Abstract
PKR (double-stranded RNA-activated protein kinase) is an important component of the innate immunity, antiviral and apoptotic pathways. Recently, our group found that palmitate, a saturated fatty acid, is involved in apoptosis by reducing the autophosphorylation of PKR at the Thr451 residue, however, the molecular mechanism by which palmitate reduces PKR autophosphorylation is not known. Thus, we investigated how palmitate affects the phosphorylation of the PKR protein at the molecular and biophysical levels. Biochemical and computational studies show that palmitate binds to PKR, near the ATP-binding site, thereby inhibiting its autophosphorylation at Thr451 and Thr446. Mutation studies suggests that Lys296 and Asp432 in the ATP binding site on the PKR protein are important for palmitate binding. We further confirmed that palmitate also interacts with other kinases, due to the conserved ATP-binding site. A better understanding of how palmitate interacts with the PKR protein, as well as other kinases, could shed light onto possible mechanisms by which palmitate mediates kinase signaling pathways, that could have implications on the efficacy of current drug therapies that target kinases.
INTRODUCTION
The double-stranded (ds) RNA-activated protein kinase (PKR) is a ubiquitously expressed serine/threonine kinase that is up-regulated upon interferon (IFN) production during mammalian innate immune response (1). The enzyme is normally in its latent form, and is activated upon binding of double-stranded RNA (dsRNA), triggering dimerization and autophosphorylation (2). Activated PKR can phosphorylate the α-subunit of eukaryotic translation initiation factor (eIF2α) at Ser 51, thereby decreasing the initiation of viral translation by inhibiting the guanidine nucleotide exchange activity of the eIF1 heterotrimeric complex (3). PKR can also phosphorylate the regulatory subunit B56α on protein phosphatase 2A (PP2A) and thereby enhance the enzymatic activity of the PP2A-B56α trimeric complex, which can lead to dephosphorylation of eIF-4E and translational arrest (4). In addition, PKR is broadly involved in diverse cellular processes, such as cellular differentiation, apoptosis, cell growth, and oncogenic transformation (5).
PKR is a 551-amino acid enzyme consisting of a N-terminal double-stranded RNA binding domain (dsRBD) and a C-terminal kinase domain connected by an 80 residue unstructured linker. The dsRBD contains two conserved dsRBD motifs responsible for dsRNA recognition. The NMR structure of the dsRBD revealed two motifs that adopt the canonical αβββα fold linked by a ~20 amino acid sequence or linker, however dsRBM1 has more backbone motion than dsRBM2 (6, 7). The x-ray crystal structure of the kinase domain of PKR complexed with eIF2α was recently resolved (8). The kinase domain adopts a bilobal structure typical of protein kinases, with a smaller N lobe and a larger C lobe. The N lobe is involved in the back-to-back dimerization of the PKR kinase domain and the C lobe is responsible for substrate recognition. ATP binds the cleft between the two lobes and the binding mode required to mediate phosphoryl transfer is highly conserved across kinases (9, 10). The two phosphorylation sites in the activation loop of the C lobe (Thr446 and Thr451) are critical for full catalytic activation of PKR as well as substrate binding (2, 8, 11).
A large body of biochemical and biological evidence revealed that PKR has both pro- and anti-apoptotic roles. The first evidence of a pro-apoptotic role of PKR was performed in PKR-overexpressed HeLa cells using a recombinant vector (12). It was further demonstrated that PKR, triggered by viruses, induces apoptosis (13–15). In the absence of viral infection, PKR has been generally recognized as a pro-apototic factor that phosphorylates eIF2α, resulting in the inhibition of protein synthesis. Transcription factors, such as nuclear factor kappa-B (NF-κB), activating transcription factor 3 (ATF-3), and p53 also have been implicated in mediating PKR-induced apoptosis (16). In contrast, other studies have shown that PKR plays anti-apoptotic roles in regulating tumor development and tumor-cell apoptosis (17–20). A recent study suggests that PKR may act as a tumor suppressor, preventing apoptosis mediated by the transcription factor NF-κB (20). Results from our lab support an anti-apoptotic role of PKR in human hepatocellular carcinoma cells (HepG2), by regulating the protein expression and phosphorylation levels of Bcl-2 (B-cell leukemia/lymphoma 2) (21, 22). We found the autophosphorylation of PKR is attenuated by a saturated free fatty acid (FFA), palmitate. However, the molecular mechanism by which palmitate inhibits PKR autophosphorylation is not known.
Palmitate has low aqueous solubility and is present at very low concentrations in the hydrophilic, intracellular environment. In order to fulfill required cellular functions, palmitate overcomes its low solubility by interacting with proteins. The most extensively studied palmitate-binding protein is serum albumin, which helps to transport fatty acids in vivo. Human serum albumin (HSA) contains three homologous α-helical domains and has 7 palmitate-binding sites that share similar binding features: such as a carboxyl group that can form a salt bridge or basic and polar amino acid side-chains that can form hydrogen bonds. The methylene chain on palmitate fits readily into a hydrophobic cavity between the helices of HSA (23). In contrast to serum albumin, other proteins, such as the family of fatty acid-binding proteins (FABPs), involved in fatty acid uptake, transport, and oxidation, are mostly β-sheets and have very little helical structure. However, the binding modes of palmitate to FABPs are similar to HSA, namely, FABPs also contain one or two conserved basic amino acid chains that form a hydrophobic binding pocket that can bind to the carboxyl group on palmitate (24, 25). Although they have similar binding modes, the palmitate binding affinity for HSA (5~10 nM) is about 10 to 100-fold stronger than FABPs depending on the detection method used (26–28). In addition, a nuclear receptor protein, hepatic nuclear factor 4 (HNF4), interacts with palmitate to form a hydrogen bond with arginine in its hydrophobic cavity and functions as a constitutively active transcription factor (29). Due to the structural simplicity of palmitate, its ability to bind to various types of proteins is enhanced in the presence of a hydrophobic pocket that contains amino acids that can stabilize the carboxyl group of palmitate. This raises the question of whether a hydrophobic pocket exists within the PKR protein to which palmitate may bind to modulate its kinase activity.
Here we report a novel molecular mechanism by which palmitate modulates PKR activity, and thereby regulates cellular functions. Using fluorescence techniques, we investigated the binding affinity between palmitate and the PKR protein. We performed computational docking experiments to evaluate the binding of palmitate to PKR and other randomly chosen kinases with conserved domain arrangements. The docking results suggest that palmitate locates in the kinase domain of PKR, near the ATP-binding site. Biochemical and biophysical experiments were performed to further demonstrate that palmitate binds to the kinase domain of PKR, and reduces the autophosphorylation of PKR. Finally, we also evaluate the ability of palmitate to function as an ATP competitive inhibitor to several kinases. The implications of this work, i.e. the role of palmitate on PKR signaling, and broadly on other kinases, are discussed.
EXPERIMENTAL PROCEDURES
Materials
4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid (BODIPY® FL C16), herein denoted as Bodipy-PA, and 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionic acid (BODIPY® FL) were obtained from Invitrogen.γ-33P-ATP was purchased from Perkin Elmer. Anti-PKR, anti-PKR-pThr451, and anti-PKR-pThr446 were purchased from Abcam, Sigma-Aldrich, and Novus Biologicals, respectively. Recombinant kinase proteins, Akt1 (Protein kinase B 1), MAP kinase-activated protein kinase 3 (MAPKAPK3), and Cyclin-dependent kinase 4 (CDK4), were obtained from GenWay Biotech, Inc. All other reagents used were reagent grade.
Protein expression and purification of PKR protein constructions
DNA recombinant plasmids, pET28a-pWT (30) and pET11a-WT (31) containing the full length of human PKR wild-type protein were kindly provided by Dr. Philip C. Bevilacqua’s and Dr. James L. Cole’s groups, respectively. As previously reported, wild-type PKR expressed in E. coli is phosphorylated, thus unphosphorylated protein was obtained by coexpressing PKR with phosphatases (32, 33): Phosphorylated PKR and unphosphorylated PKR are herein denoted as PKR-pWT and PKR-WT, respectively.
The plasmids were transformed into E. coli BL21(DE3) pLysS (Invitrogen) or Rosetta™ 2(DE3) pLysS cells (Novagen). Cells were grown in LB media at 37°C until the OD at 600 nm reaches 0.6 and then protein expression was induced with 1 mM IPTG for three hours at 30°C. The cells were collected by centrifugation at 4,000 rpm for 10 min and re-suspended in sonication buffer (50 mM HEPES, 500 mM NaCl, 5 % glycerol (pH 7.0), and 7 mM β–mercaptoethanol) containing protease inhibitor cocktail and phenylmethanesulfonyl fluoride (PMSF). For PKR-WT protein, the sonication buffer without NaCl was used to re-suspend the cells. The suspension solution was lysed with 100 mM lysozyme and sonicated. The lysate was centrifuged at 12,000 rpm for 25 min and the supernatant was filtered by Millex-HV filter (Millipore). Two affinity chromatography techniques using HiTrap™ Chelating HP and HiTrap™ Heparin HP columns were separately employed for purification of PKR proteins (GE Healthcare). The supernatant was loaded onto the columns and the PKR proteins were eluted with an imidazole gradient or a NaCl gradient using ÄKTA™ FPLC system (GE Healthcare). High salt contents in the fractions were removed using HiTrap™ Desalting column (GE Healthcare) and if necessary, size exclusion chromatography were performed using a Superdex 75 column (GE Healthcare) to improve purity. All purified PKR proteins were confirmed by SDS-PAGE and Western blotting analysis.
Bodipy-Palmitic acid binding assay
10 nM of Bodipy-C16 was mixed with PKR proteins in PBS buffer (pH 7.4) at room temperature. After 5 min of incubation, fluorescence polarization measurements were performed at 488nm/520nm using FluoroMax-4 (Horiba). The KD values were determined by fitting the data to the one-site quadratic binding equation using the Kaleida-Graph software;
Where FP is the fluorescence polarization value when the complex between PKR and B-PA (Bodipy-C16) is formed, FPmax is the fluorescence polarization value when the B-PA completely binds to the PKR, FP0 is the fluorescence polarization value when the PKR is free, and [B-PA]0 and [PKR]0 are the initial concentrations of Bodipy-C16 and PKR, respectively.
Palmitate treatment on HepG2 Cell and western blotting analysis
Human hepatocellular carcinoma cells were cultured in Dulbecco’s modified eagle medium (DMEM) with 10% fetal bovine serum and penicillin-streptomycin (penicillin: 10 000 U/ml, streptomycin: 10 000 μg/ml). All detailed processes were described in the reference (22). Palmitate (0.7 mM) were complexed to 2 % BSA (fatty acid free) dissolved in the DMEM medium. Palmitate treatment were performed for 24 hr and 2% BSA was used as a negative control. The HepG2 cells were washed twice with cold PBS and treated with CelLytic M cell lysis buffer (Sigma-Aldrich) supplemented with protease inhibitor cocktail (Sigma-Aldrich) for 10 min on ice. The cell lysate was clarified by centrifugation at 13 000 rpm for 10 min, and the supernatant was collected. Total protein levels were quantified by Bradford assay (Bio-Rad). 40 μg of total protein was loaded to 9 % SDS-PAGE gel, transferred to nitrocellulosemembranes, and probed with primary (anti-pThr446) and secondary antibodies. The image was analyzed using the Molecular Imager ChemiDoc XRS System from Bio-Rad.
Statistical Analysis
All experiments were independently performed at least three times, and representative results are shown. The statistical analysis was performed using a Student t-test on the SigmaPlot software, with the statistical significance set at P < 0.01.
In silico docking of palmitate
The docking experiments were performed on the X-ray crystallographic structures of phosphorylated PKR (PDB ID: 2A19 (8)), phosphorylated Akt1 (PDB ID: 3CQW (34)), and unphosphorylated CDK4 (PDB ID: 2W96 (35)) obtained from the Protein Data Bank (PDB) (36). The homology model of MAPKAPK3 was downloaded from the Swiss-Prot database (37). The MAPKAPK3 structure was derived using the unphosphorylated form of MAPKAPK2 bound to p38alpha (PDB ID: 2OZA (38)) as the homologue. Any bound ligands were removed from the initial structures followed by addition of missing hydrogen atoms and a short minimization of the hydrogen positions while harmonically restraining all heavy atoms with a force constant of 5 kcal/mol. Hydrogen atom addition and minimization were performed with CHARMM (39) using the CHARMM22/CMAP force-field (40). All subsequent docking experiments were performed on the minimized structure using anionic palmitate (C15COO− ) as the ligand. The CHARMM-based molecular dynamics docking algorithm CDOCKER (41) as implemented in Accelrys Discover Studio 2.1 was used to dock the flexible ligand within a sphere of 5Å radius around the heavy atom centers of selected side chain atoms (NZ for Lys, OD1/OD2 of Asp, ND2 of Asn, CE1/CE2/CZ of Phe, OH of Tyr, CD of Ile, CG1/CG2 of Val, CD1/CD2 of Leu, SG of Cys and ND1/NE2/CE1 of His). A rigid receptor cavity was maintained in all docking experiments. Extensive validations of CDOCKER and comparisons with other docking methods have been performed prior (41, 42). The results reflected that CDOCKER performs well even for a diverse set of test cases where success rate of predicting the correct ligand pose in the receptor cavity was >80%. This provided reasonable confidence in accepting the docking results in this study under the assumption that palmitate binds to the nucleotide binding cavity of the kinase.
RESULTS
Palmitic acid binds to PKR proteins
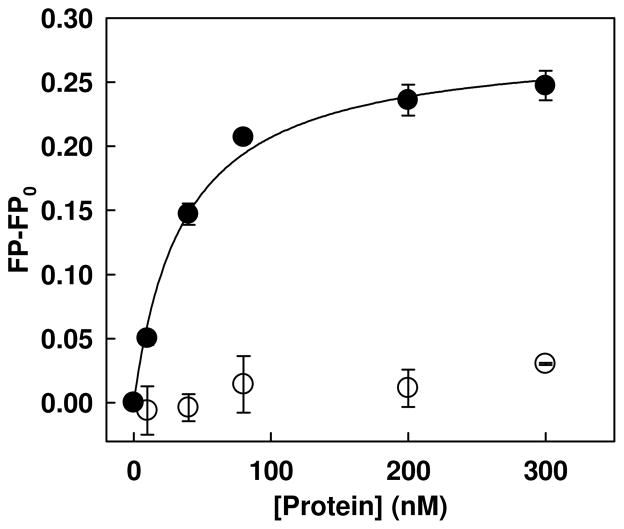
Free fatty acids (FFAs) in the plasma are transported by plasma proteins, i.e. albumin, leaving approximately 10 nmol/L of FFA unbounded (43). To determine the binding affinity of physiological level (10 nM) of unbounded FFA, palmitate, to PKR proteins, we developed a fluorescence polarization (FP)-based palmitate interaction assay using fluorescently labeled palmitate molecule, Bodipy-C16. We first assessed whether the FP-based assay is able to determine the binding constant of bovine serum albumin (BSA), which served as a positive control protein. As shown in Figure 1, the FP values of Bodipy-C16 increased gradually with increasing concentrations of BSA and eventually approached a plateau. In contrast, lysozyme did not show any significant increase, suggesting that the assay is specific to palmitic acid-binding proteins. The binding constant (KD) of BSA was determined using nonlinear regression analysis as described in the Materials and Methods section. The KD value (29.21±1.85 nM) observed is similar to the value (22 nM) reported by Burczynski’s group (44). Thus, FP-based palmitate interaction assay is well suited to quantify the interaction between Bodipy-C16 and palmitic acid-binding proteins.
Figure 1.
Fluorescence polarization measurement of the Bodipy-C16 and BSA interaction. 10 nM of Bodipy-C16 was added to PBS buffer with increasing concentrations of Lysozome (open circles) or BSA (closed circles). After 5 min of incubation, the fluorescence polarization was measured at an excitation wavelength of 488 nm and an emission wavelength of 520 nm using a spectrofluorometer. The solid lines represent fitting of the data to the quadratic binding equation described in Materials and Methods. Each error bar represents the mean of triplicates ±SD.
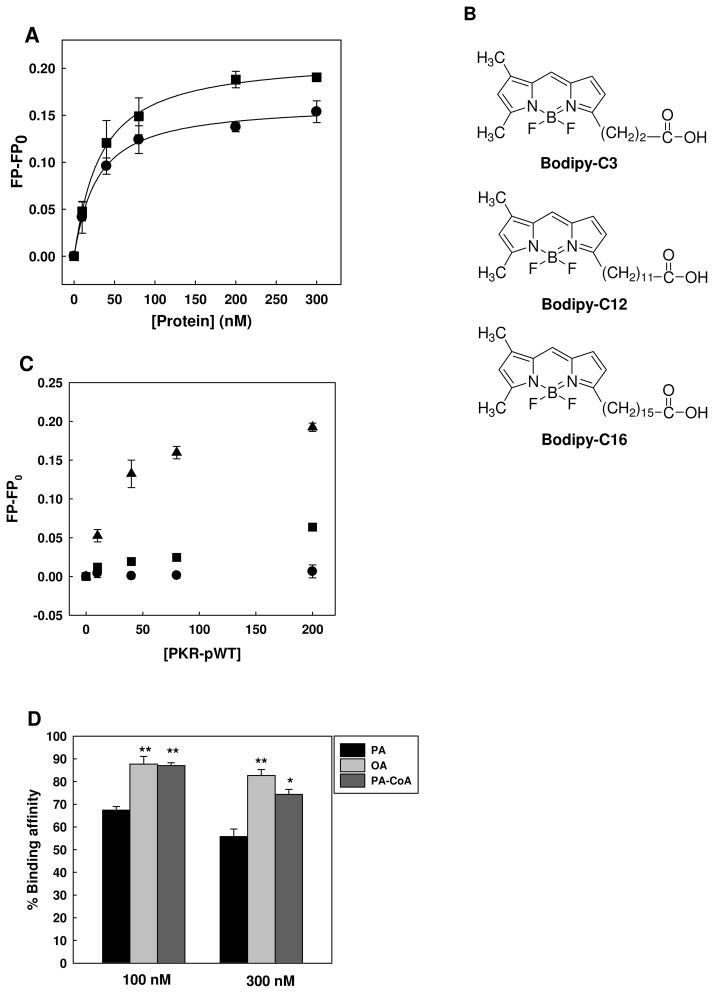
We used this FP-based assay to evaluate whether Bodipy-C16 interacts with the PKR proteins. Two recombinant PKR wild-type proteins, unphosphorylated PKR (PKR-WT) and phosphorylated PKR (PKR-pWT), were prepared for this assay. Prior to the binding assay, the phosphorylation status was assessed by immunoblot analysis against anti-PKR-pThr451 and anti-PKR-pThr446. PKR-pWT but not PKR-WT showed a phosphorylation fraction on the immunoblot (data not shown). The Bodipy-C16 binding assay was performed at 10 nM of Bodipy-C16 with increasing concentrations of the PKR proteins. As shown in Figure 2A, the FP values for both PKR-WT and PKR-pWT proteins increased significantly. Assuming a 1:1 binding stoichiometry, PKR-WT and PKR-pWT have KD values of 23.22±1.28 nM and 24.90±1.00 nM, respectively, suggesting that both have similar binding affinities to Bodipy-C16.
Figure 2.
Binding affinity of Bodipy-C16 to PKR wild-type proteins. (A) 10 nM of Bodipy-C16 was added to PBS buffer with increasing concentrations of PKR-WT (circles) or PKR-pWT (squares). After 5 min of incubation, the fluorescence polarization was measured at 488/520 nm using a spectrofluorometer. The solid lines represent fitting of the data to the quadratic binding equation described in Materials and Methods. Each error bar represents the mean of triplicates ± SD. (B) Structure comparison of Bodipy-C3, Bodipy-C12 and Bodipy-C16. (C) Binding affinity of 10 nM Bodipy-C3 (circles), Bodipy-C12 (squares), and Bodipy-C16 (triangles) with increasing concentrations of PKR-pWT in PBS buffer. After 5 min of incubation, the fluorescence polarization was measured at 488/520 nm with a spectrofluorometer. Representative data points shown are of average of three measurements and the error bars indicate the standard deviations. All data points of Bodipy-C16 were significantly different from both Bodipy-C3 and Bodipy-C12 (p<0.001). (D) Competition binding assay of fatty acids and lipid. At a constant concentration of PKR-pWT (20 nM), 5 nM of Bodipy-C16 was added to Tris buffer (20 mM Tris (pH 8.0), 100 mM NaCl) in the presence of 100 or 300 nM of palmitic acid (PA), oleic acid (OA), and palmitoyl CoA (PA-CoA). After 15 min of incubation, the fluorescence polarization was measured at an excitation wavelength of 488 nm and an emission wavelength of 520 nm with a spectrofluorometer. The percentage of binding is calculated based on the following equation: %Binding = (FP-FPnegative control)/(FPpositive control−FPnegative control)*100. * (p<0.05) and ** (p<0.001) indicate statistically different to PA.
We further addressed whether palmitic acid is a specific ligand to the PKR protein. In addition to Bodipy-C16, two more Bodipy-labeled saturated fatty acids, Bodipy-propanoic acid (Bodipy-C3) and Bodipy-lauric acid (C12), were evaluated for their binding affinity to PKR. Their structures are compared with Bodipy-C16 in Figure 2B. The structural difference is in the length of the methylene chain. As shown in Figure 2C, no significant change in FP values is observed for Bodipy-C3 (up to 200 nM) and Bodipy-C12 (up to 80 nM) with PKR-pWT and the FP values for both Bodipy-C3 and Bodipy-C12 are markedly different from Bodipy-C16. These results suggest that the PKR protein has a higher binding specificity to the long chain saturated fatty acid, palmitic acid (C16). Additionally, using a competition assay with Bodipy-C16, we tested whether other long chain unsaturated fatty acids or lipids are able to bind to the PKR protein. 100 and 300 nM of palmitic acid (a saturated fatty acid), oleic acid (an unsaturated fatty acid), and pamitoyl-CoA (a lipid) were competed with 5 nM of Bodipy-C16. We found that palmitic acid more readily displaced the Bodipy-C16 from PKR-pWT than either oleic acid or pamitoyl-CoA (Figure 2D), supporting that the PKR protein more likely interacts with a long chain saturated fatty acid, palmitic acid.
In silico docking to PKR and other kinases reveal ATP-binding cavity as a potential binding site for palmitate
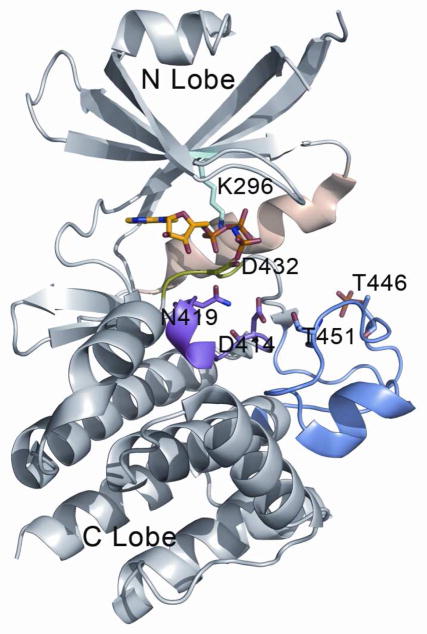
To determine potential site(s) on the PKR proteins to which palmitate may bind, we performed a preliminary search for binding cavities. A feasible approach would have been to perform blind docking of palmitate on the whole kinase monomer, but the protein surfaces change considerably upon dimerization or higher order oligomerization, which have been found to be functionally important (2, 8). Moreover, some surface loops are missing in the crystal structures of kinases, which incorporate another source of false positives while docking palmitate over the whole molecule. Alternatively, our approach was more intuitive in predicting the potential binding site. Knowing from the experiments that palmitate does bind to PKR, we searched for the single largest hydrophobic cavity on the kinase molecules. The ATP-binding cavity between the N and C lobe of the PKR kinase domain provides the largest hydrophobic pocket capable of binding palmitate consisting of a large hydrophobic chain. Other cavities were not large enough to accommodate the ligand without going through conformational changes through an induced-fit mechanism, which were not considered in this study. Figure 3 illustrates the binding mode of the nucleotide with the protein, which was obtained from the PKR crystal structure (8). The binding site consists mainly of hydrophobic residues residing on the β-sheet bundle of the N-lobe and key charged residue, such as the Lys296 (numbering as in PDB ID 2A19 for PKR), that interact with the nucleotide phosphate groups. Apart from the hydrophobic pocket formed by the β-sheet bundle of the N-lobe, there are several loop regions that play important roles in the binding and hydrolysis of ATP. One of them is the Mg2+ binding loop that contains the conserved motif DFG, the aspartic acid residue (Asp432 of PKR) being important in binding and stabilizing the ATP-Mg2+ complex. The catalytic loop containing conserved residues, such as Asp414 and Asn419, have been implicated in the catalysis of the nucleotide γ-phosphate. The activation loop contains the threonine residues (Thr446, Thr451) that undergo phosphorylation during activation of the kinase domain. Finally, the αC helix present in the N-lobe acts as an important conformational switch, changing its orientation along with the activation loop, during the inactivation-activation cycle (45). It is observed that the nucleotide binding cavity is sandwiched between the β-sheet bundle of the N-lobe, the Mg2+ binding loop, the catalytic loop and the α C helix, while the mouth of the cavity is lined by parts of the activation loop.
Figure 3.
Binding mode of AMP-PNP with PKR. Kinase domain of PKR are showed in gray, magnesium binding loop in green, catalytic loop in magenta, activation loop in blue and αC helix in wheat. Important residues in the nucleotide binding cavity are shown in sticks and bound the ATP analog adenylyl imidodiphosphate (AMP-PNP) are colored in yellow for the carbon atoms. The figure was generated using the following structures: PKR:AMP-PNP:eIF2a (PDB id = 2A19).
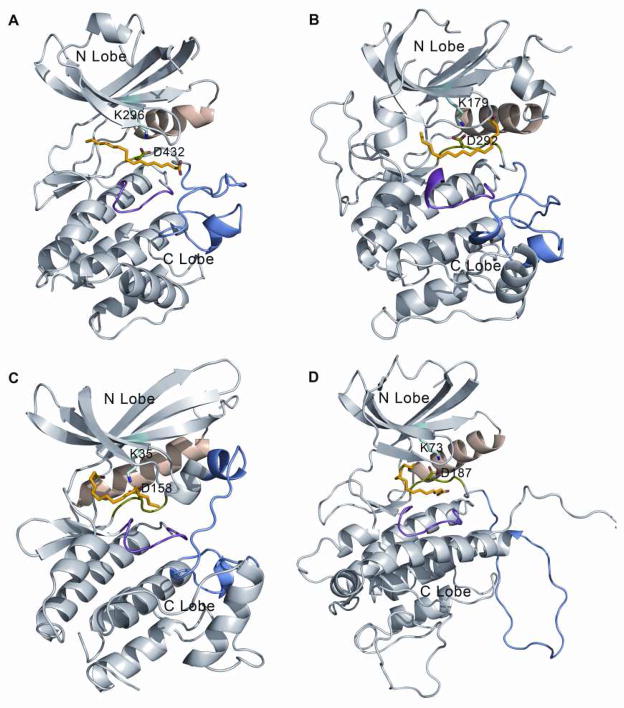
Given that the X-ray crystallographic structures of three commercial kinases, Akt1, CDK4, MAPKAPK2 as a homology model of MAPKAPK3, are available, they were also selected for the computational docking experiments. We selected several conserved residues from the nucleotide binding cavity formed by the β-sheet bundle of the N-lobe, some key residues of the Mg2+ binding loop, and some from the catalytic loop to perform our computational docking experiments (selected residues are shown in the supplementary Figure S1). Table 1 lists the docking scores obtained from CDOCKER of palmitate around each specific residue. The residues selected for docking spanned the whole cavity available between the N and C lobe of the kinase. The docking score estimates the free energy of binding between the ligand and the receptor and thus the more negative score is meant to reflect more favorable binding interactions between the two. In most cases, palmitate failed to bind to the protein receptor, and is designated as “Fail” in Table 1. The best-docked poses for all 4 protein kinases were found in the vicinity of the Lys296 or Asp432 residues of PKR and analogous residues in the other kinases. The resulting docking poses of palmitate partly occupy the nucleotide binding site and thus would be capable of replacing or inhibiting the binding of ATP to the kinase (Figure 4). Although receptor flexibility was not considered explicitly while docking palmitate to the individual kinases, the results suggest that palmitate is capable of binding to nucleotide binding sites with different side chain orientations found in the crystal structures of different kinases. Supplementary Figure S2 provides a close-up view of the aligned nucleotide binding sites of the 4 kinases which shows that receptor flexibility is considered to some extent.
Table 1.
Best binding site of palmitate in the ATP-binding cavity of the PKR, Akt1, CDK4 and MAPKAPK3 kinase domains. Best docking score (reflecting binding free-energy in kcal/mole) out of 10 ligand conformations are tabulated. Binding site for the docking experiments is defined as 5Å spheres around the side-chain heavy atoms for the tabulated residues in each case. According to the most negative docking scores, the most favorable binding site is near Lys296/Asp432 for PKR or analogous residues for the other kinases
| PKR | Akt1 | CDK4 | MAPKAPK3 | ||||
|---|---|---|---|---|---|---|---|
| K296 | −33.79 | K179 | −33.22 | K35 | −42.00 | K73 | Fail |
| D432 | −35.85 | D292 | −45.36 | D158 | Fail | D187 | −32.16 |
| I273 | −30.16 | L156 | Fail | I12 | −36.24 | L50 | −26.08 |
| V281 | Fail | V164 | Fail | V20 | Fail | V58 | Fail |
| F433 | Fail | F293 | Fail | F159 | Fail | F188 | Fail |
| V298 | Fail | L181 | Fail | V37 | Fail | L75 | Fail |
| F368 | −32.48 | Y229 | Fail | H95 | Fail | C120 | Fail |
| F421 | Fail | M281 | Fail | L147 | Fail | L173 | Fail |
| D414 | 414.68 | D274 | Fail | D140 | Fail | D166 | Fail |
| N419 | −26.60 | N279 | Fail | N145 | Fail | N171 | Fail |
Figure 4.
Best docked pose of palmitate with different kinases. Kinase domains of (A) PKR, (B) AKT1, (C) CDK4 and (D) MAPKAPK3 are shown in gray. Magnesium binding loop in green, catalytic loop in magenta, activation loop in blue and αC helix in wheat. Conserved interaction between Lys296 and Asp432 in PKR and analogous residues in other kinases are shown in sticks. Best docked poses of palmitate are also shown in sticks with carbon atoms in yellow.
Apart from the sites around Lys296 and Asp432, PKR also appears to interact with Ile273 and Phe368. Especially, the score suggests comparable binding affinities for Lys296 and Phe368 for PKR within the limits of the error estimated for predicting binding affinities in other systems, which is in the range of several kcal/mol (46). Ile273 is one of the conserved hydrophobic residues in the β-sheet bundle of the N-lobe, while Phe368 is a residue facing the dimer interface of the kinase domain. Favorable binding near the analogous sites to Ile273 of PKR was also found with CDK4 and MAPKAPK3 further supporting that Ile273 might play an important role in palmitate binding. However, favorable palmitate binding near Phe368 was only found with PKR. Palmitate also appears to bind near the catalytic loop residue, Asn419 in PKR, but such a binding mode was not observed with the other kinases. An inspection of the bound palmitate around Ile273 and Phe368 reveals that the ligand is significantly exposed to the solvent as opposed to the poses interacting with Lys296 or Asp432. This is reflected by the fact that palmitate has fewer direct interactions with protein atoms (ligand-protein atomic distance < 5Å) when bound near Ile273 and Phe368, as compared to the Lys296 or Asp432 site. While a full energetic analysis of palmitate in the two binding poses is beyond the scope of the current study, it appears that partial burial of palmitate and its long hydrophobic chain in the poses that interact with Lys296 or Asp432 may be more favorable than binding near Ile273 or Phe368 with the more significant solvent exposure. Consequently, we propose that palmitate binds near Lys296 or Asp432 as shown in Figure 4.
The accessible binding cavity around the catalytic loop is dependent, in part, on the conformation of the activation loop, which is highly flexible in all four of these kinases. Thus, the cavity itself differs between the four kinases at this region, as a result, palmitate may not be able to interact with all kinases at this site or might require an induced fit mechanism to do so, which was not considered here. It should be noted that due to the lack of rigorousness in the nature of the preliminary docking experiments conducted here, the exact mode of palmitate interactions with the kinase domain is difficult to predict with certainty. Nevertheless, consistent results for 4 different types of kinases with conserved, yet different side chain orientations of the nucleotide binding cavity (Figure S2) suggest that palmitate is binding near the ATP-binding pocket.
Bodipy-C16 binds to other kinases, likely due to the conservation of the ATP-binding site
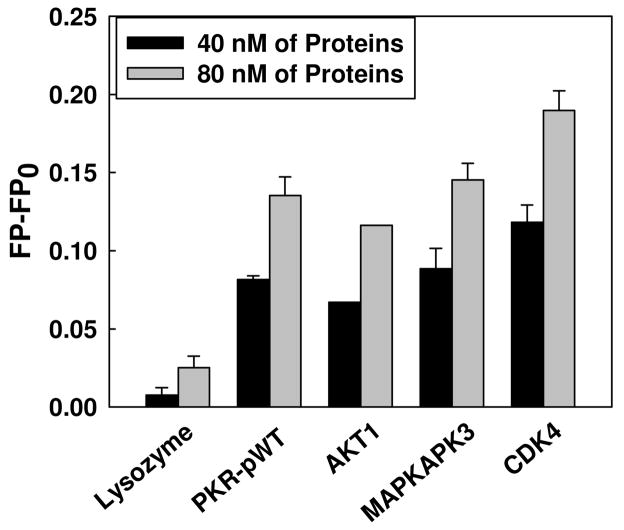
The docking experiments indicate that palmitate positions near the ATP-binding site of the three other kinases, Akt1, CDK4, and MAPKAPK3, are similar to the PKR protein. Therefore, we also evaluated whether Bodipy-PA can bind to these three kinases using the FP-based palmitate interaction assay. As shown in Figure 5, at 40 and 80 nM of kinases, the FP values of all four kinases were significantly different from the control protein (lysozyme). These results suggest palmitate binds to these kinases, likely near the conserved ATP-binding site, as shown in Figure 4.
Figure 5.
Bodipy-C16 binding to kinases. 10 nM of Bodipy-C16 was added to PBS buffer in the absence of kinase and in the presence of 40 nM and 80 nM of kinases, PKR-pWT, Akt1, MAPKAPK3, and CDK4. Lysozyme was used as a negative control under the same condition. After 5 min of incubation, the fluorescence polarization was measured at 488/520 nm using a spectrofluorometer. Representative data point shown is for the average of three measurements and the error bars indicate the standard deviations.
Bodipy-C16 binds competitively to PKR wild-type proteins in the presence of ATP, thereby preventing the autophosphorylation of PKR WT protein
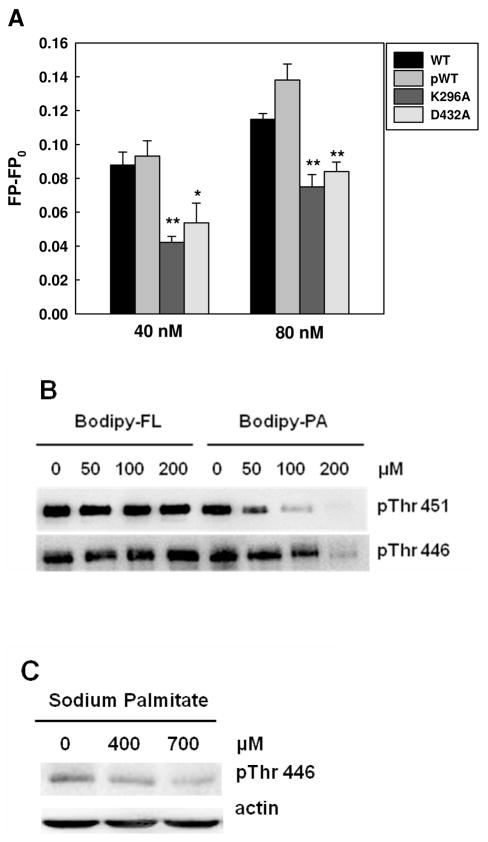
The computational docking experiments suggest that the ATP-binding pocket is the most plausible site for palmitate binding to PKR protein. To examine whether Bodipy-C16 can be located in the ATP-binding site, we mutated two amino acids (separately) with the most favorable palmitate binding residues (Lys296/Asp432) on the PKR protein (Table 1) and performed the Bodipy-C16 binding assay. At protein concentrations of 40 and 80 nM, the binding affinities of the mutant K296A and D432A proteins decreased significantly as compare to the wild-type proteins (Figure 6A). However, since the mutant proteins are not able to completely block the Bodipy-C16 binding, it suggests that the palmitic acid may not binding exactly at these residues, but near it to interfere with ATP binding to PKR.
Figure 6.
Inhibitory effects of PA on PKR phosphorylation. (A) Fluorescence polarization measurement of PKR mutants. 10 nM of Bodipy-PA was added to 40 and 80 nM of wild-type PKR proteins (phosphorylated and unphosphorylated) and PKR mutants in PBS buffer. After 5 min of incubation, the fluorescence polarization was measured at an excitation wavelength of 488 nm and an emission wavelength of 520 nm using a spectrofluorometer. * (p<0.05) and ** (p<0.001) indicate statistically different to wild-type proteins. (B) Competition assay with Western blot analysis. 2 μM of PKR-WT was incubated with Bodipy-C3 or Bodipy-C16 at the indicated concentrations in phosphorylation buffer (10 mM HEPES (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 1mM DTT) for 20 min. Autophosphorylation of WT PKR was performed by adding 100 μM of ATP at room temperature for 30 min. To stop the autophosphorylation reaction, 5X SDS-PAGE sample buffer was added to the reactions, followed by heating at 95°C for 5 min. The reactions were loaded onto 10 % Tris/glycine SDS-polyacrylamide gel and detected for PKR phosphorylation at Thr451 and Thr446 by Western blot analysis. (C) Effect of palmitate on the phosphorylated Thr446 (see (22) for Thr451 results). HepG2 cells were cultured in regular medium until reaching 90% confluency and then exposed to 400 and 700 μM for 24 hr. 2% BSA was used as a negative control. After treatment, the cells were harvested and western blot analysis was performed to detect the level of phosphorylated PKR.
To further evaluate whether Bodipy-C16 can be displaced by ATP, we performed two types of competition assays. Since the ATP binding affinity (34 μM) to PKR is much weaker (47) than Bodipy-C16 (28 nM) and high concentrations of ATP interfered with the FP-based palmitate interaction assay (data not shown), we adapted an intensity-based palmitate binding assay from a previous study that investigated the binding affinity of palmitate to FABPs (48). The fluorescence intensity method required a much higher Bodipy-PA concentration (500 nM) and the fluorescence intensity increases upon binding of Bodipy-C16 to the FABPs. We found the fluorescence intensity-based palmitate binding assay can be used to evaluate the binding affinity of palmitate to the PKR proteins (supplementary Figure S3). For our studies, we used 600 nM as the PKR concentration for the competition assay, where PKR is unable to significantly autophosphorylate in the absence of dsRNA (31). In the range of the ATP concentrations evaluated, the fluorescence intensity decreased significantly as the ATP concentration increased, suggesting that Bodipy-PA binds competitively to the PKR wild-type proteins to inhibit ATP binding (supplementary Figure S4(A)). To confirm that the Bodipy group was not altering the binding or competition, we evaluated whether radiolabeled ATP (hot ATP) bound to PKR could be displaced by unlabeled palmitate. In the absence or presence of palmitate, hot ATP was incubated with PKR-pWT. The mixture was loaded onto a ZipTip-C4 column to separate unbound ATP from the ATP-PKR complex (bound ATP). In the absence of palmitate, 16.61% of the hot ATP was bound to the PKR-pWT protein. With increasing palmitate concentration, ATP was dose-dependently displaced from binding to PKR. These results demonstrate that palmitate and ATP bind competitively to PKR. It should be noted that, in the absence of PKR, ATP did not bind to the column either in the presence or absence of palmitate (supplementary Figure S4(B)), demonstrating that ATP and palmitate do not bind to each other at these concentrations. This is further confirmation that ATP and PA competes for the kinase domain on PKR.
We further investigated whether inhibiting ATP binding by palmitate blocks the autophosphorylation reaction. Previously it was demonstrated that PKR-WT protein can be autophosphorylated in the absence of double-stranded RNA and ATP is sufficient for its autophosphorylation (31). We performed another competition assay with Bodipy-C3 (negative control) or Bodipy-C16, at constant ATP and PKR-WT concentrations. With increasing Bodipy-C16 concentration, the phosphorylation of Thr451 and Thr446, the two major autophosphorylation sites on PKR, decreased significantly, whereas Bodipy-C3 had no impact on the autophosphorylation of PKR (Figure 6B). Upon binding of palmitate to PKR the autophosphorylation is down-regulated, suggesting that palmitate acts as an ATP-binding site-directed inhibitor. These in vitro results are consistent with our computational docking results as well as our earlier results that showed palmitate decreases the cellular level of pThr451 (22). Previously we did not evaluate the effect of palmitate on pThr446. Thus we treated HepG2 cells with BSA or BSA complexed to palmitate at varying concentrations (400 and 700 μM) and found palmitate significantly decreased the level of pThr446 (Figure 6C). In the cellular experiments, the concentration required to decrease the phosphorylation level of PKR was much higher than the concentration obtained from our in-vitro binding assay. This is because most of the palmitate bound to BSA or FABP is metabolized in the liver cells. Rioux et al showed that about 1% of the palmitate exists as an unbound and non-metabolized fatty acid when rat hepatocytes were treated with 100 μM of BSA and 150 μM of palmitate (49). Thus only this small fraction of free palmitate would be able to function as a signaling molecule to down-regulates the autophosphorylation of PKR at Thr446 and Thr451, by interfering with ATP binding to the PKR protein.
DISCUSSION and CONCLUSIONS
Lipids are components of the cellular membrane and important signaling molecules. Several lipid-binding domains have been identified and recognized on kinase proteins over the years. The first lipid-binding domain was identified on the PKC kinase protein (50). The interaction of PKC with lipids by its N-terminal conserved region 1 (C1) and C2 domains at the plasma membrane is thought to induce conformational rearrangements that lead to the active form (51). Similarly, PKB/Akt has a pleckstrin homology (PH) lipid binding domain which interacts with lipids, i.e. phosphatidylinositol-3,4,5-trisphosphate. Upon lipid binding, a major conformational change occurs and the Akt protein is recruited to the plasma membrane, which enables phosphorylation of a residue in the activation segment by membrane-localized 3-phosphoinositide-dependent kinase 1 (PDK1) (52). Unlike the membrane binding kinases, PKB and Akt, Diskin R. et al. (53) recently found a novel lipid binding site on the kinase C-terminal domain of p38α MAP kinase. They showed that the binding site is able to accommodate a lipid (n-oxtyl-β-gluocopyranoside) as well as fatty acids (15(s)-hydroxyeicosatetraenoic acid and arachidonic acid) (53). The lipid-binding site is formed between the MAP kinase insert and the main segment of the C lobe. Since the lipid binding modulates the conformation of the activation loop, the authors suggest that lipids (and fatty acids) could regulate p38a MAP kinase activity. Based on these evidence, we hypothesize that palmitate may bind to the PKR protein to modulate its activity.
PKR is a major signaling enzyme that plays diverse roles in cellular functions. It is well established that PKR is activated by viral double-stranded RNA (dsRNA) and the activation of PKR regulates its dimerization and autophosphorylation (16). Furthermore, PKR is involved in signal transduction pathways such as the MAPK and NF-κB pathways (16, 54). Recently, our group proposed that PKR can also regulate the Bcl-2-JNK pathway to mediate the apoptotic response of palmitate. In this scenario, palmitate decreases the autophosphorylation level of PKR at Thr451, which down-regulates the phosphorylation of Bcl-2 at Ser70 (22) and anti-apoptosis. The latter is possible given that PKR does not have upstream kinases, thus upon autophosphorylation it is able to modulate the activity of other proteins. In this study, we address how palmitate may decrease the autophosphorylation level of PKR and suggest, based on computational and experimental results, that palmitate interacts with the ATP-binding site of WT PKR proteins to interfere with PKR autophosphorylation. This novel mechanism may provide an explanation of how palmitate reduces the autophosphorylation of PKR in HepG2 cells.
In the latent state, PKR exists in a weak monomer-dimer equilibrium with a KD of 500 μM, but the dimerization of the PKR proteins increases significantly upon either HIV TAR-RNA (HIV-1 trans-activation responsive region-RNA) binding (KD ~75 μM) or phosphorylation of PKR (KD ~20 μM) (11, 31). In addition, phosphorylated WT protein serves as a potent activator of latent PKR, increasing the autophosphorylation reaction rate by 20-fold (11). Although dimerization of PKR is very important for autophosphorylation, (0.5 μM is sufficient to induce the autophosphorylation reaction (31)), this reaction rate increases significantly by its product, phosphorylated PKR. Thus, phosphorylated PKR is clearly more potent in autophosphorylating PKR. Interestingly, we found that the binding affinity of palmitate to the phosphorylated PKR is similar to the unphosphorylated PKR. Their similar binding affinity helps to block the autophosphorylation of both the unphosphorylated WT and the phosphorylated WT. In addition, we demonstrated that palmitate binds to unphosphorylated PKR to inhibit access of ATP to the PKR protein, thereby significantly reducing the autophosphorylation reaction (supplementary Figure S4 and Figure 6A). These multiple lines of evidence support an inhibitory role of palmitate on PKR autophosphorylation.
More interestingly, the binding mode of ATP to other kinase is similar to that observed with PKR (9, 10). Recently, the structure of 15 kinases were aligned and the ATP-binding site was found to be highly conserved (55). It was computationally demonstrated that the kinases share a consensus structure that stabilizes the ATP molecule and correctly coordinates ATP for phosphotransfer. Similar to these results for ATP, our computational results suggest that palmitate also may be binding to the ATP-binding site of multiple kinases, likely near the Lys296 and Asp432 of the PKR protein, and homologous sites of the other kinases, Akt1, CDK4, and MAKAPK3, studied here. We further demonstrated that palmitate can physically interact with these other kinases based on the FP-based interaction assay. In addition, our binding studies show the mutant PKR proteins, K296A and D432A, significantly decrease the binding affinity of Bodipy-C16 to the PKR protein, suggesting that palmitate likely locates near the ATP-binding site. Based upon our results, we propose that palmitate may broadly affect many soluble kinases. In future, if one were interested in a specific signaling pathway, a kinase library could be developed with the palmitic acid binding assay to identify kinases that strongly interact with palmitate and thereby help elucidate potential kinase signaling networks mediated by palmitate. Further studies are needed to fully understand all of the interactions of palmitate with kinases and the signaling pathways mediated by palmitate.
The in silico docking experiments employed here suggest that the ATP-binding site is a potential binding site for palmitate. However, such docking methods and the resulting ligand poses have some degree of uncertainty. Further experiments with more advanced computational approaches need to be performed to conclusively determine the location of the palmitate binding to these kinases. We intend to pursue such studies in the future to follow up on the results reported here.
In summary, we uncovered how palmitate may modulate PKR phosphorylation. Palmitate is binding near the ATP-binding site, to interfere and inhibit the autophosphorylation of PKR. In addition, due to the conserved ATP-binding site, palmitate can non-specifically bind to other kinases. However, the binding affinity of palmitate to the kinases may differ, depending on both the specific configuration around the ATP-binding site and the active/inactive conformation. This novel mechanism provides potential insight into how palmitate may modulate the signaling of PKR and, more broadly, other kinases.
Supplementary Material
Acknowledgments
We thank Dr. Mark Worden’s group for the use of their FPLC machine and thank Dr. Gary J. Blanchard for valuable advices. We also thank the members of Cellular and Biomolecular laboratory, especially to Ryan Thompson and Phillip Brooks.
Abbreviations
- ATF-3
activating transcription factor 3
- Bcl-2
B-cell leukemia/lymphoma 2
- BSA
bovine serum albumin
- CD
circular dichroism
- CDK4
Cyclin-dependent kinase 4
- dsRNA
double-stranded RNA
- dsRBM
dsRNA binding motif
- eIF
eukaryotic translation initiation factor
- FABPs
fatty acid-binding proteins
- FFA
free fatty acid
- HSA
human serum albumin
- HepG2
human hepatocellular carcinoma cells
- HNF4
hepatic nuclear factor 4
- IFN
interferon
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinases
- MAPKAPK3
MAP kinase-activated protein kinase 3
- NF-κB
nuclear factor kappa-B
- NMR
nuclear magnetic resonance
- PDB
protein data bank
- PKR
double-stranded RNA-dependent protein kinase
- PP2A
protein phosphatase 2A
- TAR
trans-activation responsive region
- WT
wild-type
Footnotes
This study was supported in part by the National Science Foundation (CBET 0941055), the National Institute of Health (R01GM079688 and R21RR024439), the MUCI and the MSU Foundation and the Center for Systems Biology.
Contributor Information
Hyunju Cho, Email: chohyu11@egr.msu.edu.
Shayantani Mukherjee, Email: shayan@msu.edu.
Pratheeba Palasuberniam, Email: palasube@msu.edu.
Lisa Pillow, Email: pillowl1@msu.edu.
Betul Bilgin, Email: bilginbe@egr.msu.edu.
Catherine Nezich, Email: nezichca@msu.edu.
S. Patrick Walton, Email: spwalton@egr.msu.edu.
Michael Feig, Email: feig@msu.edu.
Christina Chan, Email: krischan@egr.msu.edu.
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annual review of biochemistry. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 3.Gale M, Jr, Katze MG. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacology & therapeutics. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Williams BR. The B56alpha regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Molecular and cellular biology. 2000;20:5285–5299. doi: 10.1128/mcb.20.14.5285-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 6.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. The EMBO journal. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. The EMBO journal. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 9.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 10.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Molecular cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 11.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular framework for the activation of RNA-dependent protein kinase. The Journal of biological chemistry. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 12.Lee SB, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 13.Takizawa T, Ohashi K, Nakanishi Y. Possible involvement of double-stranded RNA-activated protein kinase in cell death by influenza virus infection. Journal of virology. 1996;70:8128–8132. doi: 10.1128/jvi.70.11.8128-8132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kibler KV, Shors T, Perkins KB, Zeman CC, Banaszak MP, Biesterfeldt J, Langland JO, Jacobs BL. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. Journal of virology. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung MC, Chang DL, Camantigue RE, Lau AS. Inhibitory role of the host apoptogenic gene PKR in the establishment of persistent infection by encephalomyocarditis virus in U937 cells. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11860–11865. doi: 10.1073/pnas.96.21.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SH, Forman AP, Mathews MB, Gunnery S. Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene. 2000;19:3086–3094. doi: 10.1038/sj.onc.1203632. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Gunnery S, Choe JK, Mathews MB. Neoplastic progression in melanoma and colon cancer is associated with increased expression and activity of the interferon-inducible protein kinase, PKR. Oncogene. 2002;21:8741–8748. doi: 10.1038/sj.onc.1205987. [DOI] [PubMed] [Google Scholar]
- 19.Hiasa Y, Kamegaya Y, Nuriya H, Onji M, Kohara M, Schmidt EV, Chung RT. Protein kinase R is increased and is functional in hepatitis C virus-related hepatocellular carcinoma. The American journal of gastroenterology. 2003;98:2528–2534. doi: 10.1111/j.1572-0241.2003.08663.x. [DOI] [PubMed] [Google Scholar]
- 20.Delgado Andre N, De Lucca FL. Knockdown of PKR expression by RNAi reduces pulmonary metastatic potential of B16–F10 melanoma cells in mice: possible role of NF-kappaB. Cancer letters. 2007;258:118–125. doi: 10.1016/j.canlet.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Srivastava S, Mittal S, Yang X, Sheng L, Chan C. A Three Stage Integrative Pathway Search (TIPS) framework to identify toxicity relevant genes and pathways. BMC bioinformatics. 2007;8:202. doi: 10.1186/1471-2105-8-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Chan C. Repression of PKR mediates palmitate-induced apoptosis in HepG2 cells through regulation of Bcl-2. Cell research. 2009;19:469–486. doi: 10.1038/cr.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya AA, Grune T, Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. Journal of molecular biology. 2000;303:721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- 24.Chmurzynska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. Journal of applied genetics. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- 25.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nature reviews. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richieri GV, Anel A, Kleinfeld AM. Interactions of long-chain fatty acids and albumin: determination of free fatty acid levels using the fluorescent probe ADIFAB. Biochemistry. 1993;32:7574–7580. doi: 10.1021/bi00080a032. [DOI] [PubMed] [Google Scholar]
- 27.Veerkamp JH, van Moerkerk HT, Prinsen CF, van Kuppevelt TH. Structural and functional studies on different human FABP types. Molecular and cellular biochemistry. 1999;192:137–142. [PubMed] [Google Scholar]
- 28.Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry. 2000;39:7197–7204. doi: 10.1021/bi000314z. [DOI] [PubMed] [Google Scholar]
- 29.Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, Willson TM, Williams SP. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 30.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 31.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. Journal of molecular biology. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 32.Jammi NV, Beal PA. Phosphorylation of the RNA-dependent protein kinase regulates its RNA-binding activity. Nucleic acids research. 2001;29:3020–3029. doi: 10.1093/nar/29.14.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsui T, Tanihara K, Date T. Expression of unphosphorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochemical and biophysical research communications. 2001;284:798–807. doi: 10.1006/bbrc.2001.5039. [DOI] [PubMed] [Google Scholar]
- 34.Lippa B, Pan G, Corbett M, Li C, Kauffman GS, Pandit J, Robinson S, Wei L, Kozina E, Marr ES, Borzillo G, Knauth E, Barbacci-Tobin EG, Vincent P, Troutman M, Baker D, Rajamohan F, Kakar S, Clark T, Morris J. Synthesis and structure based optimization of novel Akt inhibitors. Bioorganic & medicinal chemistry letters. 2008;18:3359–3363. doi: 10.1016/j.bmcl.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 35.Day PJ, Cleasby A, Tickle IJ, O’Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C, Jhoti H. Crystal structure of human CDK4 in complex with a D-type cyclin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4166–4170. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic acids research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic acids research. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White A, Pargellis CA, Studts JM, Werneburg BG, Farmer BT., 2nd Molecular basis of MAPK-activated protein kinase 2:p38 assembly. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6353–6358. doi: 10.1073/pnas.0701679104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. Journal of computational chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of computational chemistry. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 41.Wu G, Robertson DH, Brooks CL, 3rd, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm. Journal of computational chemistry. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 42.Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. Journal of medicinal chemistry. 2004;47:45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- 43.Richieri GV, Kleinfeld AM. Unbound free fatty acid levels in human serum. Journal of lipid research. 1995;36:229–240. [PubMed] [Google Scholar]
- 44.Elmadhoun BM, Wang GQ, Templeton JF, Burczynski FJ. Binding of [3H]palmitate to BSA. The American journal of physiology. 1998;275:G638–644. doi: 10.1152/ajpgi.1998.275.4.G638. [DOI] [PubMed] [Google Scholar]
- 45.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wittayanarakul K, Hannongbua S, Feig M. Accurate prediction of protonation state as a prerequisite for reliable MM-PB(GB)SA binding free energy calculations of HIV-1 protease inhibitors. Journal of computational chemistry. 2008;29:673–685. doi: 10.1002/jcc.20821. [DOI] [PubMed] [Google Scholar]
- 47.Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45:9074–9084. doi: 10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thumser AE, Storch J. Characterization of a BODIPY-labeled fluorescent fatty acid analogue. Binding to fatty acid-binding proteins, intracellular localization, and metabolism. Molecular and cellular biochemistry. 2007;299:67–73. doi: 10.1007/s11010-005-9041-2. [DOI] [PubMed] [Google Scholar]
- 49.Rioux V, Lemarchal P, Legrand P. Myristic acid, unlike palmitic acid, is rapidly metabolized in cultured rat hepatocytes. The Journal of nutritional biochemistry. 2000;11:198–207. doi: 10.1016/s0955-2863(00)00065-6. [DOI] [PubMed] [Google Scholar]
- 50.Takai Y, Kishimoto A, Iwasa Y, Kawahara Y, Mori T, Nishizuka Y. Calcium-dependent activation of a multifunctional protein kinase by membrane phospholipids. The Journal of biological chemistry. 1979;254:3692–3695. [PubMed] [Google Scholar]
- 51.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. The Biochemical journal. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS biology. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diskin R, Engelberg D, Livnah O. A novel lipid binding site formed by the MAP kinase insert in p38 alpha. Journal of molecular biology. 2008;375:70–79. doi: 10.1016/j.jmb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Barber GN. The dsRNA-dependent protein kinase, PKR and cell death. Cell death and differentiation. 2005;12:563–570. doi: 10.1038/sj.cdd.4401643. [DOI] [PubMed] [Google Scholar]
- 55.Knight JD, Qian B, Baker D, Kothary R. Conservation, variability and the modeling of active protien kinases. plos one. 2007;2:e982. doi: 10.1371/journal.pone.0000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.