generation of insertions in exo1Δ strains occurs by the canonical NHEJ pathway.
In our earlier experiments, we showed that templated insertions occur in tdp1Δ strains, but not in tdp1Δ pol4Δ strains. To examine whether Pol4p was involved in the filling synthesis during NHEJ in exo1Δ strains, we analyzed the sequences of rejoined plasmids recovered from the pol4Δ strain and a pol4Δ exo1Δ double mutant. Daley et al. (19) reported that Pol4 is required for end-joining of 3′-extensions only if gaps need to be filled on both strands. For the junctions arising from the HindIII substrate with 5′-extensions, no significant difference was observed in the repair of pol4 deleted strain (95.5% accurate, 3% carrying insertions) or pol4 exo1 double deleted strain (95% accurate, 3% carrying insertion, Figure 3C). These findings indicate that Pol4 is required for the inaccurate repair of 5′-cohesive extensions in exo1Δ strains.
Exo1 is involved in NHEJ repair of blunt ends
Consistent with earlier reports (33,34), we found that wild-type strains showed lower repair efficiency with blunt ends (SmaI-cut plasmid) than with 3′- or 5′-extensions (Figure 1). The repair efficiency of blunt ends in the wild-type was only ∼10–20% of that seen with 5′- or 3′-extensions. The exo1Δ strain also showed inefficient repair with a slight decrease in efficiency compared with wild-type (Figure 1). This result is opposite of what we recently reported with tdp1Δ strains (23), where deletion of TDP1 leads to an increase in the efficiency of repairing plasmid DNA carrying a blunt end (see also Figure 6A). Thus, exo1Δ and tdp1Δ strains have similar effects on the accuracy of repairing plasmid DNA with 5′-extensions; they have different effects on the efficiency of repairing plasmid DNA with blunt ends.
Figure 6.
EXO1 is not required for tdp1Δmediated stimulation of joining blunt ends. (A) YCplac111 digested with SmaI was transfected into WT, exo1Δ, tdp1Δ or exo1Δ tdp1Δ strains. Repair frequencies were normalized with transfection of undigested YCplac111. The results shown are the mean of at least three independent transfections; error bars indicate SEM. (B) Plasmids were recovered from 100 independent colonies from wild-type, exo1Δ, tdp1Δ and exo1Δ tdp1Δ mutant strains. Data from the wild-type and single mutants were as presented in Figure 4B. As in Figure 4B, each dot represents a single isolate with the indicated deletion size, and accurately repaired junctions are not presented.
We next examined the accuracy of the repair junctions generated with SmaI digested plasmid DNA. Figure 4A shows the results obtained from the analysis of 100 colonies from wild-type and exo1Δ strains. The overall frequency of deletions was the same for both strains (61/100 repaired accurately). Interestingly, the size of the deletions differed between the two strains (Figure 4B). The median deletion size for wild-type cells was eight nucleotides, whereas the median deletion was 2 nt in the exo1Δ strain. Results of the spectrum of deletions seen in a tdp1Δ strain is also shown in Figure 4B (median deletion of 8 nt). It is clear that the exo1Δstrain has a greater frequency of deletions of 1–2 nucleotides, whereas both the wild-type and tdp1Δ strains have few small deletions and more isolates with deletions ≥10 nt.
Figure 4.
Repair of plasmid with blunt ends leads to smaller deletions in exo1Δ cells. (A) After transformation with YCplac111 DNA linearized with SmaI, plasmids were isolated from cells with the indicated genotypes. A total of 100 plasmids were analyzed from each genotype. The total numbers of accurately repaired and misrepaired plasmids are indicated. The number of misrepaired plasmids was not statistically different between any of the four genotypes examined. (B) The isolates carrying deletions of a given size is shown on the dot plot for wild-type, exo1Δ, tdp1Δ, pol4Δ and exo1Δ pol4Δ mutants. Each dot represents a single isolate with the indicated deletion size, and accurately repaired junctions are not presented. The median deletion size is shown as a solid gray line, and was calculated based on the colonies carrying deletions. Colonies with accurately repaired junctions are not included in the calculation of the median and are not shown on the dot plot.
Since we had observed that POL4 was required for the stimulation of joining blunt ends in a tdp1Δ strain, we were interested in examining whether a deletion of POL4 affected the size of deletions recovered with blunt ends. A pol4Δ strain had a minimal effect on the frequency of isolates with deletions (55/100 accurately repaired, P = 0.47 compared with wild-type), and the median deletion size was 8 nt (Figure 4B). Interestingly, the pol4Δ exo1Δ double mutant strain showed a large effect on the overall frequency of repair junctions with deletions. For the double mutant, 89/100 junctions exhibited accurate repair compared with 61/100 repaired accurately for the wild-type strain (P < 0.0001). Taken together these results show that the recovery of repaired plasmids with deletions can occur by two separable pathways, an EXO1-dependent pathway and a POL4-dependent pathway. Elimination of both pathways largely prevents the recovery of plasmids with deletions.
Genetic analysis of Tdp1 and Exo1 in end-joining pathways
As described earlier, we demonstrated that tdp1Δ strains carry out inaccurate end-joining with plasmids carrying 5′-extensions. Since exo1Δ strains exhibit a similar phenotype, we tested the genetic interaction between tdp1Δ and exo1Δ. There was no difference in efficiency of transformation of exo1Δ and tdp1Δ exo1Δ strains with HindIII linearized plasmid (data not shown). However, the two mutants showed an additive effect in the frequency of formation of junctions with templated insertions. The overall frequency of accurate repair of HindIII linearized DNA in an exo1Δ strain was 77 versus 67% in a tdp1Δ exo1Δ strain (Figure 5A, P = 0.034). A similar although greater effect was seen with EcoRI digested DNA (Figure 5B, P = 0.008). While there is a small increase in the number of deletions in tdp1Δ exo1Δ double mutants, the increase in deletion frequency is not significant (P = 0. 2 for the comparison of HindIII digested DNA in exo1Δ versus tdp1Δ exo1Δ strains, P = 0.4 for the comparison of EcoRI digested DNA in exo1Δ vs. tdp1Δ exo1Δ strains). As was observed with both single mutants, there was no significant effect with DNA carrying a 3′-extension (Pst1 digested, Figure 5C). We conclude that tdp1Δ and exo1Δ exert partly independent and additive effects on the accuracy of NHEJ.
Figure 5.
Deletion of EXO1 along with TDP1 results in an additive increase in additions. The accuracy of repair of plasmids of linearized DNA with 5′-extensions (HindIII, A and EcoRI, B) or 3′-extensions (PstI, C) was determined in exo1Δ tdp1Δ deleted strains. Results with wild-type and exo1Δ are the data presented in figure 2, and the data from the tdp1Δ deleted strain was published earlier (23). The overall level of inaccurate repair for exo1Δ tdp1Δ was compared with exo1Δ using Fisher’s exact test. For the HindIII digested DNA, P = 0.034; and for the EcoRI digested DNA, P = 0.008. The difference between any of the mutants with plasmid linearized with PstI was not significant.
Deletion of TDP1 or EXO1 have differing effects on joining blunt ends, with tdp1Δ strain exhibiting elevated repair while the exo1Δ strain changing the spectrum of repair junctions. Figure 6A shows the results obtained with repair efficiency for both single mutants and the tdp1Δ exo1Δ double mutant. Both tdp1Δ and tdp1Δ exo1Δ strains show elevated levels of transformation with SmaI digested DNA compared with wild-type (or the exo1Δ single mutant). However, the spectrum of deletions in the tdp1Δ exo1Δ double mutant is like the exo1Δ single mutant, with a median deletion size of 2 nt (Figure 6B). Deletion of EXO1 is epistatic to deletion of TDP1 for efficiency of repair, while the epistasis relationship is reversed for the median deletion size. As described in the discussion, we rationalize these results and propose that TDP1 acts at an early step in the NHEJ pathway, and that EXO1 has the potential to act at a subsequent step.
DISCUSSION
The major conclusion from this work is that yeast Exo1 plays a role in end-processing during non-homologous end-joining. The phenotypes of exo1Δ mutants are similar to what we observed for tdp1Δ mutants with substrates bearing cohesive ends, but differ for substrates with blunt ends. Like tdp1Δ mutants, exo1Δ mutants exhibit templated additions with substrates with 5′-overhangs, and neither mutant significantly affects the efficiency or accuracy of repair of plasmids terminating with 3′-overhangs. We found that exo1Δ mutants show smaller deletions with blunt ended substrates, and that deletion of both EXO1 and POL4 largely eliminates deletions that are recovered with blunt end substrates.
In this article we have used a plasmid repair assay, in which cells are transfected with linearized plasmid, and recovery of transformants depends on recircularization of the plasmid, predominantly by the NHEJ pathway. This assay has been used to demonstrate the role of yeast genes in NHEJ (5,6,35,36). While this assay does not completely reconstitute the conditions occurring when the substrates are broken chromosomes, it has the advantage of allowing the use of substrates with different types of DNA ends. The major alternate approach is to express homing endonucleases such as HO or Sce-I. Since all known homing endonucleases lead to ends with 3′-extensions, broken ends with 5′-extensions or blunt ends cannot be studied with this approach. The mutants studied here lead to phenotypes only with DNA with 5′-extensions or blunt ends; therefore, rare cutting nucleases have not been appropriate tools.
We recently proposed that Tdp1 controls the accuracy of NHEJ by its 3′-nucleosidase activity. We suggested that Tdp1 frequently removes a nucleoside leaving a 3′-phosphate. The 3′-phosphate blocks polymerases (and other DNA metabolic enzymes) until the phosphate is removed. Strong evidence for this model comes from the observation that over-expression of Tpp1, a phosphatase that is specific for 3′-PO4 ends (37,38) leads to the same types of errors seen when Tdp1 activity is absent (23). Generation of additions in the absence of Tdp1 occurs through the canonical NHEJ pathway since additions are not seen when yKu80 or DNA ligase IV is absent. Furthermore, DNA polymerase IV is also required for the templated additions. The additions are specific for 5′-extensions, and no change in accuracy was noted for DNA ends carrying 3′-extensions.
We found that the same properties seen with tdp1Δ mutants are also seen with exo1Δ mutants. Transformation of DNA ends with 5′-extensions lead to templated insertions at the repair junctions in exo1Δ mutants. The insertions require yKu80 and DNA polymerase IV. While deletion of either TDP1 or EXO1 greatly increases the frequency of insertions at the repair junctions, the majority of junctions formed exhibit accurate repair. The insertions do not occur when the substrate DNA has a 3′-extension. Furthermore, the effects of deletion of Exo1 and Tdp1 are at least partly additive (Figure 5). These observations, summarized in the model shown in Figure 7, suggest that Tdp1 acts relatively early, and blocks the ability of polymerase IV to extend the end with a 5′-extension. If Tdp1 fails to prevent nucleotide insertions by polymerase, we suggest that Exo1 can remove the added nucleotides. This likely occurs after synapsis, and may arise form the generation of 5′-flaps during synapsis. This reaction of Exo1 is likely different from the resection of ends during homologous recombination because Ku has been shown to inhbiit Exo1-mediated synapsis (39). The added nucleotides would be removed by either direct 5′→3′-exonuclease activity or through 5′-flap endonuclease activity of Exo1 (40). Although we suggest that Exo1 acts after synapsis, we have no direct evidence that synapsis is required for Exo1 processing. This model rationalizes both the effects of exo1Δ single mutants, and the additive effect of tdp1Δ exo1Δ double mutants on accuracy.
Figure 7.
A model for the roles of Exo1 in accurate joining of cohesive ends. DNA ends with 5′-extensions are potentially substrates for filling in by Pol4 or another DNA polymerase. We previously suggested that the filling reaction might be largely prevented in the presence of active Tdp1, due to the removal of a nucleoside, and the generation of a 3′-phosphate (this reaction is not shown for simplicity). If Tdp1 does not act, and a DNA polymerase fills in the extension, synapsis and isomerization can lead to a structure with a 5′-flap. The flap can be removed by either the 5′→3′-exonuclease or the flap endonuclease activity of Exo1, leading to the recovery of an error-free product. The filling-in reaction cannot be directly reversed by Exo1, since it does not have 3′→5′-exonuclease activity.
While tdp1Δ and exo1Δ have similar effects on NHEJ with DNA substrates with cohesive ends, the mutants have distinct effects on substrates with blunt ends. While tdp1Δ mutants result in enhanced recovery of repaired plasmids that had been linearized with SmaI, there is no enhanced repair with this substrate seen in exo1Δ single mutants. For repair of blunt plasmid DNA, exo1Δ is epistatic to tdp1Δ. Therefore, the enhanced recovery of repaired plasmid in tdp1Δ strains does not require Exo1. However, for the size of deletions recovered in the mutants, the epistatic relationship is reversed, with tdp1Δ exo1Δ mutants showing the same reduced median deletion size as the exo1Δ single mutant. These observations suggest that the two genes perform distinct functions in processing blunt ends. We propose that Tdp1 inhibits processing of the 3′-end of blunt ends. This inhibition reduces the efficiency of the overall repair of blunt ends. The major determinant of the deletion size comes from the 5′→3′-resection by Exo1. Since exo1Δ tdp1Δ mutants show the same elevated efficiency of repair as tdp1Δ single mutants, we suggest that the processing of the 5′-ends by Exo1 is not critical for repair efficiency.
A striking finding from our work is that in the absence of both Exo1 and Pol4, almost all of the plasmids bearing blunt ends are repaired in a manner that does not lead to the deletion of any nucleotides. We would propose that these plasmids arise from a failure to process both the 5′- and 3′-ends. A plausible corollary is that efficient joining of blunt ends requires an obligatory intermediate with either a 3′- or 5′-tail. This hypothesis is consistent with the observation that joining of blunt ends is partially Ku independent and inefficient. However, joining of plasmids with either 3′- or 5′-extensions is more efficient in yeast than joining of blunt ends, even when the ends are not cohesive. The joining of mismatched ends requires canonical end-joining functions [see (31) for an example of the requirement of Ku70 for joining mismatched ends]. Therefore, end processing is likely to be the critical determinant in joining blunt ends. An interesting possibility is that end-processing occurs before recruitment of Ku to blunt DNA ends. Alternately, Ku binding inhibits the initial processing if the substrate DNA carries blunt ends.
An earlier examination of exo1Δ mutants did not observe a major effect on the efficiency of NHEJ (31). Since that study did not examine the effects of accuracy in detail, nor did it examine joining of DNA with blunt ends, it is not surprising that the effects we describe here were not detected.
The precise ordering of events that occur with different types of DNA ends will require the development of systems for introducing defined broken ends into cells. At present, rare cutting homing endonucleases such as Sce-1 or HO generate ends with 3′-extensions. Recent work by Lewis and colleagues have demonstrated the ability to use blunt cutting restriction enzymes such as PvuII in yeast (41), and other restriction enzymes generating 5′-extensions have been successfully applied to yeast for some time (42–44). It is interesting to note that expression of PvuII or EcoRV is much more poorly tolerated in yeast than expression of EcoRI (41). This suggests that efficient resection of blunt ends may be relatively deficient in yeast cells.
In conclusion, we have shown that Exo1 participates in some NHEJ reactions in yeast. Exo1 is required for accurate repair of 5′-extensions, and influences the nature of deletions in DNA with blunt ends. An important next step will be understanding how different end-processing factors are recruited to DNA ends, and how choices among different end-processing functions are regulated in physiological contexts.
FUNDING
National Institutes of Health, grant CA82313 and core grant CA21765; American Lebanese Syrian Associated Charities (ALSAC). Funding for open access charge: American Lebanese Syrian Associated Charities.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lieber M. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 3.Morio T, Kim H. Ku, Artemis, and ataxia-telangiectasia-mutated: signalling networks in DNA damage. Int. J. Biochem. Cell Biol. 2008;40:598–603. doi: 10.1016/j.biocel.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 5.Feldmann H, Driller L, Meier B, Mages G, Kellermann J, Winnacker EL. HDF2, the second subunit of the Ku homologue from Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27765–27769. doi: 10.1074/jbc.271.44.27765. [DOI] [PubMed] [Google Scholar]
- 6.Teo SH, Jackson SP. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 8.Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat. Struct. Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 9.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl Acad. Sci. USA. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis LK, Storici F, Van Komen S, Calero S, Sung P, Resnick MA. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166:1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu. Rev. Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 13.Darroudi F, Wiegant W, Meijers M, Friedl AA, van der Burg M, Fomina J, van Dongen JJ, van Gent DC, Zdzienicka MZ. Role of artemis in DSB repair and guarding chromosomal stability following exposure to ionizing radiation at different stages of cell cycle. Mutat. Res. 2007;615:111–124. doi: 10.1016/j.mrfmmm.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Kurosawa A, Adachi N. functions and regulation of artemis: a goddess in the maintenance of genome integrity. J. Radiat. Res. 2010 doi: 10.1269/jrr.10017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Picher AJ, Garcia-Diaz M, Bebenek K, Pedersen LC, Kunkel TA, Blanco L. Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res. 2006;34:3259–3266. doi: 10.1093/nar/gkl377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase mu. Nat. Struct. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 18.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 19.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 20.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc. Natl Acad. Sci. USA. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human artemis. J. Biol. Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 23.Bahmed K, Nitiss KC, Nitiss JL. Yeast Tdp1 regulates the fidelity of nonhomologous end joining. Proc. Natl Acad. Sci. USA. 2010;107:4057–4062. doi: 10.1073/pnas.0909917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manivasakam P, Schiestl RH. High efficiency transformation of Saccharomyces cerevisiae by electroporation. Nucleic Acids Res. 1993;21:4414–4415. doi: 10.1093/nar/21.18.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Decottignies A. Capture of extranuclear DNA at fission yeast double-strand breaks. Genetics. 2005;171:1535–1548. doi: 10.1534/genetics.105.046144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardo B, Ma E, Marcand S. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae. Genetics. 2006;172:2689–2694. doi: 10.1534/genetics.105.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haviv-Chesner A, Kobayashi Y, Gabriel A, Kupiec M. Capture of linear fragments at a double-strand break in yeast. Nucleic Acids Res. 2007;35:5192–5202. doi: 10.1093/nar/gkm521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrero VA, Symington LS. Extensive DNA end processing by exo1 and sgs1 inhibits break-induced replication. PLoS Genet. 2010;6:e1001–e1007. doi: 10.1371/journal.pgen.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair. 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl Acad. Sci. USA. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis LK, Resnick MA. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 2000;451:71–89. doi: 10.1016/s0027-5107(00)00041-5. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann G, Lindahl T, Schar P. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hegde V, Klein H. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 2000;28:2779–2783. doi: 10.1093/nar/28.14.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mages GJ, Feldmann HM, Winnacker EL. Involvement of the Saccharomyces cerevisiae HDF1 gene in DNA double-strand break repair and recombination. J. Biol. Chem. 1996;271:7910–7915. doi: 10.1074/jbc.271.14.7910. [DOI] [PubMed] [Google Scholar]
- 37.Deshpande RA, Wilson TE. Identification of DNA 3′-phosphatase active site residues and their differential role in DNA binding, Mg2+ coordination, and catalysis. Biochemistry. 2004;43:8579–8589. doi: 10.1021/bi049434n. [DOI] [PubMed] [Google Scholar]
- 38.Karumbati AS, Deshpande RA, Jilani A, Vance JR, Ramotar D, Wilson TE. The role of yeast DNA 3′-phosphatase Tpp1 and rad1/Rad10 endonuclease in processing spontaneous and induced base lesions. J. Biol. Chem. 2003;278:31434–31443. doi: 10.1074/jbc.M304586200. [DOI] [PubMed] [Google Scholar]
- 39.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010 doi: 10.1038/emboj.2010.193. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 41.Westmoreland JW, Summers JA, Holland CL, Resnick MA, Lewis LK. Blunt-ended DNA double-strand breaks induced by endonucleases PvuII and EcoRV are poor substrates for repair in Saccharomyces cerevisiae. DNA Repair. 2010;9:617–626. doi: 10.1016/j.dnarep.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett CB, Lewis AL, Baldwin KK, Resnick MA. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl Acad. Sci. USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes G, Rine J. Regulated expression of endonuclease EcoRI in Saccharomyces cerevisiae: nuclear entry and biological consequences. Proc. Natl Acad. Sci. USA. 1985;82:1354–1358. doi: 10.1073/pnas.82.5.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis LK, Westmoreland JW, Resnick MA. Repair of endonuclease-induced double-strand breaks, in Saccharomyces cervisiae: essential role for genes associated with nonhomologous end-joining. Genetics. 1999;152:1513–1529. doi: 10.1093/genetics/152.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]