Abstract
Posttranslational modifications of histones are coupled in the regulation of the cellular processes involving chromatin such as transcription, replication, repair, and genome stability. Recent biochemical and genetic studies have clearly demonstrated that many aspects of chromatin, and not just posttranslational modifications of histones, provide surfaces that can interact with effectors and the modifying machineries in a context-dependent manner, all as a part of the “chromatin signaling pathway”. Here, we have reviewed recent findings on the molecular basis for the recruitment of the chromatin-modifying machineries and their diverse and varied biological outcomes.
Introduction
Although every cell within our body bears the same genetic information and the same set of genes, only a small subset of genes is transcribed in a given cell at a given time. The molecular mechanism underlying this cell/stage-specific transcriptional control has been the subject of intense study for many years. The genetic information encoded in our DNA is packaged within nucleosomal arrays forming what is referred to as chromatin. A nucleosome contains ~146 bp of DNA wrapped twice around an octamer composed of two copies of histones H3 and H4, and two copies of histones H2A and H2B. Initial structural studies using electron microscopy demonstrated that nucleosomes are found in arrays forming a series of beads on a string, with the beads being the individual nucleosomes, and the string the linker DNA (Kornberg, 1974; Kornberg and Lorch, 1999). Histone proteins contain a flexible amino-terminal tail and a characteristic histone fold, a globular domain that mediates substantial interactions between histones to form the nucleosome. High resolution X-ray crystallography demonstrated that histone N-terminal tails protrude outward from the nucleosome (Luger et al., 1997), and biochemical studies have confirmed that such histone tails are extensively, posttranslationally modified (Bhaumik et al., 2007).
Reported posttranslational modifications of histones so far include acetylation, phosphorylation, methylation, monoubiquitination, sumoylation and ADP ribosylation (Berger, 2007; Bernstein et al., 2007; Campos and Reinberg, 2009; Kouzarides, 2007; Shilatifard, 2006; Weake and Workman, 2008; Workman and Kingston, 1998). For almost every modification identified, there are also machineries involved in their removal (Bhaumik et al., 2007). Histone modifications can have a variety of functions; they can change the charge of a residue to disrupt protein-DNA, protein-protein, and nucleosome-nucleosome interactions; and they can form binding surfaces for a variety of proteins (Campos and Reinberg, 2009; Kouzarides, 2007). Histone modifications that form binding surfaces for other proteins have been implicated in several epigenetic processes. For example, HP1 can bind to the histone H3 tail methylated on lysine 9 as part of the process of stable maintenance of heterochromatic silencing. Polycomb (Pc) and Eed proteins can bind to histone H3K27-methylated tails at different stages of the cell cycle to help maintain the silencing of developmental genes during differentiation (Hansen et al., 2008; Kouzarides, 2007; Margueron et al., 2009). However, histone modifications are not always epigenetic. The addition and removal of histone acetylation and phosphorylation can be transient events that are associated with the initiation and repression of genes. For example, histone H2B monoubiquitination is added, then quickly removed during the process of gene activation (Henry et al., 2003). Histone H3K4 methylation is associated with counteracting Pc silencing of developmental genes when implemented by Trithorax, but is also found on many housekeeping genes where its function is unknown. Thus, the same modification can have both epigenetic and non-epigenetic functions.
Recent studies have demonstrated that the posttranslational modifications of histones do not represent a code and are no different than the posttranslational modifications associated with any other proteins in the cell (Lee et al., 2010; Schreiber and Bernstein, 2002; Sims and Reinberg, 2008). The posttranslational modifications of histones are part of signaling pathways, and their readout is context-dependent with the biological outcomes dictated by many variables. In this review, we analyze recent reports on the different molecular mechanisms of recruitment and biological outcomes for histone-modifying machineries. These studies demonstrate that a combination of many factors, including DNA elements, protein-protein interactions, stage and origin of cells, and posttranslational modifications on transcription factors and histones, regulate the diverse biological outcome associated within the “chromatin signaling pathway.”
The coactivator and corepressor models of recruitment
Although it was known for decades that histone acetylation was associated with actively transcribed genes, it was not known if histone acetylation was a consequence of transcription or was involved in the activation process. Purification of a histone acetyltransferase from the transcriptionally active macronucleus of the ciliated protozoan Tetrahymena thermophila identified a protein homologous to yeast Gcn5, previously described as a coactivator of transcription (Berger et al., 1992; Brownell et al., 1996; Marcus et al., 1994). Coactivators were postulated to be factors required for bridging sequence-specific binding proteins and the basal transcriptional machinery (Berger et al., 1990; Pugh and Tjian, 1990). Subsequently, other previously known coactivators were shown to be histone acetyltransferases or to participate in complexes with histone acetyltransferases, including CBP/p300, SRC-1 and ACTR (Bannister and Kouzarides, 1996; Chen et al., 1997; Ogryzko et al., 1996; Spencer et al., 1997). Both Gcn5, as part of the SAGA histone H3 acetyltransferase complex, and Esa1, as part of the NuA4 histone H4 acetyltransferase complex, were shown to be recruited by certain acidic activators (Utley et al., 1998). Importantly, another Gcn5-containing complex, the ADA complex, did not interact with these activators, demonstrating specificity in the recruitment of different histone-modifying activities by distinct activators.
Concurrent with the discovery of a histone acetyltransferase as a transcriptional activator was the discovery of a known transcriptional repressor, Rpd3, as a histone deacetylase (Taunton et al., 1996). Rpd3 and related enzymes are found to act as corepressors with sequence-specific transcriptional binding factors such as nuclear hormone receptors that can recruit SMRT/NCOR histone deacetylase complexes (Alland et al., 1997; Hassig et al., 1997; Heinzel et al., 1997; Kadosh and Struhl, 1997; Nagy et al., 1997). The same repressor can recruit different deacetylase complexes. For example, Ikaros is a critical regulator of hematopoiesis that can interact with both the SIN3A and Mi-2-NuRD deacetylase complexes to repress lineage-specific genes (Kim et al., 1999; Koipally et al., 1999). Together, these studies not only demonstrated a functional role for histone-modifying activities as key regulators of transcription, but also provided the first paradigm of how histone-modifying activities could be recruited to chromatin (Figure 1).
Figure 1. Coactivator and Corepressors are histone-modifying enzymes.
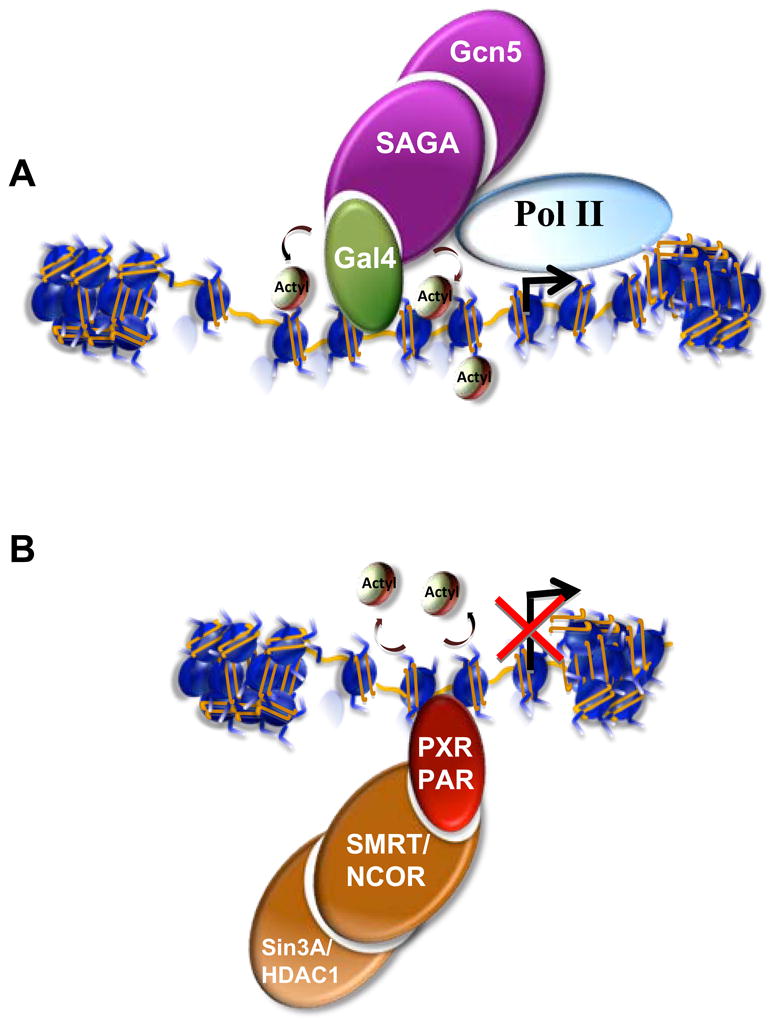
(A) The histone acetyltransferase, Gcn5, as part of the SAGA complex, is recruited to genes by a variety of transcriptional activators such as the sequence-specific binding protein Gal4. Acetylation of histone H3 through recruitment of Gcn5 by Gal4 synergizes with nucleosome remodeling activities leading to the recruitment of RNA Pol II. (B) The histone deacetylase HDAC1 can be recruited to genes as part of the SMRT/NCOR complexes by sequence-specific DNA-binding proteins such as Retinoic acid receptors (RXR/RAR). Deacetylation of histones facilitates reassembly of compact chromatin for transcriptional repression.
Recruitment of histone modifiers by RNA Polymerase II
While histone acetylation and other histone modifications can function at the promoter during transcriptional activation, histone modifications can also have roles in the body of genes. The trimethylation of histone H3 on lysine 36 is mediated by the Set2 enzyme in yeast and animals, and this modification peaks in the middle and the 3 end of genes associated with the elongating RNA polymerase II (Krogan et al., 2003b; Shilatifard, 2006). Purification of the Set2 enzyme from yeast identified the large subunit of Pol II, Rpb1, as an interactor of Set2 (Krogan et al., 2003b; Xiao et al., 2003). This interaction depends on the phosphorylation of Rpb1’s C-terminal repeat domain on Serine 2, a marker of elongating Pol II. Thus, Set2 is recruited to gene bodies through an interaction with elongating Pol II (Figure 2).
Figure 2. Recruitment of histone-modifying activities by RNA Pol II.
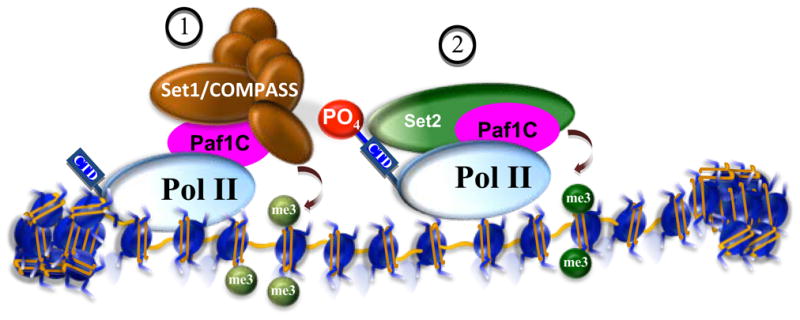
The histone H3K4 methyltransferase, Set1, as part of COMPASS, is recruited to genes through interaction with the Polymerase-associated factor (Paf1) complex (1) (Krogan et al., 2003a; Wood et al., 2003). H3K4 trimethylation is found at the start sites of the transcription of active genes, as well as “poised” genes with stalled RNA Pol II (Gilmour, 2009). Upon release of RNA Pol II by phosphorylation of its C-terminal domain (CTD) at serine 2 (P-Ser2), numerous factors associate with the phosphorylated C-terminal domain including the histone H3K36 methyltransferase Set2 (Xiao et al., 2003) (2). Set2 can be recruited through interactions with both the CTD of Pol II and the Paf1 complex (Krogan et al., 2003b; Xiao et al., 2003).
In contrast to the direct physical interaction between Set2 and RNA Pol II, other histone-modifying activities can be recruited subsequently to transcription initiation through the interaction with the Polymerase-associated factor (Paf1) complex (Krogan et al., 2003a; Ng et al., 2003a; Wood et al., 2003). The Paf1 complex is associated with elongating Pol II. It was found in a global proteomic analysis in S. cerevisiae (GPS) to be required for proper H3K4 methylation (Krogan et al., 2003a; Wood et al., 2003). Paf1 directly interacts with COMPASS (Complex of Proteins Associated with Set1), the sole H3K4 methyltransferase in yeast, and Paf1 is required for the recruitment of COMPASS to chromatin (Wood et al., 2003). These early studies in yeast set the paradigm that the Paf1 complex plays a role as a “platform” for the requirement of histone-modifying machinery to the elongating Pol II (Gerber and Shilatifard, 2003) (Figure 2). We now know that the Drosophila and mammalian Paf1 complexes also function as a platform for the recruitment of Set1/TRX/MLL-containing complexes in Drosophila and human cells (Tenney et al., 2006; Wang et al., 2008).
In addition to the recruitment of histone-modifying activities to chromatin, complexes can be recruited to chromatin, but remain in a relatively inactive state. The Paf1 complex appears to regulate several activities in this manner. Histone H2B can be monoubiquitinated by the Rad6/Bre1 E2/E3 ubiquitin ligase complex. This monoubiquitination is needed for the higher methylation states of H3K4 and H3K79, and Paf1 is required for H2B monoubiquitination (Krogan et al., 2003a; Wood et al., 2003). However, both Rad6/Bre1 and Dot1 appear to be recruited independently of Paf1 (Wood et al., 2003). Paf1 stimulates the enzymatic activity of Rad6/Bre1, perhaps post-recruitment of these complexes to chromatin. The subsequent monoubiquitination of H2B stimulates the activity of Set1/COMPASS and Dot1 to generate the trimethylation of H3K4 and H3K79 (Dover et al., 2002; Krogan et al., 2003a; Wood et al., 2003). Several recent mammalian studies have also confirmed the generality of these early observations made in yeast regarding the role of the Paf1 complex in the regulation of histone H2B monoubiquitination by Rad6/Bre1 and H3K4 methylation by Set1/COMPASS and H3K79 methylation byDot1 (Kim et al., 2009; McGinty et al., 2008; Pavri et al., 2006; Zhu et al., 2005).
Recruitment by the preceding histone modifications
Many of the posttranslational modifications of histones can enhance chromatin binding by other proteins through a variety of protein domains: bromodomains, found in several transcriptional activators can preferentially bind peptides with acetylated lysines; 14-3-3 and forkhead-domains can bind phosphorylated serines and threonines; chromodomains, MBT repeats, and PHD fingers can discriminate among lysines that are mono-, di- or trimethylated; and tudor domains can recognize methylated arginine or lysine residues (Maurer-Stroh et al., 2003; Taverna et al., 2007; Yaffe and Elia, 2001). The importance of the interaction between histone modifications and proteins containing these modules is exemplified by the occurrence of mutations within these domains in human disease (Matthews et al., 2007; Pena et al., 2006). Modifications can help form a landing platform for proteins and their complexes to aid in recruitment to chromatin through recognition of the modified residue. One example of this is the recruitment of deacetylase complexes to the body of transcriptionally active genes that require the chromodomain-containing protein Eaf3. Eaf3 preferentially binds to H3K36 di- and trimethylated states. While Eaf3 is a component of both histone acetyltransferase and deacetylase complexes, loss of Eaf3 leads to an increase in acetylation in the body of genes, which is proposed to have the effect of opening up the chromatin structure to allow “cryptic” transcription (Carrozza et al., 2005; Keogh et al., 2005) (Figure 3).
Figure 3. Recruitment of histone-modifying activities by preexisting histone modifications.
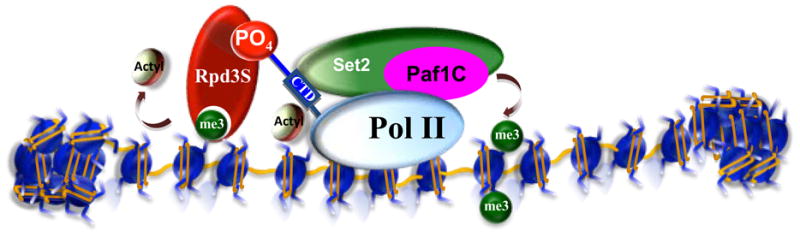
Methylation of histone H3K36 in the body of actively transcribed genes (see Figure 2) can be bound by the chromodomain-containing protein, Eaf3, as part of the Rpd3S deacetylase complex. This process helps to reposition nucleosomes in the wake of transcribing RNA Pol II. In the absence of Rpd3S, nucleosomes are misplaced, resulting in the exposure of cryptic start sites that can lead to the formation of aberrant transcription. Thus, recruitment of Rpd3S by a histone modification placed by passing RNA polymerase complexes can maintain transcriptional fidelity. However, at most genes, Rpd3S can be recruited directly through the interaction with the Serine 5 phosphorylated form of the RNA Pol II CTD (Govind et al., 2010, F. Robert, unpublished data). These findings indicate that the relative contribution of H3K36 methylation and CTD phosphorylation on the recruitment of Rpd3S can be gene specific and context dependent.
An important issue is the relative contribution of histone modifications in recruitment of other factors to chromatin. Since Eaf3 is also a component of the NuA4 histone acetyltransferase complex, one might expect both of the Eaf3 complexes to compete for binding to H3K36-methylated nucleosomes. However, Rco1, a component of Rpd3S, but not of NuA4, interacts with nucleosomes (with or without histone modifications) and is required for the interaction of Rpd3S with nucleosomes methylated at H3K36; indicating that both histone modification-dependent and independent mechanisms are important for targeting Rpd3S to the coding regions of transcribed genes (Li et al., 2007). Recently, Rpd3S has been demonstrated to be recruited to genes through interactions between the Rco1 subunit and the serine 5 phosphorylated form of the CTD of RNA Pol II. This finding suggests that the interaction with H3K36 methylated nucleosomes by Eaf3/Rco1 is subsequent to the initial recruitment of the complex by the CTD of RNA Pol II (Govind et al., 2010). Supporting these observations, genome wide profiling of Rpd3S in a Set2 deletion by Robert and colleagues demonstrated that H3K36 methylation had a modest effect on the recruitment of Rpd3S to transcribed regions (F. Robert, unpublished data).
Animals have at least two homologs of Eaf3, its ortholog Mrg15, and a paralog MSL3. Mrg15 participates in the NuA4-like Tip60 complex as well as the histone deacetylase complexes (Kusch et al., 2004; Lee et al., 2009; Moshkin et al., 2009; Spain et al., 2010). MSL3 is part of an H4K16-specific histone acetyltransferase complex. MSL3 can interact with H3K36-methylated nucleosomes, and mediates acetylation in the ORFs of transcribed genes (Sural et al., 2008). Although a balance of histone acetylation/deacetylation in ORFs is probably important for such processes as enhancing transcription elongation and maintaining transcription fidelity, how the MRG15 and MSL3 complexes are precisely targeted to the promoters or gene bodies likely involves factors other than just H3K36me3 binding. Rpd3S has not been extensively characterized in metazoans, although complexes with MRG15 and histone deacetylases have been reported (Lee et al., 2009; Tominaga et al., 2003; Yochum and Ayer, 2002). Importantly, the metazoan homologs of Rco1, PHF12 in mammals and CG3815 in Drosophila, are found in Rpd3S-like complexes, although the relative contribution of this subunit to the recruitment of Rpd3S by direct interaction with RNA Pol II, or indirect interaction with the H3K36 methylation deposited by Set2, remains to be determined.
Direct recruitment by DNA sequences
In the coactivator model, the DNA bound transcription factors recruit histone-modifying complexes. However, the most direct way to recruit a histone-modifying activity to particular genes is through recognition of specific DNA sequences by the histone-modifying complexes. This process is perhaps at the heart of, and the basis for, all recruitment to chromatin and can be considered the main step in the epigenetic regulation as well (Berger et al., 2009). Here, we review the role of DNA elements that form target sites for histone-modifying complexes that extend sequence-dependent recruitment mechanisms beyond the simple coactivator/corepressor model.
The PRE in Polycomb-mediated repression
The identification of the Polycomb Group of genes (PcG) (Ringrose and Paro, 2004) was made by the discovery of mutations in several genes in Drosophila that led to the ectopic expression of the Hox genes and a transformation of segmental identity. The PcG genes are required for the repression of Hox genes in regions where these genes have not been activated. The repression can be stably maintained for several cell generations after the transcription factors that initially determined cell identity are no longer present, constituting a form of epigenetic memory (Berger et al., 2009). Cloned regions near the Hox genes were shown to confer proper repression of reporter genes in a PcG-dependent manner, thus being known as Polycomb responsive elements (PRE). PREs are like enhancers in that they can function from large distances from genes and are sometimes found in introns of the genes they regulate. However, unlike canonical enhancer-binding proteins, PcG proteins are expressed ubiquitously and are downstream of the decision of whether a target gene is to be activated or repressed (Ringrose and Paro, 2004).
PcG proteins are found in at least three distinct complexes (Schuettengruber et al., 2007; Schwartz and Pirrotta, 2007; Simon and Kingston, 2009; Wang et al., 2004). Polycomb repressive complex 2 (PRC2) contains the Suppressor of zeste 12 (Su(z)12), extra sex combs (ESC) as well as the histone methyltransferase Enhancer of zeste (E(z)), or EZH1 and EZH2 in mammals) that implements H3K27 methylation. H3K27 methylation has been shown to form a binding site for the Polycomb protein (Pc), a component of the Polycomb repressive complex 1 (PRC1). PRC1 also includes the E3 ubiquitin ligase RING, Posterior Sex Combs (PSC, or Bmi-1 in mammals) and Polyhomeotic (PH). A third complex, the PHO-repressive complex, consists of Pleiohomeotic (PHO, similar to YY1 in mammals) and Sfmbt (MBTD1 in mammals), a protein that can recognize H4K20 mono- and dimethylated states. Of the known PcG proteins, PHO, which bears several zinc fingers, is clearly a sequence-specific DNA-binding protein and is required for recruitment of PRC2 to chromatin in Drosophila. However PHO binding is not sufficient for recruitment, and other factors have been implicated in PRC2 localization in Drosophila. While statistically significant, enrichment of sequences such as the PHO/YY1 consensus can be found in several PREs, a sequence-based definition of PREs has remained elusive (Ringrose et al. 2003).
One outstanding question is whether PREs exist in mammals. Although PRC2 is conserved from flies to mammals, YY1 appears to be the only mammalian homolog of the Drosophila transcription factors implicated in sequence-specific binding to elements commonly found within Drosophila PREs. However, Jarid2, a protein with very weak DNA-binding activity was recently identified as a component of PRC2 that is required for recruitment to chromatin. The mechanism by which Jarid2 recruits PRC2 is not known (Landeira et al., 2010; Li et al., 2010; Pasini et al., 2010; Peng et al., 2009) [for review please see (Herz and Shilatifard, 2010)]. Unlike other PRC2 components, Jarid2 shows obvious tissue-specific differences in expression, being particularly highly expressed in embryonic stem cells. Additionally, Jarid2 seems to have different effects on different genes, perhaps reflecting the diversity in the PRC2 complexes (Herz and Shilatifard, 2010). A full understanding of the role of Jarid2 in the regulation of PRC2 activity and gene expression will require further detailed and comprehensive biochemical and in vivo studies using organisms amenable to genetic manipulations.
Recently, Kingston and colleagues identified a 1.8 kb region that conferred PcG responsiveness at the HoxD locus on a reporter gene (Woo et al., 2010). This region contained YY1 binding sites (Wang et al., 2004), as well as an evolutionarily conserved sequence within the HoxD locus (Beckers and Duboule, 1998). In addition to these two features, this putative mammalian PRE contains a CpG island (Illingworth and Bird, 2009). In mammals, PRC2-binding is highly correlated with the presence of CpG islands, and CpG islands are largely predictive of the presence of PRC2, suggesting that CpG islands behave as PREs in mammalian cells (Ku et al., 2008). CpG islands are regions enriched for CpG dinucleotides that are associated with promoters of ~ 60–70 % of genes (Illingworth and Bird, 2009). Most CpG dinucleotides in the mammalian genome are cytosine-methylated, but when they are sufficiently clustered together in islands, they are typically unmethylated, except during transcriptional silencing associated with imprinting, X inactivation, or silencing of tumor suppressor genes during oncogenesis. Genes that are silenced during development are not associated with the CpG methylation of their promoters, even if associated with CpG islands (Baylin and Bestor, 2002; Illingworth and Bird, 2009). Therefore, it is likely that PcG function, without DNA methylation, is the major repressor of many developmentally regulated genes, perhaps through recognition of CpG islands; however PcG proteins can also synergize with DNA methylation machineries during silencing associated with imprinting and X inactivation (Illingworth and Bird, 2009). How is PRC2 targeted to CpG islands? CpG-rich sequences can be recognized by proteins bearing a CXXC domain, a type of zinc finger that can recognize CpG sequences. However, CXXC domains are not found within PRC2 components, suggesting a novel domain recognizes these sequences, or that other motifs found within these islands help recruit PRC2.
PRE vs. TRE
While Hox genes can be silenced with PcG complexes, they can be activated with the help of the Trithorax group of proteins (TrxG) (Eissenberg and Shilatifard, 2009). Trithorax, like its mammalian counterpart MLL, is an H3K4 methyltransferase. H3K4 and H3K27-methylated nucleosomes are usually found to occur in a mutually exclusive pattern throughout the genome. This can perhaps be explained by the competition for the same recruitment sites by the enzyme complexes that implement these modifications. Experimental evidence indicates that PREs and Trithorax recruitment elements (TREs) substantially overlap (Ringrose and Paro, 2007). DNA-binding factors that are important for recognizing TREs include the GAGA factor (also known as Trl), which recognizes GA-rich sequences (Farkas et al., 1994), and Pho and Zeste, although other combinations of factors are likely to be important for recruitment (Schwartz et al., 2006). In mammals, the TRX homologs MLL1 and MLL2 have CXXC domains within their polypeptide, suggesting that the CpG character within a PRE may constitute a distinct sequence that helps recruit the competing TrxG activity to the PREs/TREs. Interestingly, recent work from several laboratories has shown that the CXXC domains are involved in targeting distinct types of chromatin-modifying activities to chromatin, confirming the importance of the CpG islands in recruiting histone-modifying activities to chromatin while also raising the question of which other features determine the specificity demonstrated for these diverse activities?
CpG islands recruit multiple modifiers
The CXXC domain binding of CpG was originally discovered in MBD1, a protein isolated for its homology to the methylated CpG DNA-binding protein, MeCP2, but also having three zinc fingers of the CXXC type (Cross et al., 1997). Subsequently, it was determined that while two of the CXXC domains within MBD1 bind methylated DNA, many CXXC domains are specific for unmethylated CpG sequences (Birke et al., 2002; Lee et al., 2001), including a third CXXC domain within MBD1 (Jorgensen et al., 2004) and the DNA methyltransferase DNMT1 (Pradhan et al., 2008). Thus, multiple chromatin modifiers could potentially be recruited by CpG-rich sequences. Two recent studies have tested this idea and found a critical role for CXXC domains in recruiting histone-modifying activities to chromatin (Blackledge et al., 2010; Thomson et al., 2010).
In one test of the role of the CXXC domains in targeting CpG islands, Bird and colleagues studied the recruitment of CXXC1 (Cfp1), a component of mammalian Set1/COMPASS, the major H3K4me3 in mammals (Lee and Skalnik, 2005; Miller et al., 2001; Thomson et al., 2010; Wu et al., 2008). Bird and colleagues performed genome-wide profiling of CXXC1 in mouse brain. They found CXXC1 to be localized at 80% of the CpG islands, 90% of which were enriched for H3K4me3. Half of the CXXC1-negative CpG islands were previously reported to be sites of Polycomb and H3K27me3 occupancy. Comparing CXXC1 occupancy at Xist, a gene transcribed on only one of two X chromosomes in females, CXXC1 was found to associate exclusively with the transcribed, unmethylated, CpG copy. Together, these findings suggest that CpG islands could help recruit mammalian Set1/COMPASS through CXXC1’s affinity for unmethylated CpG sequences.
To experimentally test the role of CpG islands in recruiting CXXC1, Bird and colleagues created two ES cell lines carrying a promoterless construct with eGFP and puromycin sequences that contain CpG dinucleotide densities similar to those found in CpG islands. In one cell line, the artificial CpG island was targeted to the 3 end of Nanog, while in the other, the construct was targeted to the 3 end of Mecp2, an X-linked gene. When located adjacent to Nanog, the cassette remained free of CpG methylation, despite lacking detectable Pol II. H3K4me3 and CXXC1 occupancy tracked CpG density within the cassette and around the insertion site. Interestingly, the cassette inserted next to the Mecp2 gene showed two-thirds CpG methylation, but still was found to be bound by CXXC1. Bisulfite sequencing of the CXXC1 and H3K4me3 ChIP DNA demonstrated that only a third of the immunoprecipitated cassette was CpG-methylated, strongly suggesting that unmethylated CpG sequences are recruitment sites for mammalian Set1/COMPASS. However, point mutations within the CXXC domain of CXXC1 would be required to demonstrate the direct role of this protein in binding CpG and the ensuing regulation of H3K4 methylation by mammalian Set1/COMPASS.
While Set1/COMPASS is the major H3K4 trimethylase in mammalian cells (Wu et al., 2008), in other studies it has been demonstrated that the loss of CXXC1 leads to an increase in global H3K4 trimethylation levels in ES cells (Tate et al., 2009). Based on this observation, Skalnik and colleagues have suggested that CXXC1 functions by restricting Set1/COMPASS methyltransferase activity. In contrast, Bird and colleagues find that H3K4me3 levels are reduced at the CpG islands in the absence of CXXC1. It will be interesting to learn where in the genome H3K4me3 is increasing upon loss of CXXC1.
An important finding by Bird and colleagues is that H3K4 methylation implemented by mammalian Set1/COMPASS is independent of Pol II and transcription, while findings in yeast have shown that transcription is required for proper H3K4 trimethylation (Krogan et al., 2003a; Ng et al., 2003b; Shilatifard, 2006). One possible explanation for this apparent difference is that the interaction of Pol II with CpG islands might be transient and not as easily detectable as the product of the process, histone H3K4 trimethylation. Therefore, sensitive RNA-seq methods, such as global run-on sequencing GRO-seq (Core et al., 2008), could reveal transcription within these CpG islands, thus explaining the association of CXXC1 and mammalian COMPASS at these CpG islands.
Interestingly, the yeast homolog of CXXC1, Cps40, has a PHD finger in common, but lacks a CXXC domain. However, the Drosophila homolog of CXXC1, CG17446, contains both the PHD and CXXC domains. CpG islands have not been studied in Drosophila, although their existence has been predicted (Takai and Jones, 2002). It is notable that Drosophila Trithorax, unlike its mammalian counterpart MLL, lacks a CXXC domain, further demonstrating that although these H3K4 methyltransferase complexes are largely conserved in composition and function, some aspects of their recruitment can differ, perhaps reflecting differences in the size or complexity of the genome in which they are found.
Another class of histone-modifying enzymes bearing a CXXC domain is KDM2A/B. KDM2A/B is a histone demethylase that preferentially uses H3K36me2 as a substrate (Tsukada et al., 2006). H3K36me2 is a modification that has previously been linked to gene silencing (Bender et al., 2006), and can recruit histone deacetylases (Li et al., 2009; Youdell et al., 2008); suggesting that removal of H3K36me2 could facilitate the formation of open chromatin. Klose and colleagues tested the role of the CXXC domain in targeting KDM2A. First, they tested the DNA-binding specificity of KDM2A’s CXXC domain and found that it preferentially binds to unmethylated CpG sequences (Blackledge et al., 2010). Genome-wide profiling demonstrated that KDM2A was highly enriched at annotated CpG islands. Major peaks of KDM2A binding not corresponding to CpG islands were shown to have a high CpG content with little DNA methylation as assessed by bisulfite sequencing. Since most CpGs outside of the CpG islands are unmethylated, Klose and colleagues likely found novel CpG islands that were previously unnoticed due to the statistical criteria used in the annotation process. Sites of strong KDM2A binding were also found to be depleted for H3K36me2, suggesting that KDM2A localization to CpG islands results in active demethylation of H3K36me2. Indeed, knockdown of KDM2A results in increased H3K36me2 at some CpG islands, however, very little alteration in transcription was reported when KMD2A levels were reduced by RNAi, suggesting that H3K36 dimethylation at CpG islands at the promoters does not have a major transcriptional regulatory role. KDM2A and the highly related KDM2B have been shown to be highly concentrated in nucleoli of cells where they can repress ribosomal RNA transcription (Frescas et al., 2007; Tanaka et al., 2010). Interestingly, rDNA accounts for 20% of the unmethylated CpG sequences in the mouse genome (Bird et al., 1985), suggesting that KDM2A and KDM2B are targeted in part through CpG recognition for rDNA transcription by RNA Pol I.
The studies by the Bird and Klose groups were both in agreement that recruitment to unmethylated CpG islands via the CXXC domain was uncorrelated with transcriptional activity, suggesting that unmethylated CpG content is sufficient for recruitment. Since MBD1 and the DNA methyltransferase DNMT1 also have CXXC fingers that recognize unmethylated CpG, it will be important to ask how various CXXC-associated activities compete or coexist with each other on the same site and how this is regulated during development. It would not be surprising if future studies in this regard will find that the interactions of the CXXC finger-containing proteins with their target site are context-dependent and require other cellular signals for proper function. The studies by the Bird and Klose groups should stimulate investigations into the role of CpG islands in transcription and the function of histone-modifiying activities in this process.
Same modification, different complexes and distinct outcomes
One of the best studied histone modifications is the methylation of H3 on lysine 4 (H3K4). The first methyltransferase complex for H3K4 to be identified was COMPASS (Complex of Proteins Associated with Set1) in yeast (Krogan et al., 2002; Miller et al., 2001; Roguev et al., 2001; Shilatifard, 2006). The original interest in purifying COMPASS was based on the similarity between yeast Set1 and the human MLL protein involved in leukemia, and indeed, human MLL forms a COMPASS-like complex with methyltransferase activity for H3K4 (Eissenberg and Shilatifard, 2009; Hughes et al., 2004; Miller et al., 2001; Shilatifard, 2006). While yeast has just one methyltransferase for H3K4, humans have at least six methyltransferases that can methylate H3K4, including MLL1-4 and Set1A/B which are found in COMPASS-like complexes (Cho et al., 2007b; Eissenberg and Shilatifard, 2010; Hughes et al., 2004; Lee and Skalnik, 2008; Lee et al., 2007a; Shilatifard, 2008; Wu et al., 2008). COMPASS-like complexes are defined as having a Set1 or MLL-related protein, and the core subunits Cps60/Ash2, Cps30/Wdr5, and Cps50/Rbbp5.
In addition to the common subunits, the Set1 and MLL complexes each have unique subunits. MLL1 and MLL2, which are related to Drosophila Trithorax (TRX), interact with the tumor suppressor Menin, which helps target MLL1/2 to Hox and other targets for transcription activation (Hughes et al., 2004; Wang et al., 2009). MLL3 and MLL4, which show high similarity to Drosophila trithorax-related (TRR), contain NCOA6, PTIP and PA-1 which may help target MLL3/4 to hormone-responsive genes, together with UTX, an H3K27 histone demethylase (Cho et al., 2007b; Issaeva et al., 2007; Patel et al., 2007). Two subunits found in the SETD1A and SETD1B complexes, but not in the MLL1-4 complexes, are CXXC1 and WDR82 (Lee and Skalnik, 2008; Lee et al., 2007a; Wu et al., 2008). These subunits are homologous to the COMPASS subunits Cps40 and Cps35 (Miller et al., 2001). Cps35/WDR82 has a unique role in mediating the crosstalk between histone H2B ubiquitination and H3K4 methylation (Lee et al., 2007b; Wu et al., 2008; Zheng et al., 2010).
The COMPASS and MLL1-4 complexes can be recruited by different mechanisms to chromatin, with varying functional consequences associated with H3K4 methylation. COMPASS can be recruited to the actively transcribed genes through its interaction with the Paf1 complex and RNA Pol II, which is sufficient for H3K4 monomethylation (Gerber and Shilatifard, 2003; Krogan et al., 2003a; Wood et al., 2003). The Paf1 complex may also be involved in recruiting Rad6/Bre1 through a direct Paf1/Bre1 interaction leading to ubiquitinated H2B within this region (Wood et al., 2003). Cps35/WDR82 interacts with COMPASS in a histone H2B monoubiquitination-dependent manner, and this interaction on chromatin converts COMPASS to a trimethylation competent complex (Lee et al., 2007b; Zheng et al., 2010). Deletion of COMPASS subunits or mutation of H3K4 to alanine has little effect on transcription levels in yeast (Miller et al., 2001), consistent with the recruitment and stimulation of COMPASS activity following gene activation (Krogan et al., 2003a; Ng et al., 2003b).
In contrast to COMPASS, the MLL complexes can function as transcriptional activators. Loss of MLL leads to loss of H3K4 methylation and activation of transcription at Hox and other loci (Hughes et al., 2004; Wang et al., 2009). MLL1-4 complexes appear to be co-activators of nuclear receptors (Cho et al., 2007a; Dreijerink et al., 2009; Eissenberg and Shilatifard, 2010; Goo et al., 2003). H3K4 methylation by the MLL complexes could activate genes through recruitment of H3K4me3-binding proteins. For example, the basal transcription factor can be directly recruited by H3K4me3 via the Taf3 subunit’s PHD finger (Vermeulen et al., 2007). Alternatively, H3K4me3 at the promoter could help recruit histone acetyltransferase complexes or nucleosome remodeling complexes, which harbor subunits with PHD fingers capable of recognizing the H3K4me3 state (Kouzarides, 2007; Ruthenburg et al., 2007). Thus, the timing and mechanism of recruitment to a gene can influence the biological readout of H3K4 methylation (Wang et al., 2009).
Noncoding RNAs as recruitment vehicles for histone modifiers
In addition to protein and DNA-based recruitment of histone modifiers, noncoding RNAs have recently been shown to be major regulators of chromatin and transcription. While RNAs are well-known integral components of ribosomes and spliceosomes for the translation and splicing of mRNAs, the role of RNA molecules in transcriptional regulatory complexes has only recently gained widespread notice. One of the first noncoding RNAs associated with transcriptional regulation was the Xist RNA, whose transcription is required for somatic silencing of an X chromosome in mammalian females, part of a process of dosage compensation to equalize expression from X-linked genes between XY males and XX females (Brockdorff et al., 1992; Brown et al., 1991). Inactivation of the X chromosome is associated with accumulation of repressive histone marks and the compaction of the X chromosome into the Barr body (Chow and Heard, 2009; Senner and Brockdorff, 2009). Silencing initiates and spreads in cis from the site of Xist transcription to encompass most of the X chromosome. The ability of certain forms of Xist RNA to interact with PRC2 complexes provides a molecular explanation for the requirement of Xist for H3K27 methylation and gene silencing (Zhao et al., 2008).
Equalization of transcription between males and females of Drosophila also involves noncoding RNAs. roX1 and roX2 are transcribed from the X chromosome in males and are required for the twofold increased expression of the male X chromosome to equalize transcription with the two X chromosomes from females. Unlike Xist, roX RNAs can act in trans (Kelley et al., 1999). They form a ribonucleoprotein complex with the MSL proteins that includes the histone acetyltransferase, MOF. MOF mediates H4K16 acetylation in the transcribed region of genes, which is proposed to facilitate the decompaction of the X chromosome to facilitate higher rates of transcription elongation by Pol II (Smith et al., 2001). The targeting of the roX-MSL complex to its X-linked targets is comprised of two steps; 1) recognition of high affinity sites for the MSL1-MSL2 subunits, and 2) spreading to nearby actively transcribed genes (Alekseyenko et al., 2008). The nature of the high affinity sites is unknown, perhaps consisting of low affinity sites within a specific context (Alekseyenko et al., 2008; Fauth et al., 2010; Straub et al., 2008). Spreading from high affinity sites to nearby genes could be facilitated by the MLE subunit, an RNA helicase whose closest mammalian homolog has been shown to interact with RNA Pol II (Nakajima et al., 1997). MLE’s presence in the roX-MSL complex is dependent on the integrity of the RNA component (Ilik and Akhtar, 2009; Smith et al., 2000). Thus, roX RNAs could function in MSL spreading by interacting with the RNA helicase MLE, which in turn associates with RNA Pol II at active genes, allowing the roX-MSL complex to spread from one active gene to another. The enrichment of the MSL complexes in the middle and the 3 end of the transcribed genes could be facilitated by interactions between the chromodomain of the MSL3 subunit and H3K36me3 implemented by Pol II-associated Set2 (Sural et al., 2008). Placing high affinity binding sites for MSL1-MSL2 on autosomes allows a similar spreading to nearby autosomal genes, which becomes even more pronounced if particular spliced forms of roX RNAs are ectopically expressed (Park et al., 2005).
While the roX RNAs are associated with the upregulation of transcription, it is more common for noncoding RNAs to recruit repressive histone-modifying activities similarly to Xist. For example, Air and Kcnq1ot1 are noncoding RNAs expressed from the paternal chromosome that are required for silencing of neighboring genes in cis (Mancini-Dinardo et al., 2006; Sleutels et al., 2002). Both have been shown to associate with the H3K9 methyltransferase, G9a, while Kcnq1ot1 is also associated with PRC2 (Nagano et al., 2008; Pandey et al., 2008). Although Xist, Air, and Kcnq1ot1 all work to silence genes in cis, it is becoming apparent that noncoding RNAs can silence genes on other chromosomes. HOTAIR, a noncoding RNA transcribed from the HOXC cluster, recruits PRC2 to silence genes in the HOXD cluster (Rinn et al., 2007). Separate regions of HOTAIR interact with the PRC2 and LSD1/CoREST complexes (Tsai et al., 2010). LSD1 is an H3K4 demethylase that can demethylate H3K4me2, but not H3K4me3, and associates with the CoREST histone deacetylase complex (Lee et al., 2005; Shi et al., 2004). In addition to a role in developmental regulation, HOTAIR is also implicated in promoting metastasis. HOTAIR is frequently upregulated in metastatic tumors and its high expression is associated with a poor prognosis (Gupta et al., 2010). Importantly, PRC2 components are required for the matrix invasiveness of cells ectopically expressing HOTAIR, suggesting that HOTAIR is mistargeting PRC2 components when over-expressed (Gupta et al., 2010).
The potential regulatory scope of noncoding RNAs is just being realized. Genome-wide analyses have identified over 1000 “large intergenic noncoding RNAs” (lincRNAs) based on signatures of H3K4me3 at transcription start sites and H3K36me3 in transcribed regions (Guttman et al., 2009). RNA immunoprecipitation with PRC2 antibodies identified 24% of the expressed noncoding RNAs as interactors (Khalil et al., 2009). RNAi knockdown of six of these lincRNAs showed significant overlap between genes upregulated by the knockdown of PRC2 components, but no significant overlap among the six noncoding RNAs (Khalil et al., 2009). One of these, TUG1, is induced upon DNA damage in a P53-dependent manner and is required for the repression of a set of genes involved in cell cycle regulation (Guttman et al., 2009; Khalil et al., 2009). Subsequent studies aimed at finding P53-induced noncoding RNAs identified lincRNA-p21 as a gene required for repression of genes in the P53 pathway (Huarte et al., 2010). While lincRNA-p21 is located near p21, which is also a repressor in the P53 pathway, their gene targets do not significantly overlap. LincRNA-p21 interacts with PRC2 similarly to many other lincRNAs, but it also interacts with hnRNP-K (Huarte et al., 2010). hnRNP-K, when interacting with P53, can activate genes. When associating with lincRNA-p21, hnRNP-K mediates repression of genes; and hnRNP-K localization to these targets requires intact lincRNA-p21 (Huarte et al., 2010). The corepression of genes by hnRNP-K and lincRNA-p21 provides a hint as to why so many different noncoding RNAs associate with PRC2 for transcriptional repression. Distinct noncoding RNAs could form unique scaffolds for histone-modifying complexes to associate with gene-specific targeting factors. P53 alone is involved in the upregulation of approximately 30 noncoding RNAs under a variety of conditions and cell types, indicating that the gene regulatory potential for these large noncoding RNAs is enormous (Huarte et al., 2010). Recent work suggests that short RNA transcripts from the PRC2 target genes can form short hairpin structures that directly recruit PRC2 (Kanhere et al., 2010), and small RNAs transcribed from LINE elements on the mammalian X chromosome could play a key role in X inactivation (Chow et al., 2010), indicating that both large and small RNAs contribute to these silencing events.
Multiple targeting modes within a complex
For many of the complexes we ve discussed here, several mechanisms exist for recruiting the same complex. PRC2, for example, can be recruited by its previously deposited H3K27me3 mark for epigenetic memory, it can be recruited through DNA sequences such as PREs, and it can be recruited through interacting with noncoding RNAs. The opposing activity of PRC2 is mediated in part by MLL, which also displays multiple modes of targeting to chromatin. For example, MLL complexes can be recruited through MLL’s CXXC domain to CpG islands (Ayton et al., 2004; Cierpicki et al., 2010). Translocation of the MLL gene with a variety of other genes results in chimeric proteins comprised of an N-terminal portion of MLL that includes the CXXC domain. Mutations in MLL’s CXXC that prevent DNA-binding, cause an increase in DNA methylation at the Hoxa9 locus, reduced Hoxa9 expression and reduced transformation of bone marrow cells by the MLL-AF9 chimera (Ayton et al., 2004; Bach et al., 2009; Cierpicki et al., 2010). It has also recently been proposed that MLL can be recruited to genes through a direct interaction between its CXXC domain and the PAF complex. Remarkably, it was found that only MLL1, and not other Set1-related proteins, could interact with the PAF complex (Milne et al., 2010; Muntean et al., 2010). At this time, it is unclear how these data can be reconciled with demonstrated functional differences for the Set1 and MLL complexes in mammals (Wang et al., 2009; Wu et al., 2008).
Another domain that may be important for targeting MLL to chromatin is its third PHD finger (PHD3), which specifically recognizes H3K4me3 (Chang et al., 2010). MLL also contains AT hooks, which are found in proteins that bind AT-rich sequences (Reeves and Nissen, 1990), although their requirement for recruitment of MLL to Hox or other loci is n’t known. Aside from domains that bind DNA sequences or histone modifications, MLL could also be recruited to genes through the coactivator model, due to its interaction with Menin-LEDGF and their recruitment by nuclear hormone receptors (Dreijerink et al., 2009; Eissenberg and Shilatifard, 2010).
Summary and future perspectives
Histone modifications play an indisputably important role in transcription and other DNA-templated processes. However, the identity of the enzyme and the mechanism of recruitment can influence the effect of the modification. This is exemplified by the diverse functions of the highly related Set1/COMPASS and MLL COMPASS-like complexes, whose biological roles are reflected in part by their mode of recruitment. Another example is the differing role of H3S10P at the FOSL1 gene, which when mediated early during activation by MSK1 at the promoter, or later at the enhancer by Pim1, leads to distinct downstream events (Zippo et al., 2009). Furthermore, histone-modifying enzymes can themselves be multi-faceted transcription factors, with only one aspect being the ability to modify histones. For example, distinct phenotypes are found for loss of MLL and loss of just the histone-modifying Set domain within MLL, the latter of which yields viable offspring (Terranova et al., 2006). Thus, rather than simply “writing or erasing” a code that is waiting to be “read”, histone-modifying activities are integral components of gene regulatory networks in a larger “chromatin signaling pathway”. A better understanding of the multiple modes of recruitment of these histone-modifying activities is therefore essential for understanding the gene regulatory processes in which they are engaged. The growing number of noncoding RNAs involved in this process provides a challenging, yet promising, avenue for future research. Identification of the factors that bind to these RNAs could help us better understand how histone-modifying enzymes are targeted for gene activation or repression.
Figure 4. Recruitment by DNA sequences.
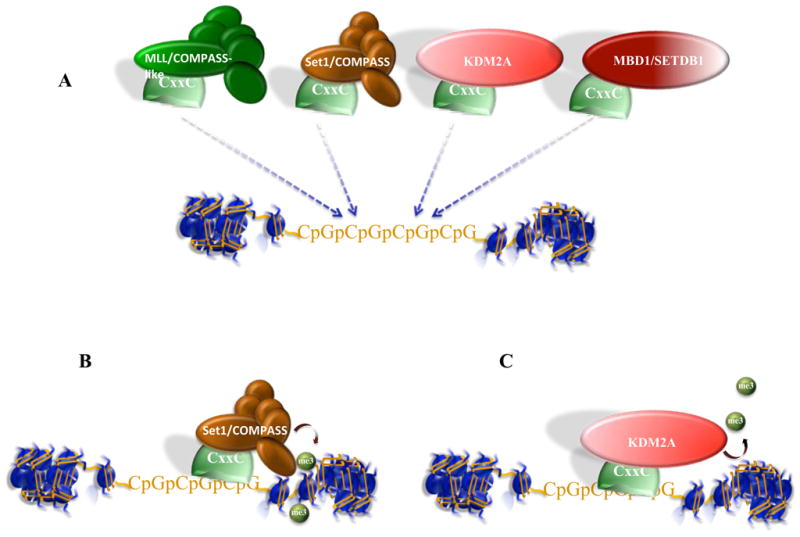
In humans, CpG islands are found near the promoters of 60–70 % of genes (Illingworth and Bird, 2009). While cytosines within CpG dinucleotides are frequently methylated, in CpG islands cytosines are largely unmethylated. (A) A number of proteins bearing a zinc finger of the CXXC class preferentially bind unmethylated CpG islands. These proteins are associated with a variety of histone-modifying activities including the H3K4 methyltransferases, MLL and Set1/COMPASS, and the H3K36 demethylase, KDM2A (Clouaire and Stancheva, 2008). MBD1 has two zinc fingers that bind methylated DNA and a third of the CXXC class that binds unmethylated DNA. MBD1 is associated with H3K9 methyltransferase activities and is thought to help coordinate silencing by DNA and histone methylations (Sarraf and Stancheva, 2004). (B) A recent study demonstrated that CpG islands were major sites of recruitment of CXXC1 (Cfp1) and H3K4 methylation via mammalian COMPASS, independent of the transcriptional status of the nearby gene (Thomson et al., 2010). (C) In another recent manuscript, KDM2A, an H3K36 demethylase, was found to be targeted to CpG islands via its CXXC motif. Since H3K36 methylation can recruit histone deacetylases (Figure 3), removal of H3K36 methylation in CpG islands could maintain CpG islands in an open, permissive state (Blackledge et al., 2010).
Figure 5. The same histone modification implemented by different complexes can have different consequences.
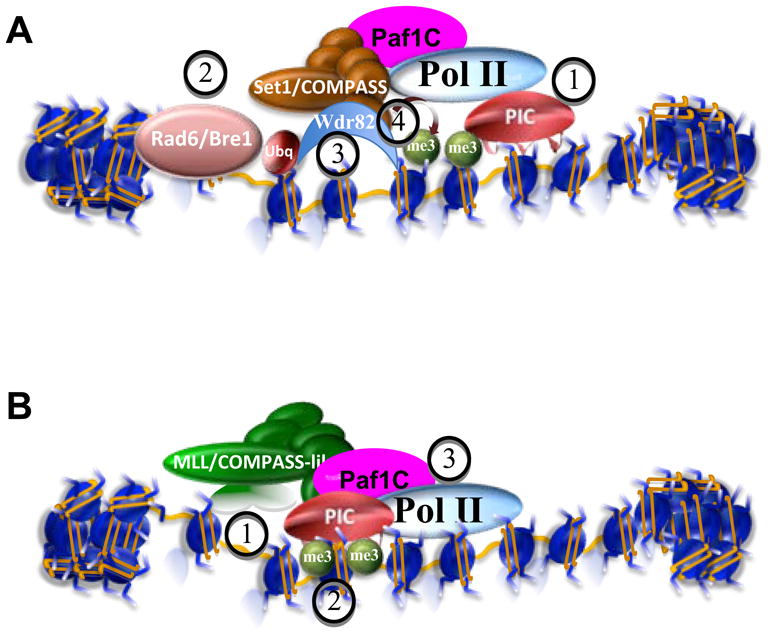
(A) Mammalian Set1/COMPASS is recruited to chromatin of transcriptionally active genes through interaction with the Paf1 Complex (1) (Krogan et al., 2003a), where it is only capable of implementing H3K4 monomethylation. Upon H2B monoubiquitination (H2Bub) by Rad6/Bre1 (2), WDR82 (Cps35 in yeast) interacts with chromatin and associates with COMPASS (3), enabling the complex to trimethylate H3K4 (4), whose presence at promoters is a mark of actively transcribed genes (see Figure 2). (B) MLL is part of a COMPASS-like complex that shares many subunits, but importantly lacks WDR82, and therefore, its methyltransferase activity is likely to be independent of H2Bub. MLL, like its Drosophila homolog Trithorax, is well known for its role in activating Hox genes. Recruitment of MLL via Menin and sequence-specific binding factors could help activate transcription of its target loci by mediating trimethylation of H3K4 (1) which can form a binding site for TAF3, a component of TFIID in the pre-initiation complexes (PIC) (2), thereby, mediating recruitment or stabilization and the basal machinery (PIC) and RNA Pol II (3) (Vermeulen et al., 2007; Wang et al., 2009). Thus, H3K4 trimethylation implemented by the MLL complexes is instructive for transcription (Wang et al., 2009), while H3K4 trimethylation implemented by Set1/COMPASS may be recruited subsequent to initiation of transcription by PIC and RNA Pol II through the H2Bub-WDR82 pathway (Wu et al., 2008; Wang et al., 2009).
Acknowledgments
We thank Laura Shilatifard for Editorial assistance. The studies in the Shilatifard laboratory are supported in part by grants from the National Institute of Health: R01GM069905, R01CA150265, and R01CA89455.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, Kuroda MI. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene. 2009;28:815–823. doi: 10.1038/onc.2008.443. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- Beckers J, Duboule D. Genetic analysis of a conserved sequence in the HoxD complex: regulatory redundancy or limitations of the transgenic approach? Dev Dyn. 1998;213:1–11. doi: 10.1002/(SICI)1097-0177(199809)213:1<1::AID-AJA1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Berger SL, Cress WD, Cress A, Triezenberg SJ, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30:958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chang PY, Hom RA, Musselman CA, Zhu L, Kuo A, Gozani O, Kutateladze TG, Cleary ML. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400:137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007a;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007b doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci. 2008;65:1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Meehan RR, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Dreijerink KM, Lips CJ, Timmers HT. Multiple endocrine neoplasia type 1: a chromatin writer’s block. J Intern Med. 2009;266:53–59. doi: 10.1111/j.1365-2796.2009.02115.x. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Fauth T, Muller-Planitz F, Konig C, Straub T, Becker PB. The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome. Nucleic Acids Res. 2010;38:3209–3221. doi: 10.1093/nar/gkq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A. The JARID2-PRC2 duality. Genes Dev. 2010;24:857–861. doi: 10.1101/gad.1921610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Ilik I, Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 2009;6:113–121. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Ben-Porath I, Bird AP. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Mol Cell Biol. 2004;24:3387–3395. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003a;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003b;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007a;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Voo KS, Skalnik DG. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone Crosstalk between H2B Monoubiquitination and H3 Methylation Mediated by COMPASS. Cell. 2007b;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3. Mol Cell Biol. 2009;29:1401–1410. doi: 10.1128/MCB.01643-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, Allis CD. Multiple Interactions Recruit MLL1 and MLL1 Fusion Proteins to the HOXA9 Locus in Leukemogenesis. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003a;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003b;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Park Y, Oh H, Meller VH, Kuroda MI. Variable splicing of non-coding roX2 RNAs influences targeting of MSL dosage compensation complexes in Drosophila. RNA Biol. 2005;2:157–164. doi: 10.4161/rna.2.4.2473. [DOI] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan M, Esteve PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–10009. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Senner CE, Brockdorff N. Xist gene regulation at the onset of X inactivation. Curr Opin Genet Dev. 2009;19:122–126. doi: 10.1016/j.gde.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Caruso JA, Swaminathan A, Pile LA. Drosophila SIN3 Isoforms Interact with Distinct Proteins and Have Unique Biological Function. J Biol Chem. 2010 doi: 10.1074/jbc.M110.130245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Okamoto K, Teye K, Umata T, Yamagiwa N, Suto Y, Zhang Y, Tsuneoka M. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010;29:1510–1522. doi: 10.1038/emboj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CM, Lee JH, Skalnik DG. CXXC finger protein 1 contains redundant functional domains that support embryonic stem cell cytosine methylation, histone methylation, and differentiation. Mol Cell Biol. 2009;29:3817–3831. doi: 10.1128/MCB.00243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Leung JK, Rookard P, Echigo J, Smith JR, Pereira-Smith OM. MRGX is a novel transcriptional regulator that exhibits activation or repression of the B-myb promoter in a cell type-dependent manner. J Biol Chem. 2003;278:49618–49624. doi: 10.1074/jbc.M309192200. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wang P, Bowl MR, Bender S, Peng J, Farber L, Chen J, Ali A, Zhang Z, Alberts AS, Thakker RV, et al. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol Cell Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Current opinion in cell biology. 2001;13:131–138. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Wyrick JJ, Reese JC. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol. 30:3635–3645. doi: 10.1128/MCB.00324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Wyrick JJ, Reese JC. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Molecular and cellular biology. 2010;30:3635–3645. doi: 10.1128/MCB.00324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]


