Abstract
This study was designed to control plant fertility by cell lethal gene Barnase expressing at specific developmental stage and in specific tissue of male organ under the control of Cre/loxP system, for heterosis breeding, producing hybrid seed of eggplant. The Barnase-coding region was flanked by loxP recognition sites for Cre-recombinase. The eggplant inbred/pure line (‘E-38') was transformed with Cre gene and the inbred/pure line (‘E-8') was transformed with the Barnase gene situated between loxp. The experiments were done separately, by means of Agrobacterium co-culture. Four T0 -plants with the Barnase gene were obtained, all proved to be male-sterile and incapable of producing viable pollen. Flowers stamens were shorter, but the vegetative phenotype was similar to wild-type. Five T 0 -plants with the Cre gene developed well, blossomed out and set fruit normally. The crossing of male-sterile Barnase-plants with Cre expression transgenic eggplants resulted in site-specific excision with the male-sterile plants producing normal fruits. With the Barnase was excised, pollen fertility was fully restored in the hybrids. The phenotype of these restored plants was the same as that of the wild-type. Thus, the Barnase and Cre genes were capable of stable inheritance and expression in progenies of transgenic plants.
Keywords: eggplant, male sterility, Barnase gene, Cre gene, Cre/loxP system
Introduction
Eggplant (Solanum melongena L.) is one of the most popular vegetables in Asia and the Mediterranean basin. In these areas, hybrid varieties have been widely grown for many years, because heterosis has significantly enhanced productivity, as well as disease and stress resistance. To obtain hybrid seeds, two inbred lines should be mutually crossed. This procedure, however, is extremely time-consuming and labor-intensive. In contrast, the utilization of male sterile line is a more efficient way. For this, a suitable restorer system in the male parent is indispensable for acquiring seed-sets, as seeds from F1 hybrids are much sought after for producing economically feasible products. To date, several cytoplasmic male sterility (CMS) systems have been extensively studied in eggplant (Fang et al., 1985; Isshiki and Kawajiri, 2002). However, these could not be successfully applied to the production of hybrid seed and heterosis breeding due to several limitations, such as the instability of male sterility and the absence of agronomically suitable CMS/restorer system.
Genetic engineering offers the opportunity to introduce nuclear male sterility (NMS) into a wide range of plant species. Various pollination-control mechanisms are based on the genetic engineering of nuclear male sterility and its restoration had been reported and had emerged as tangible options for development of male sterile/restorer lines (Mariani et al., 1990; Rosellini et al., 2001; Jagannath et al., 2001, 2002; Bayer and Hess, 2005). Several strategies have been reported as producing NMS-plants in term of blocking pollen development by tissue-specific transgene expression(Mariani et al., 1990; Cho et al., 2001; Burgess et al., 2002),or altering specific metabolite levels in pollen development, such as sugars (Goetz et al., 2001), jasmonic acid (McConn and Browse, 1996; Stintzi and Browse, 2000) and flavonols (Fischer et al., 1997; Mayer et al., 2001).MS line sterility can be attributed to a newly introduced gene, Barnase, which encodes a special enzyme capable of cleaving RNA molecules in the cells, thereby leading to cell death. This ribonuclease is derived from Bacillusamyloliquefaciens (Hartley, 1988). To ensure that only male-flower parts are affected, Barnase should be linked to a special promoter activating the gene only in specific cells responsible for the development of the male flower. As a result, either no pollen or no viable pollen is produced. This strategy had been successfully applied in obtaining male-sterile plants (Denis et al., 1993; Burgess et al., 2002; Luo et al., 2005). To date, most researchers have concentrated on creating RF lines containing a gene for an active substance (Barstar), which neutralizes Barnase (Jagannath et al., 2001, 2002; Bisht et al., 2004, 2007), and most crosses of Barnase x Barstar were still male sterile and weak expression of the transgene in vegetative tissues led to yield reduction in Brassica juncea (Jagannath et al., 2002).
Cre/loxP is a site-specific recombination system from phage P1, which was introduced in the 1980s (Sternberg and Hamilton, 1981;Sauer and Henderson, 1988). It is based on the ability of the P1 bacteriophage cyclization recombination (Cre) recombinase gene to affect recombination between pairs of loxp sites, if the lox sites flank a DNA segment in a cis arrangement and are oriented in the same direction Cre recombinase mediates excision or circularization of the very segment (Dale and Ow, 1990, 1991; Russell et al., 1992).This principle has been utilized to develop different technologies, including marker gene deletion(Gleave et al., 1999; Hoa et al., 2002; Wang et al., 2005) and transgene integration (Srivastava and Ow, 2002). This system can also be used in hybrid breeding programs. Bayer and Hess (2005) succeeded in restoring male fertility by the removal of Barnase via Cre/loxP site-specific recombination in model-plant tobacco.
In this study, a novel method was designed to create an MS line with Barnase and a restore line with Cre/loxP thereby substituting Barstar. Eggplants were transformed with either Cre or Barnase under the control of the tapetum-specific promoter TA29 from tobacco flanked by two identical orientation loxp sites, respectively. The latter(with Barnase) should be male sterile, since tapetum is essential for pollen formation. When crossed, the former (with Cre) could cause the latter to lose the Barnase gene in tapetum, whereby F1 male viability could be restored, with the subsequent use of the F1 hybrid seeds in producing eggplants. In this work we demonstrate tapetum-specific expression of a Barnase transgene causing pollen sterility, and the restoration of male fertility by the same transgene via site-specific recombination using Cre/loxP.
Materials and Methods
Agrobacterium tumefaciens strain and plasmid
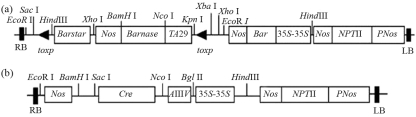
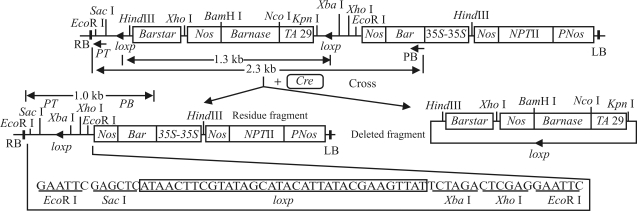
The expression vector of pCABARTABn and pBINPLUSCre (Figure1) were constructed and transferred into Agrobacterium tumefaciens strain EHA105 by freeze-thaw method (Song et al., 2004). The pCABARTABn vector contains the Barnase gene under the control of the tapetum-specific promoter TA29 from tobacco, flanked by two identical orientation loxp sites independently, and the Bar gene [conferring phosphinothricin (PPT)-resistance] driven by the CaMV35S promoter from pCABar and the nptII gene, both of which serve as plant selection markers.The Barstar gene itself is promoterless. The pBINPLUSCre vector contains Cre driven by a CaMV35S promoter with a 5'- untranslated leader sequence from alfalfa mosaic virus RNA4 (designated as CaMV 35S/AMV an enhancer element from pBI525) and the nptII gene.
Figure 1.
Sketch map of plant expression vector of Barnase and Cre gene. a: pCABARTABn. b: pBINPLUSCre.
Plant materials
Seeds of the two inbred/pure line eggplant varieties (Solanum melonggena L.) ‘E-8' and ‘E-38'were provided by the Vegetable Varieties and Genetic Improvement Research Center, South China Agricultural University. After surface sterilization in 70% ethanol for 50 s, and then in 1.2% sodium hypochlorite (20%v/v Clorox Ultra) for 15 min, and rinsing 3 times for 5 min with sterile water, the seeds were placed on 1/2MS medium with 30 g.L-1 of sucrose and 6.5 g.L-1 of agar. Cultures were incubated in the dark until germination (about 5 d after inoculation), and then kept under a 16 h/8 h light/dark period at 25 ± 0.5 °C. The hypocotyls of 9~11-day-old seedling were excised and used as explants.
Eggplant transformation and plant regeneration
‘E-8'was transformed with the Barnase gene, and ‘E-38' with the Cre gene. The explants were dipped for 10~15 min into Agrobacterium tumefaciens solution of an OD600 of 0.5.They were then pre-conditioned on a differentiation medium [MS+6-BA 2.0 mg.L-1 + IAA 0.1 mg.L-1 +ZT 2.0 mg.L-1 + sucrose (30 g.L-1) + agar (6.5 g.L-1), pH 5.8] for 2 days, and co-cultured for 4~5 days on differentiation medium. Subsequently, the hypocotyl explants were washed with MS liquid medium containing 500 mg.L-1cefotaxime, blotted dry and transferred to selection pressure medium [MS+ 6-BA (2.0 mg.L-1) + IAA (0.1 mg.L-1) + ZT (2.0 mg.L-1) + PPT(15 mg.L-1) + Cb (500 mg.L-1) + 30 g.L-1 sucrose + 6.5 g.L-1 agar, pH 5.8] in the case of Barnase gene transformation, and another [MS+6-BA 2.0 mg.L-1 + IAA 0.1 mg.L-1 + ZT 2.0 mg.L-1 + Km (65 mg.L-1) + Cb (500 mg.L-1) + sucrose (30 g.L-1) + agar (6.5 g.L-1), pH 5.8] in the case of Cre gene transformation. PPT-resistant and Km-resistant adventitious buds were induced after 25 days. These were cultured on differentiation medium for a 2~3 days subculture, to be then excised and transferred to a rooting medium (MS+IAA 0.5 mg.L-1 + Cb 200 mg.L-1). About four weeks later, the adventitious buds had already rooted. The plantlets were then transplanted to soil for further analysis.
Transgenic plants were identified by Southern blotting and Northern blotting analysis.
DNA and RNA extraction
Total genomic DNA was extracted employing CTAB methods (Doyle and Dovle, 1990).
Total RNA was extracted by means of an RNA extraction Trizol Kit (Takara.com).
Polymerase chain reaction
Total genomic DNA was extracted from the leaves of eggplant (transformed and untransformed) shoots using the CTAB method, and served for Southern blotting. Two pairs of primers for Cre and Barnase gene were designed from their coding region respective. The forward and reverse primer sequences for Barnase were pBn-1:5'-GCAGAATTCACCA TGGCACAGGTTATCAAC-3' and pBn-2:5'-CCCCTCGGATCCGTTATCTGATCTTT GTA-3' respectively; those for Cre were pCre-1:5'-GACCATGGCTCCCAAGAAGAAGAGAAAGG TAATGTCCAATTTACTGACCG-3' and pCre-2:5'-CCCCTCGGATCCGTTATCTGATCTTT GTA-3'; those for Bar were pBar1:5'-CCGCTCGAGTCTACCATGAGC CCAGAAC-3' and pBar2 5'-CCGCTCGAGATCAGAT CTCGGTGACG GG-3', and finally those for the deleted fragment were pBar1and pT: 5'-AAGGCGATTAAGTTG GGTAACGCC AG-3'.
The PCR reactions were carried out in a 25 μL volume containing 2.5 μL of a 10 x PCR buffer (Takara), 0.5 μL of 10 mmol.L-1dNTPs (Takara), 0.5 μL of two 20 μM primer, 0.5 μL of 5U/ μL Taq polymerase (Takara), 20 μL of distilled H2O and 1 μL (20-50 ng) of a DNA template. Reactions were carried out in a Peltier thermal cycler (Bio-Rad, USA) as follows: for Cre, one cycle of 5 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 1 min at 55 °C, 2 min at 72 °C, and one final cycle of 10 min at 72 °C; for Barnase, one cycle of 3 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 1 min at 52 °C, 2 min at 72 °C, and one final cycle of 10 min at 72 °C; for Bar and the deleted fragment, one cycle of 5 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 56 °C, 2 min at 72 °C, and one final cycle of 10 min at 72 °C. The products were stored at 4 °C, and separated in 1.2% agarose with an electrophoresis systems (Bio-Rad,sub-cell model 192, USA). Bands were recorded using the Chemi Doc system (Bio-Rad, USA).
Southern blot analysis
Genomic DNA of the transformants (T0) and untransformed control plants were digested with EcoR [The digestion reaction was carried out in a 10 μL volume containing 2 μg of DNA (about2 μL), 2 μL of a 10 x buffer, 2 μL of EcoR and 4 μL of ddH2O at 37 °C for 10 to12 h]. Southern blot analysis was carried out using 15 μg of genomic DNA. The digested genomic DNA was separated on a 0.8% agarose gel and transferred to positively charged nylon membranes according to manufacturer's instructions (Boehringer Mannheim Com). The Barnase and Cre gene fragment labeled with (DIG) dCTP by using random labeling (Boehringer Mannheim Com), were used as probes. Hybridization was carried out by DNA labeling and a Detection Kit (Boehringer Mannheim Com.), the nylon membranes being washed with 2 x SSC, 0.1% SDS; 1 x SSC, 0.1% SDS; 0.5 x SSC and 0.1% SDS at 65 °C for 15 min respectively.
Northern blot analysis
Hybridization was carried out by DNA labeling and a Detection Kit (Boehringer Mannheim Com.). Approximately 12 μg of the total RNA was run on a 1.2% denaturing agarose gel containing formaldehyde and then transferred onto positively charged nylon membrane (Boehringer Mannheim Com). The DIG-labeled Barnase and Cre gene fragment were used as probes.
Floral organ morphology and pollen viability testing
The difference between transgenic and non-transgenic plants, especially as regards floral organ morphology, was carefully checked. Pollen grains, collected between 8 and 9 a.m. were stored in a Petri dishes, which were then placed in a refrigerator. On initiating the experiment, one drop of the freshly prepared medium was placed on a cover slip, and small quantities of pollen grains dispersed therein. The cover slip was then mounted on a cavity slide and the margins of the cover slip were smeared with wax. After ten hours the preparations were microscopically examined. More than a hundred pollen grains per germination were observed and the percentage of germination was calculated on the basis of these observations. The culture medium was 1.0% sucrose supplemented with 1.0% boric acid, 5 mg.L-1 GA3 and 1.0% agar. The hanging drop method was then applied in experiments, with exception of those where sugar-agar constituted the culture medium base.
Pollen vigor was tested by the TTC staining method, whereby pollen was placed into a 1% TTC solution for 30 min for microscopic examination.
Pollination
Unopened flower buds about 3-4 cm in length were sliced open lengthwise and emasculated with forceps. Mature pollen from the donor plant was transferred by brushing anthers onto the stigmas of emasculated plants. Pollinated flowers were labeled and bagged with small plastic bags to prevent uncontrolled cross-pollination. Self-pollination was achieved by covering intact flowers with small plastic bags prior to opening.
Analysis of seed germination frequency and segregation ratios
Backcrossed T1 seeds from individual T0 plants were surface-sterilized and germinated on non-selective media according to the procedures described above. Seed germination frequency for each progeny plant was calculated as the percentage of seeds germinated versus seeds inoculated. The apices of seedlings were excised and placed onto an MS medium with the appropriate selective pressure (15 mg.L-1 PPT). Rooting and survival of plantlets were recorded. Segregation ratios were calculated in terms of resistance/sensitivity(R/s) to the selective agent and subsequently correlated with the male sterility/fertility phenotype when the T1 plants came to flowering. Segregation data for each event was subjected to statistical analysis (χ 2 test at 95% confidence limit) to determine fitness.
Analysis of heterosis of F1 crossing of transgenic male sterile plants with Cre-expressing transgenic plants
The F1 crosses of transgenic male-sterile plants with Cre-expressing transgenic plants, ‘E-8', ‘E-38'and F1 (‘E-8' x ‘E-38') were transplanted to greenhouse. Plants growth, fruit, seeds and yield were all analyzed.
Results
Regeneration and detection of transgenic plants
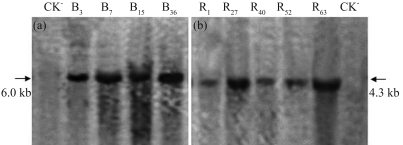
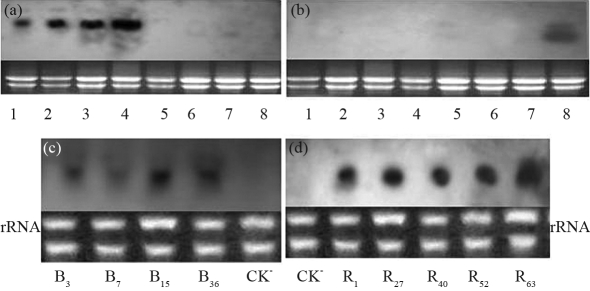
Most of the explants were incapable of differentiating, and turned yellow, although some Kmr and PPTr adventitious bud rosettes were differentiated from the cut end of hypocotyl explants after 25 days of culture on a selective medium. Five Kan-resistant buds and four PPT- resistant buds were finally obtained at last, the Barnase transgenic plants (B3, B7,B15, B36) and the Cre transgenic plants (R1, R27, R40, R52, R63) being screened out by Southern blotting (Figure 2). The results demonstrated that Cre and Barnase genes had been integrated into the genome of the respective transgenic lines, the target gene being all single copy in the transgenic plants. These transgenic plants were propagated. After shoot rooting had been induced on the rooting medium, these were then transplanted into vermiculite. The transgenic and nontransgenic plants were detected by Northern blotting. The transgenic plant R63 and B3 were chosen randomly for analyzing Cre and Barnase gene expression in the different parts of the plant. The results showed that the Cre gene was expressed in the flower, stem, leaf and root of the transgenic plant R63, but not in those of the non-transgenic plant (Figure 3a), whereas Barnase gene was only expressed in inflorescence of the transgenic plant B3, but not in the root, stem and leaf of B3, and with no expression whatsoever in the non-transgenic plants (Figure 3b). In order to analyze the expression level of Cre and Barnse gene among different transgenic plants, all the four transgenic lines with the Barnase gene and five transgenic lines with the Cre gene were identified by Northern blotting, the results demonstrating that expression levels were different in various transgenic plants (Figure 3c, d). These results showed that the target genes were expressed in transgenic plants, but not in non-transgenic plants, although Cre gene expression was established, and expression the Barnase gene occurred only in the inflorescence of the transgenic plants.
Figure 2.
Southern-blot analysis of transgenic plants. (a) Southern blot of Barnase transgenic plants; CK_ showing a non-transgenic plant (E-8); B3, B7, B15, B36 showing Barnase transgenic plants; (b) Southern blot of Cre transgenic plants; CK- showing a non-transgenic plant (E-38); R1, R27, R40, R52, R63 showing Cre transgenic plants.
Figure 3.
Northern blotting analysis of transgenic plants. (a) Cre gene expression in different parts of transgenic and non-transgenic plants; lanes 1-4 showing the flower, stem, leaf and root of a transgenic plant (R63); lanes 5-8 showing the flower, stem, leaf and root of a non-transgenic plant(E-38). (b) Barnase gene expression in different parts of transgenic and non-transgenic plants; lanes 1-4 showing the root, stem, leaf and flower of a non-transgenic plant (E-8); lanes 5-8 showing the root, stem, leaf and flower of a transgenic plant (B3); (c) Barnase gene expression in the flower of four different transgenic plants (B3, B7, B15 and B36); CK-: a non-transgenic plant (E-8); (d) Cre gene expression in the leaf of five different transgenic plants (R1, R27, R40, R52 and R63); CK-: a non-transgenic plant (E-38).
Floral organ morphology and pollen vigor of transgenic plants
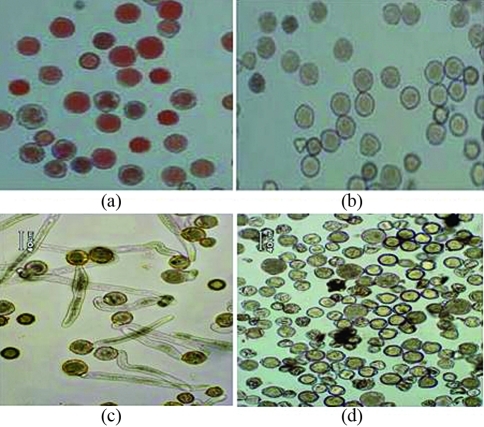
Compared with CK plants, the transgenic plants with Barnase gene showed no morphological differences in leaves, shape or height except for the inflorescences in greenhouse culture. Transgenic plants either produced no pollens or only a small amount of non-viable pollens incapable of germinating, thereby indicating male- sterility. In contrast to the red pollen of the wild-type, pollen grains from male-sterile plants displayed a grayish color by TTC testing (Figure 4). The staminal length of male-sterile plant was shorter than that of wild-type , while the anthers of the male-sterile plants were shrunken (Figure 5).
Figure 4.
Testing the viability of pollen from a Barnase transgenic plant(B3) and a non-transgenic plant. (a) TTC testing of pollen from a nontransgenic plant (E-8). (b) TTC testing of pollen from a Barnase gene transgenic plant (B3). (c) showing the germination of pollen from a nontransgenic plant (E-8). (d) showing the germination of pollen from a Barnase gene transgenic plant (B3).
Figure 5.
Stigmata, anthers and flowers of a Barnase transgenic plant and a non-transgenic plant. Panel1 showing the flower of a transgenic MS plant (B7), 2 and 4 the flower of a transgenic MS plant (B3), and 3 and 5 showing the flower of a non-transgenic plant (E-8).
Compared with the wild-type, the phenotypes of all the five transgenic T0-plants with the Cre gene showed normal phenotypes and were self-pollinated so as to produce homozygous progenies.
Crossing of male-sterile transgenic with non-transgenic plants
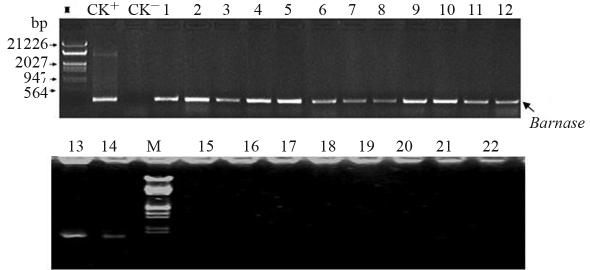
Four male-sterile T0 plants (B3, B7, B15 and B36), when crossed with wild-type plants (‘E-8',CK) were capable of producing normal fruit with normal seeds. In PPTr and PPTs plants, the ratio in the progeny showed 1:1, after spraying greenhouse seedlings with 1~2 leaves, two or three times with 15 mg.L-1 PPT (Table 1).The PPTr plants in the progenies showed male sterility and could not produce fruits after self-pollination. About 50% plants in progenies were fertile. The progeny plants were identified by PCR using Barnase gene primers (Figure 6), and the results revealing that all the male-sterile plants contained the Barnase gene, whereas fertile ones did not. From these results,it is possible to deduce that the Barnase gene could be stably inherited and expressed in the progeny of transgenic plants. At the same time, if transgenic male sterile plants are used to produce F1 seed in the future, 100% male-sterile lines may be obtained by spraying PPT to eliminate fertile plants.
Table 1.
Segregation profiles among T1 progenies of transgenic plants crossed with nontransgenic plants E-8 .
| Transgenic line | Number of seeding on selection | Number of PPT-resistant plants | Number of PPT-susceptible plant | χ20.05 = 3.84 |
| B3-1 | 90 | 47 | 43 | 0.1 |
| B7-1 | 110 | 54 | 56 | 0.01 |
| B15-1 | 95 | 49 | 46 | 0.04 |
| B36-1 | 100 | 52 | 48 | 0.09 |
Figure 6.
PCR detection of Barnase-related MS and male-fertile plants in the progeny of a Barnase transgenic plant (B3) crossed with a non-transgenic (E-8). M: Marker; lane 1 showing positive CK (pCABARTABn); lanes 2-14 showing male-sterile plants among progenies of B3 x E-8; lanes 15-22 showing male fertile plants among progenies of B3 x E-8.
When T0 transgenic plants with Cre gene (R1, R27, R40, R52, R63) were self-pollinated, they could produce normal fruit with normal seeds. The progenies of R63 from self-pollination were randomly chosen for analyzing Cre gene inheritance. The result showed that 99 plants were Kan-resistant, 36 plants were Kan-susceptible among 135 T1 plants of R63 with a R:S ration of 3:1(χ2 = 0.12 < χ20.05 = 3.84).
Crossing of male-sterile with Cre-expressing plants restored fertility in hybrids
The Barnase gene was flanked by loxP sites in the same orientation (Figure 1). Thus, Cre -mediated recombination should lead to Barnase excision, whereby the crossing of male-sterile plants with Cre-expressing plants would give rise to fertile progeny. Four Barnase-expression lines (B3, B7, B15 and B36) were crossed with R63, and B7 were crossed with R27, 60 plants from each combination being transferred to the field. All the F1-plants were fully self-fertile, thus capable of producing normal fruits (Figure 7), besides displaying the same phenotype and flower morphology as wild F1-plants (‘E-8' x ‘E-38',nontransgenic plant). F1-plants of two combinations (B3 x R63 and B7 x R27) used to analyze the heterosis, showed this phenomenon in plant height, fruit length and weight, when compare to ‘E-8' and ‘E-38' (Table 2), thereby indicating that transgenic male-sterile plants and Cre-expressing plants may be applied to future eggplant-heterosis breeding.
Figure 7.

Fruit in F1 of a cross between a male-sterility plant (B3) and a Cre-expressing plant (R63).
Table 2.
Comparison of heterosis in hybrid combination with that of other varieties.
| Material | Plant height (cm) | Per fruit weight (g) | Fruit length (cm) | Total Yield (kg) |
| E-8 (CK1) | 86.3 ± 4.29b | 195.6 ± 5.42b | 23.7 ± 1.18b | 13.6 ± 1.23b |
| E-38 (CK2) | 90.7 ± 3.16b | 207.4 ± 4.33b | 25.3 ± 1.65b | 15.7 ± 1.35b |
| E-8 x E-38(CK3) | 120.8 ± 3.13a | 245.5 ± 4.21a | 30.8 ± 1.35a | 25.4 ± 1.62a |
| B3 x R63 | 123.5 ± 3.02a | 243.6 ± 3.87a | 30.5 ± 1.52a | 25.7 ± 1.27a |
| B7 x R27 | 122.6 ± 3.34a | 242.8 ± 4.05a | 30.3 ± 1.63a | 24.8 ± 2.06a |
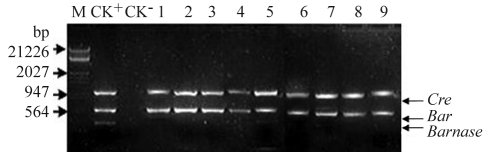
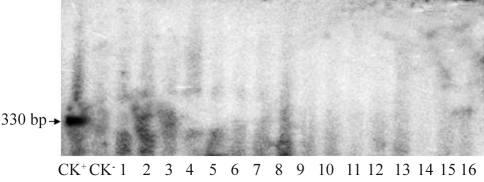
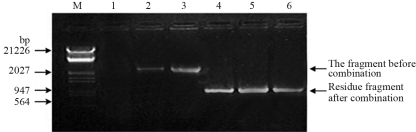
Thirty F1-plants surviving kanamycin and PPT selection from each of five crosses were randomly chosen for further analysis. Barnase, Bar and Cre gene were analyzed in early flower buds by PCR. The result showed that, apart from the impossibility amplifying Barnase gene, Cre and Bar gene bands were present in the flower of F1-plants (Figure 8; Table 3). At the same time in all 30 F1-plants from B3 x R63 detected by Southern blotting using a Barnase gene fragment as probe, there were no signs of hybridization (Figure 9). The recombination residue fragment was analyzed deeply by PCR. The expected fragment should be about 2.3 kb before recombination and about 1.3 kb fragment should be deleted after recombination and a fragment of about 1.0 kb could be residual in F1 plants distributed differently between transgenic male -sterile plants and transgenic Cre plants (Figure 10). Thus the transgenic male-sterile plant (B3), three F1 plants without the Barnase gene (B3 x R63) and the pCABARTABn expression vector , were identified though PCR, the results showing that about 2.3 kb of the band had been amplified in the B3 plants and pCABARTABn (positive control), whereas only the 1.0 kb residual fragment was only amplified in F1 plant (Figure 11). PCR products and Southern blotting results confirmed the site-specific nature of recombination and the excision of the Barnase gene.
Figure 8.
PCR detection of progenies of Barnase , Bar and Cre in progenies of transgenic plant crossings. M: Markers; Ck+: showing positive CK (pCABARTABn + pBINPLUSCre); CK_ showing a non-transgenic plant (E-8 x E-38); Lanes 1-9 showing a F1 plant from crossing a transgenic male-sterile plant (B3) and a transgenic plant with Cre gene (R63).
Table 3.
PCR analysis of Barnase x Cre progenies.
| cross | Sequence analysis by PCR | Barnase parent | Cre parent | 1 | 2 | F1 | Plant 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| B3 x R63 | Barnase | ++ | - | - | - | - | - | - | - | - | - | - | - |
| Cre | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Bar | ++ | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| B7 x R63 | Barnase | ++ | - | - | - | - | - | - | - | - | - | - | - |
| Cre | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Bar | + | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| B15 x R63 | Barnase | ++ | - | - | - | - | - | - | - | - | - | - | - |
| Cre | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Bar | + | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| B36 x R63 | Barnase | ++ | - | - | - | - | - | - | - | - | - | - | - |
| Cre | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Bar | + | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| B7 x R27 | Barnase | ++ | - | - | - | - | - | - | - | - | - | - | - |
| Cre | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Bar | + | - | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| CK (E-8 x E-38) | Barnase | - | - | - | - | - | - | - | - | - | - | - | - |
| Cre | - | - | - | - | - | - | - | - | - | - | - | - | |
| Bar | - | - | - | - | - | - | - | - | - | - | - | - | |
Ck; notransgenic plant; + showing presence; - showing absence.
Figure 9.
Southern blotting for Barnase gene detection in F1 of a crossing between a male-sterile plant (B3) and a Cre-expressing plant (R63). Lane Ck+ showing positive CK (Barnase gene PCR product); lane CK_ showing a non-transgenic plant (E-8 x E-38); lanes 1-16 showing F1 progeny from the crossing of a transgenic male-sterile plant (B3) and a Cre transgenic plant (R63).
Figure 10.
Sequence structure and site characteristics before and after Cre-mediated recombination.
Figure 11.
PCR analysis of the change in fragment size after recombination. M: λ DNA/EcoRV +Hind marker; lanes lane 1 showing negative control (F1 of E-8 x E-38, non-transgenic plant); lane2 showing pCABARTABn; lane 3 showing a transgenic male-sterile plant (B3); lanes 4-6 showing sterile gene-deleted F1 plants (B3 x R63).
Discussion
Up to now there have been many reports on the male sterility of eggplant (Jasmin, 1954; Fang et al., 1985; Phatak and Jaworski, 1989; Phatak et al., 1991; Isshiki and Kawajiri, 2002; Tian et al., 2004). Even so, there is the lack of an appropriate male-sterile line for heterosis breeding and the production of F1 seeds. Genetic engineering may offer a new opportunity to introduce nuclear male sterility (NMS) to eggplant.
Previous studies of Cre-mediated recombination in plants focused on the removal of selectable marker genes or the production of transgenic plants for single copies of marker genes (Dale and Ow, 1991; Bayley et al., 1992; Srivastava et al., 1999; Hoa et al., 2002). Tapetum- specific expression of a Barnase transgene was used to introduce male sterility ,as was shown earlier by Mariani et al. (1990). In the this study, we demonstrated that the use of the Cre/loxP- system is another trait of considerable impact in plant breeding, restoring transgenic pollen fertility. The transgenic T0-plants showed the expected male-sterile phenotype. Reciprocal cross between transgenic and non-transgenic, T0-plants clearly demonstrated male-sterility without affecting female fertility, and stable inheritance of the phenotype. Co-segregation of Barnase and Bar gene provided an easy way to obtain male-sterile plants by crossing with wild-type plants via PPT selection in progeny. A transgenic line with constitutive Cre expression served as fertility restorer. The absence of phenotypic differences in Cre-homozygous plants when compared with the wild-type, supports earlier reports to this effect in transgenic tobacco (Odell and Russel, 1994; Ow and Medburry, 1995). In contrast it was observed that high-level expression of Cre may affect growth in transgenic tomato, petunia, and tobacco (Que et al., 1998; Coppoolse et al., 2003).The Barnase-coding region was flanked by a loxP site Cre-mediated site-specific recombination restored fertility in the hybrids by excision of Barnase and all progenies showed male fertility, thus demonstrating the high efficiency of the system. The one hundred percent efficiency of Cre in the excision of single-copy marker genes has also been reported by Gleave et al. (1999). although in other studies recombination efficiencies were no than 45% (Bayley et al. 1992) or about 50% (Dale and Ow, 1991).
Luo et al. (2000) used the FLP/frt site-specific recombination system for restoring fertility in Arabidopsisthaliana transgenic plants. They observed 100% recombination efficiency in parents with a single copy of the frt-flanked transgene per haploid genome, thereby infering that this occurs irrespective of the recombinase used. Luo et al. (2000) postulated that alterations in recombination sites, such as DNA methylation, possibly hinder the binding of the recombinase protein to its target site. In other systems using transgenes, such as Barnase/Barstar, crop loss due to partial sterility cannot be ruled out. In Brassica juncea, most of the Barnase-Barstar crosses were still male-sterile and weak expression of the transgene in vegetative tissues resulted in yield reduction (Jagannath et al., 2002). In our system we detected no phenotypic effects other than those implied in fully restoring pollen fertility. Thus, the use of the Cre/loxP site-specific recombination system, as demonstrated with model plant tobacco (Bayer and Hess, 2005), appears to be the most promising in hybrid breeding programs with agronomically important crop plants.
Footnotes
Associate Editor: Márcio de Castro Silva Filho
References
- Bayer M., Hess D. Restoring full pollen fertility in transgenic male-sterile tobacco (Nicotiana tabacum L. ) by Cre-mediated site-specific recombination. Mol Breed. 2005;15:193–203. [Google Scholar]
- Bayley C.C., Morgan M., Dale E.C., Ow D.W. Exchange of gene activity in transgenic plants catalyzed by the Cre-lox site-specific recombination system. Plant Mol Biol. 1992;18:353–361. doi: 10.1007/BF00034962. [DOI] [PubMed] [Google Scholar]
- Bisht N.C., Jagannath A., Gupta V., Burma P.K., Pental D. A two gene-two promoter system for enhanced expression of a restorer gene (barstar) and development of improved fertility restorer lines for hybrid seed production in crop plants. Mol Breed. 2004;14:129–144. [Google Scholar]
- Bisht N.C., Jagannath A., Burma P.K., Pradhan A.K., Pental D. Retransformation of a male sterile barnase line with the barstar gene as an efficient alternative method to identify male sterile-restorer combinations for heterosis breeding. Plant Cell Rep. 2007;26:727–733. doi: 10.1007/s00299-006-0274-7. [DOI] [PubMed] [Google Scholar]
- Burgess D.G., Ralston E.J., Hanson W.G., Heckert M., Ho M., Jenq T., Palys J.M., Tang K., Gutterson N. A novel, two-component system for cell lethality and its use in engineering nuclear male-sterility in plants. Plant J. 2002;31:113–125. doi: 10.1046/j.1365-313x.2002.01330.x. [DOI] [PubMed] [Google Scholar]
- Cho H.J., Kim S., Kim M., Kim B.D. Production of transgenic male sterile tobacco plants with the cDNA encoding a ribosome inactivating protein in Dianthus sinensis L. Mol Cell. 2001;11:326–333. [PubMed] [Google Scholar]
- Coppoolse E.R., de Vroomen M.J., Roelofs D., Smit J., van Gennip F., Hersmus B.J.M., Nijkamp H.J.J., van Haren M.J.J. Cre recombinase expression can result in phenotypic aberrations in plants. Plant Mol Biol. 2003;51:263–279. doi: 10.1023/a:1021174726070. [DOI] [PubMed] [Google Scholar]
- Dale E.C., Ow D.W. Intra- and inter site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene. 1990;91:79–85. doi: 10.1016/0378-1119(90)90165-n. [DOI] [PubMed] [Google Scholar]
- Dale E.C., Ow D.W. Gene transfer with subsequent removal of the selection gene from the host genome. Proc Natl Acad Sci USA. 1991;88:10558–10562. doi: 10.1073/pnas.88.23.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis M., Delourme R., Gourret J.P., Mariani C., Renard M. Expression of engineered nuclear male sterility in Brassica napus (genetics, morphology, cytology, and sensitivity to temperature) Plant Physiol. 1993;101:1295–1304. doi: 10.1104/pp.101.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:12–14. [Google Scholar]
- Fang M.R., Mao R.C., Xie W.H. Breeding cytoplasmically male sterile lines of eggplant. Acta Hort Sinica. 1985;12:261–266. [Google Scholar]
- Fischer R., Budde I., Hain R. Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant J. 1997;11:489–498. [Google Scholar]
- Gleave A.P., Mitra D.S., Mudge S.R., Morris B.A.M. Selectable marker-free transgenic plants without sexual crossing: Transient expression of cre recombinase and use of a conditional lethal dominant gene. Plant Mol Biol. 1999;40:223–235. doi: 10.1023/a:1006184221051. [DOI] [PubMed] [Google Scholar]
- Goetz M., Godt D.E., Guivarc'h A., Kahmann U., Chriqui D., Roitsch T. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley R.W. Barnase and barstar, expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol. 1988;202:913–915. doi: 10.1016/0022-2836(88)90568-2. [DOI] [PubMed] [Google Scholar]
- Hoa T.T.C., Bong B.B., Huq E., Hodges T.K. Cre/lox site-specific recombination controls the excision of a transgene from the rice genome. Theor Appl Genet. 2002;104:518–525. doi: 10.1007/s001220100748. [DOI] [PubMed] [Google Scholar]
- Isshiki S., Kawajiri N. Effect of cytoplasm of Solanum violaceum Ort. on fertility of eggplants (Solanum melongena L.) Sci Hort. 2002;93:9–18. [Google Scholar]
- Jagannath A., Bandyopadhyay P., Arumugam N., Gupta V., Burma P.K., Pental D. The use of a spacer DNA fragment insulates the tissue-specific expression of a cytotoxic gene (barnase) and allows high-frequency generation of transgenic male sterile lines in Brassica juncea L. Mol Breed. 2001;8:11–23. [Google Scholar]
- Jagannath A., Arumugam N., Gupta V., Pradhan A.K., Burma P.K., Pental D. Development of transgenic barstar lines and identification of a male sterile (barnase)/restorer (barstar) combination for heterosis breeding in Indian oilseed mustard (Brassica juncea) Curr Sci. 2002;82:46–52. [Google Scholar]
- Jasmin J.J. Male sterility in Solanum melongena L. : Preliminary report on a functional type of male seterility in eggplants. Proc Am Soc Hort Sci. 1954;63:443. [Google Scholar]
- Luo H., Lyznik L.A., Gidoni D., Hodges T.K. FLP-mediated recombination for use in hybrid plant production. Plant J. 2000;3:423–430. doi: 10.1046/j.1365-313x.2000.00782.x. [DOI] [PubMed] [Google Scholar]
- Luo H., Kausch A.P., Hu Q., Nelson K., Wipff J.K., Fricker C.C.R., Owen T.P., Moreno M.A., Lee J.-.Y., Hodges T.K. Controlling transgene escape in GM creeping bentgrass. Mol Breed. 2005;16:185–188. [Google Scholar]
- Mariani C., De Beuckeleer M., Truettner J., Leemans J., Goldberg R.B. Induction of male sterility in plants by a chimeric ribonuclease gene. Nature. 1990;347:737–741. [Google Scholar]
- Mayer M.J., Narbad A., Parr A.J., Parker M.L., Walton N.J., Mellon F.A. Rerouting the plant phenyl propanoid pathway by expression of a novel bacterial enoyl-coa hydratase/lyase enzyme function. Plant Cell. 2001;13:1669–1682. doi: 10.1105/TPC.010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Browse J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an arabidopsis mutant. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell J.T., Russell S.H. Use of site-specific recombination systems in plants. In: Homologous Recombination and Gene Silencing in Plants. The Netherlands: Kluwer Academic Publishers; 1994. pp. 219–270. [Google Scholar]
- Ow D.W., Medburry S.L. Genome manipulation through site-specific recombination. Crit Rev Plant Sci. 1995;14:239–261. [Google Scholar]
- Phatak S.C., Jaworski C.A. UGA 1-MS male sterile eggplant germplasm. Hort Sci. 1989;24:1050. [Google Scholar]
- Phatak S.C., Liu J., Jaworski C.A., Sultanbawa A.F. Functional male sterility in eggplant: Inheritance and linkage to the purple fruit color gene. J Hered. 1991;82:81–83. [Google Scholar]
- Que Q., Wang H., Jorgensen R.A. Distinct patterns of pigmentations suppression are produced by allelic sense and antisense chalcone synthase transgenes in petunia flowers. Plant J. 1998;13:401–409. [Google Scholar]
- Rosellini D., Pezzotti M., Veronesi F. Characterization of transgenic male sterility in alfalfa. Euphytica. 2001;118:313–319. [Google Scholar]
- Russell S.H., Hoopes J.L., Odell J.T. Directed excision of a transgene from the plant genome. Mol Gen Genet. 1992;234:49–59. doi: 10.1007/BF00272344. [DOI] [PubMed] [Google Scholar]
- Sauer B., Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.Y., Ding J.G., Cao B.H., Lei J.J., Song M. Construction of the expression vectors for artifial male sterility gene and fertility restoring gene. J Southwest Agric Univ. 2004;3:249–253. [Google Scholar]
- Srivastava V., Ow D.W. Biolistic mediated site-specific integration in rice. Mol Breed. 2002;4:345–349. [Google Scholar]
- Srivastava V., Anderson O.D., Ow D.W. Single-copy transgenic wheat generated through the resolution of complex intergration patterns. Proc Natl Acad Sci USA. 1999;96:11117–11121. doi: 10.1073/pnas.96.20.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Sauer B., Hoess R. Bacteriophage P1 cre gene and its regulatory region evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- Stintzi A., Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA. 2000;19:10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S.B., Wang Y.Q., Liu F.H., Luo Z.G., Pi W., Chen Y.K., Liu J.S. Development of eggplant functional male sterile lines and its utilization; 2004. p. 91. [Google Scholar]
- Wang Y., Chen B., Hu Y., Li J., Lin Z. Inducible excision of selectable marker gene from transgenic plants by the Cre/lox site-specific recombination system. Transgenic Res. 2005;14:605–614. doi: 10.1007/s11248-005-0884-9. [DOI] [PubMed] [Google Scholar]