Abstract
The Plant Organelles Database (PODB) was launched in 2006 and provides imaging data of plant organelles, protocols for plant organelle research and external links to relevant websites. To provide comprehensive information on plant organelle dynamics and accommodate movie files that contain time-lapse images and 3D structure rotations, PODB was updated to the next version, PODB2 (http://podb.nibb.ac.jp/Organellome). PODB2 contains movie data submitted directly by plant researchers and can be freely downloaded. Through this organelle movie database, users can examine the dynamics of organelles of interest, including their movement, division, subcellular positioning and behavior, in response to external stimuli. In addition, the user interface for access and submission has been enhanced. PODB2 contains all of the information included in PODB, and the volume of data and protocols deposited in the PODB2 continues to grow steadily. Moreover, a new website, Plant Organelles World (http://podb.nibb.ac.jp/Organellome/PODBworld/en/index.html), which is based on PODB2, was recently launched as an educational tool to engage members of the non-scientific community such as students and school teachers. Plant Organelles World is written in layman's terms, and technical terms were avoided where possible. We would appreciate contributions of data from all plant researchers to enhance the usefulness of PODB2 and Plant Organelles World.
Keywords: 3D structure, Database, Image, Movie , Organelle dynamics, PODB
Introduction
The field of plant organelle research has expanded recently because of advances in in vivo imaging analysis techniques and the increase in the amount of genome sequence information available for various plant species (The Arabidopsis Initiative 2000, Goff et al. 2002, Yu et al. 2002, Tuskan et al. 2006, Jaillon et al. 2007, Mochida and Shinozaki 2010). In particular, the utilization of fluorescent probes, such as grren fluorescent protein (GFP), has contributed greatly to our understanding of plant organelle dynamics in living cells. Therefore, databases that compile image data of visualized organelles have been constructed (Cutler et al. 2000, Damme et al. 2004, Brown et al. 2005, Koroleva et al. 2005, Li et al. 2006, Mano et al. 2008), and are a useful ‘omics’ tool in systems biology analysis (Mano et al. 2009).
The Plant Organelles Database (PODB) is a specialized database that facilitates our understanding of organelle dynamics, including organelle functions, biogenesis, differentiation, movements and interactions with other cellular components. The first version of PODB, which was launched in 2006, has been widely used as a data source for plant organelle research (Mano et al. 2008). Initially, PODB consisted of three units: the organellome database, which was a compilation of static image data; the functional analysis database, which was a collection of protocols for plant organelle research; and external links to other relevant websites. The unique feature of PODB was that researchers in plant biology produced and directly contributed all of the imaging data and protocols presented. Using PODB, users were readily able to examine organelle dynamics. All organelles are motile, and their number, size, morphology, movement, subcellular position and interaction with other subcellular structures change in a dynamic fashion. Since the dynamic nature of organelles supports the integrated functioning of the entire plant (Hayashi and Nishimura 2009), we reasoned that it would be useful to incorporate movie data into our online database. Thus, PODB2, the revised version of PODB, contains movie data that may serve as a useful tool to expand our knowledge of plant organelle dynamics. Users of the organelle movie database can examine the dynamics of organelle(s) of interest, such as their movement, division, subcellular position(s) and response to external stimuli.
Because PODB2 was designed for use by scientists, it is difficult for members of the non-scientific community, such as students and school teachers, to understand the data presented in this website. To address this, we launched a new website, Plant Organelles World, in March 2010. This website is based on PODB2, and allows users to view images and movies of organelles. However, Plant Organelles World is written in layman's terms, and we avoided the use of technical language where possible. In addition, we include basic information on the importance of organelles and electron micrographs that show the ultrastructure of organelles in Plant Organelles World.
Here we describe the novel features of PODB2 and emphasize its improvements over the previous version, and introduce Plant Organelles World. PODB2 and Plant Organelles World can be freely accessed online at http://podb.nibb.ac.jp/Organellome and http://podb.nibb.ac.jp/Organellome/PODBworld/en/index.html, respectively. We expect that PODB2 and Plant Organelles World will enhance the understanding of plant organelles among members of the scientific and non-scientific community, respectively.
Contents of PODB2
PODB2 is publicly accessible, available at http://podb.nibb.ac.jp/Organellome/. In addition to containing all the information included in PODB (i.e. collections of static images, protocols and external links), PODB2 also houses an organelles movie database. Therefore, PODB2 consists of four individual units; namely, the organelles movie database, the organellome database, the functional analysis database and a compilation of external links to pertinent databases and homepages (Fig. 1A). Users can access each database page by clicking on links on the homepage (http://podb.nibb.ac.jp/Organellome/; red arrows in Fig. 1A) and at the top of each page in the database (blue arrow in Fig. 1A). The organellome database, the functional analysis database and the external links have previously been described in detail (Mano et al. 2008). Below, we introduce the new content of PODB2 (i.e. the organelles movie database) and highlight improvements over the previous version.
Fig. 1.
The graphical user interface for the homepage of PODB2 and the external links page. (A) Four individual parts, the organelles movie database, the organellome database, the functional analysis database and a compilation of external links, are included in PODB2. Users can access the three databases and external links by clicking on each term located on the updated homepage (red arrows) and at the top of each page in the database (blue arrow). (B) On the external links page, each link is categorized into seven subgroups. Each link is accessed by clicking on the square button to the left of the category name. The Japanese flag icon represents the link to the website that is depicted in Japanese.
The organelles movie database
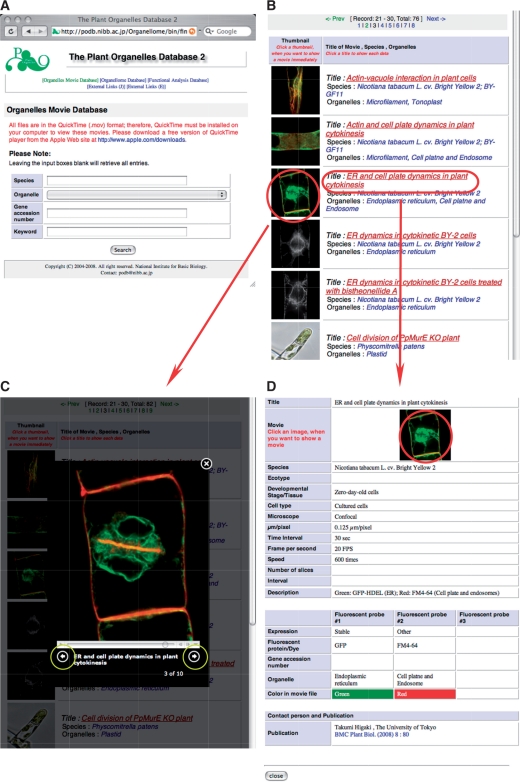
Like the organellome database, the organelles movie database is a fully searchable database. All movies are deposited in QuickTime format; therefore, the free application QuickTime Player (Apple Inc.) must be installed on a computer to view the movies. Users may access specific data via the ‘Search’ page (Fig. 2A), using four query parameters: species, organelle name, gene accession number and keyword. More complex searches can be performed by combining these different search parameters.
Fig. 2.
The graphical user interface of the organelle movie database. (A) A search window provides four query parameters. Leaving the input boxes empty retrieves all entries. (B) A results window consists of a table of thumbnail images of organelles as well as annotations. Clicking on the image prompts a movie to run in the same window, without showing a detailed information page (C). Clicking on the title (in red) allows access to the detailed information page (D), and clicking the forward or reverse arrow buttons, which are marked by yellow circles (C), allows users to view the following or previous movie without returning to the results page. (D) Clicking on the thumbnail in an information page (encircled in red) enlarges the movie as in (C) in this detailed information page.
Entries matching the query are displayed as a table with thumbnails of an image and the title of a movie, as well as the species and the name of the organelle(s) (Fig. 2B). Users can instantly view a movie on the same page by clicking on the thumbnail (Fig. 2C). The forward and reverse arrows (encircled in yellow in Fig. 2C) allow the following and previous movies, respectively, to be displayed without returning to the results page. Clicking on a movie title will open a page with data that relate to the movie (Fig. 2D), such as the plant material, experimental conditions and imaging parameters used, and the contact person (usually the scientist who provided the movie). Users can view the movie included on each data page by clicking on a thumbnail at the top of the page (red circle in Fig. 2D).
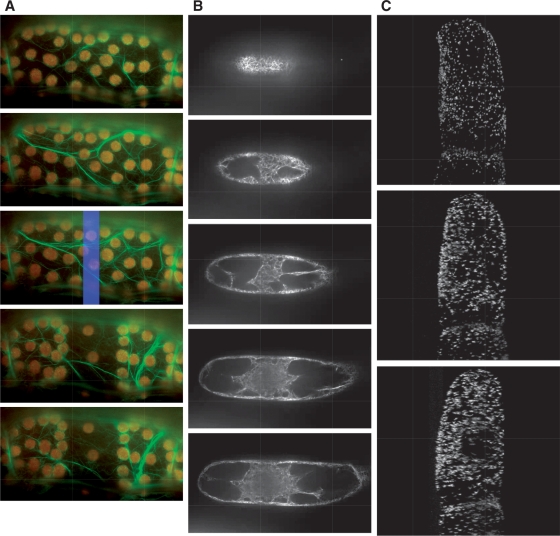
Currently, the organelles movie database houses three types of movie data: (i) time-lapse movies that show organelle movement (Fig. 3A and Supplemental Movie 1); (ii) serial images derived from different focal planes (Fig. 3B and Supplemental Movie 2); and (iii) movies that display the rotation of 3D structures (Fig. 3C and Supplemental Movie 3). For example, in these movies, users can observe the dynamic avoidance movement of chloroplasts in Arabidopsis petiole cells (Fig. 3A and Supplemental Movie 1; Kadota et al. 2009). In addition, as shown by serial images of microfilaments in suspension-cultured tobacco cells taken at different focal planes (Fig. 3B and Supplemental Movie 2), the spatial position of microfilaments can be visualized and these data can be used for stereological analysis (Higaki et al. 2007). Moreover, 3D structures that can be rotated, such as the one shown in Fig. 3C and Supplemental Movie 3 (Mano et al. 1999), enable a clear understanding of the stereological arrangement of organelles. In this way, users can query PODB2 to examine the dynamics of organelles, such as movement, division, subcellular position(s) and behavior in response to external stimuli at various developmental stages in different plant species.
Fig. 3.
Examples of submitted movies. Each image shows a snapshot from a movie that was downloaded from PODB2. From the top to the bottom, time proceeds (A and C), or the focal plane changes from the surface to the bottom of the cell (B). These movies are available as Supplementary Movies 1, 2 and 3, respectively, at PCP online. (A) Avoidance movement of chloroplasts in a petiole cell of an Arabidopsis adult leaf expressing GFP–talin. Green and red signals show GFP-labeled actin filaments and chloroplasts, respectively. The blue rectangle in the third image represents the photo-irradiation area. (B) Serial images of microfilaments, which are visualized with GFP–ABD2, in tobacco suspension-cultured cells taken at different focal planes at metaphase during cytokinesis. (C) Stereological arrangement of GFP-labeled peroxisomes in suspension-cultured tobacco cells.
The Plant Organelles World Website
We believe that it is both essential and important to promote curiosity in plant biology among members of the non-scientific community. Therefore, we launched a new website, Plant Organelles World, which was written in lay terminology, in March 2010. The first version was prepared in Japanese, and the English version was launched 4 months later. Each page in Plant Organelles World contains a link to change the display between Japanese and English. In addition, the link to PODB2 on each page of Plant Organelles World, and the addition of an ‘Organelles World’ link at the top of every page in PODB2 (blue arrow in Fig. 1A), allows users to toggle between these two websites.
Plant Organelles World contains four units of content: static images, movies, electron micrographs and images of the mutants that have defect(s) in organelle function(s). Some data were the same as those presented in PODB2. Electron micrographs and images of mutant plants with defects in organelles were added because we anticipated that users would be interested in this subject. As for PODB2, all movies are deposited in Plant Organelles World in QuickTime format; therefore, the free application QuickTime Player (Apple Inc.) must be installed on a computer to view the movies.
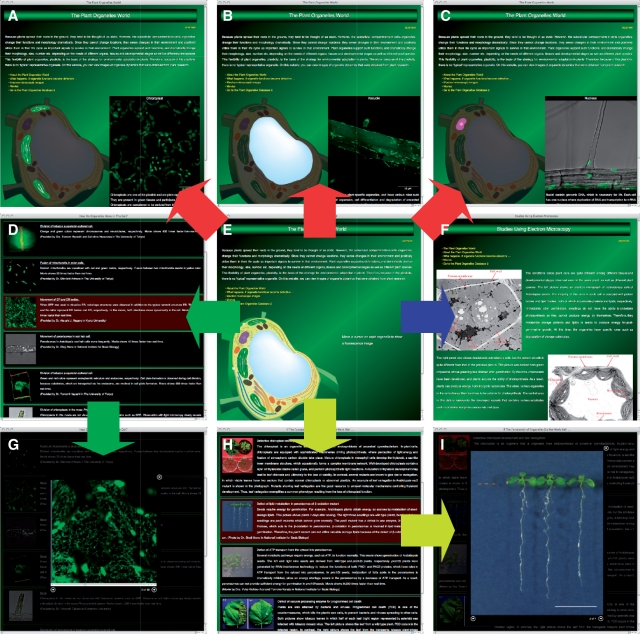
The homepage (Fig. 4E; http://podb.nibb.ac.jp/Organellome/PODBworld/en/index.html) contains a link to each unit, and also exhibits a fluorescence image of each organelle, which can be displayed by moving a cursor onto each organelle depicted in a cartoon of the plant cell (flowchart in red in Fig. 4). Using this system, users can readily navigate to an image and brief caption of an organelle (Fig. 4A–C).
Fig. 4.
Plant Organelle World graphical user interface. On the main window (E), users can view images of organelles by moving a cursor onto each organelle in the cell, as shown by the flowchart in red. (A–C) Representative organelles, i.e. chloroplasts, a vacuole and a nucleus. From the homepage, the user can move to pages containing movies (D and G; flowchart in green), electron microscopy images (F; flowchart in blue) and images or movies showing mutant plants that have a defect in organelle function (H and I; flowchart in yellow). To view the enlarged movie or image (G and I), the user clicks on the row containing the thumbnail and description of interest on the movie (D) or mutant plant (H) pages.
From the homepage, users can access pages for movies (flowchart in green in Fig. 4), electron micrographs (flowchart in blue in Fig. 4) and mutant plants (flowchart in yellow in Fig. 4). The entries in the movie page are displayed as a table with thumbnails of the images and titles of the movies, as well as a brief caption and the name(s) of scientist(s) who submitted the data (Fig. 4D). Users can view a movie in the same window by clicking on a thumbnail image (Fig. 4G). By clicking the forward and reverse arrows on the bottom of the movie window, the next and previous movies, respectively, can be displayed without closing the movie window.
The electron micrograph page contains images that were acquired by electron microscopy. The user can view the ultrastructure of plant cells and organelles, and appreciate the dynamic nature of plant organelles, even within the same tissue (Fig. 4F).
On the page that presents information on mutant plants that have defect(s) in organelle function(s), the entries are displayed as a table with thumbnails of an image, the title, a brief caption and the name(s) of the submitters (Fig. 4H). Users can view an image or movie in the same page by clicking on a thumbnail image (Fig. 4I). By clicking the forward and reverse arrow buttons on the bottom of the movie window, the next and previous images and movies, respectively, can be displayed without closing the movie window. On this page, users can view the morphology and phenotype of plants that have defective organelle function(s).
Redesign of Pages in PODB2
We improved almost all of the pages by redesigning the layout, debugging the programs, and developing compatibility with some web browsers such as Safari, Internet Explorer and Firefox. Whereas the homepage of the previous version contained a long introduction to the databases and a description of how to submit data, PODB2 has a concise introduction on the homepage, and the detailed information is presented on separate pages (Fig. 1A). Some fields were deleted on each page of the organellome database, as these fields were important identifiers for the deposition of data in the database, but not for the interface with users.
In PODB, two external links pages were prepared: one contained external links to both Japanese and English web pages, and the other contained external links to only English web pages. In PODB2, all links were presented in one group as a tree structure instead of relying on tabs, as in the previous version (Fig. 1B). Web pages that are in Japanese are marked with the Japanese flag icon (Fig. 1B). All links in the previous version were classified into three categories, i.e. Transcriptomics links, Proteomics links and Other links, under tabs on each page. To comply with the revised layout of external links in PODB2, these three categories were replaced with the following categories: Genomics, Transcriptomics, Proteomics, Organelles, Images, Analytical Tools and Others, and all links were categorized into these groups (Fig. 1B). Depending on requests from users, we will revise and add categories, and include additional links.
As stated above, the addition of a new ‘Organelles Movie Database’ link at the top of every page included in PODB2 allows users to access the ‘Search page’ of the organelles movie database directly (blue arrow in Fig. 1A).
In addition, the submission site of the organellome database in the previous version was improved to handle the submission of both static image files to the organellome database and movie files to the organelles movie database. Users who wish to contribute static images and movie data can submit data using the same submission site.
Data Submission
All data in PODB2 and Plant Organelles World were acquired and submitted by researchers in plant biology, and we welcome all submissions. We would like contributors to submit movie files to the organelles movie database via the web-based data submission system. Only registered users are able to log into the submission site using a username and password, which are common to both the organellome database for static image files and the organelles movie database. Therefore, contributors who already have a username and password for the organellome database in PODB can log into PODB2 using the same username and password. Researchers who have not yet registered with our site are asked to complete a one-time registration before submitting data. An application form can be downloaded as an MS Word file from the ‘Submit data to the Organelles Movie Database’ page (http://podb.nibb.ac.jp/Organellome/bin/expMovie.php) or ‘Submit data to the Organellome Database’ page (http://podb.nibb.ac.jp/Organellome/bin/expPicture.php). The username and password will be issued after the contributor has sent an application form to podb@nibb.ac.jp.
For movie file submission, we request that contributors prepare movie files in QuickTime format (.mov) and that they install the free QuickTime Player (Apple Inc.) application, as this application is required to confirm that a submitted movie can be viewed correctly in our system during the submission process. In addition to the movie file, one high-resolution image file [generated in Tiff format (.tif or .tiff)] is needed for submission and will be used to create the thumbnail.
To maintain the accuracy of the data deposited, submitted data are evaluated for quality prior to addition to the database. If the movie data submitted are considered to be of insufficient quality, such as low resolution, different file types and unsuitable file size (maximum 10 Mbyte in size), by the PODB steering committee members, we may ask the contributing researcher to make revisions and resubmit the material. Corrections, revisions or updates of submitted movie data can be made at any time, but users will not be able to correct, revise or update the data by themselves. In such cases, users should contact podb@nibb.ac.jp.
With regard to Plant Organelles World, we do not have the web-based submission system for this website, unlike PODB2. Therefore, we ask submitters to send data with a simple description directly. In addition, we select image and movie data from PODB2 whose subject users will be interested in, and use them to update Plant Organelles World. In such cases, we request an additional caption from submitters. On requests from users, we will add the additional data and/or contents.
PODB2 welcomes the submission of static image data to the organellome database, and of protocols to the functional analysis database in addition to movie data to the organelles movie database. Since its inception in 2006, the volume of static image data and protocols submitted to these databases has been increasing exponentially. However, we hope to continue to expand our databases, as the information contained therein, including bioimaging data such as 3D structures, will advance the state of plant organelle research and enhance the value of our database. Therefore, we would appreciate contributions of data and protocols from all plant researchers.
Discussion
Live cell images, which can be used to track the subcellular localization of labeled organelles over time and to show the subcellular position of organelles, portray the dynamic nature of plant organelles more clearly than do static images. Therefore, we reasoned that the incorporation of movie data would enhance the value of the database. Whereas PODB contained static image data, PODB2 was updated to include movie data.
PODB2
Although several databases other than PODB2 gather image data of plant organelles, most databases deal only with static data, and not with movie data (Cutler et al. 2000, Damme et al. 2004, Brown et al. 2005, Koroleva et al. 2005, Li et al. 2006). Although a wealth of movie data of plant organelles is presented on the websites of individual laboratory pages, these are not large-scale data. For example, ‘The Illuminated Plant Cell’ (http://www.illuminatedcell.com/; Mathur 2007) and ‘Plant Cell Dynamics’ (http://deepgreen.stanford.edu/) are well-organized and informative websites that provide both static image data and movie data for several organelles. In comparison with these two websites, PODB2 (i) presents movie data of organelles in various tissues at different developmental stages, even for the same organelle; (ii) contains movie data from various plant species; and (iii) shows the behavior of organelles in response to external stimuli.
The organelles movie database currently contains >80 movies. Of these movies, the majority of data in PODB2 are derived from Arabidopsis and tobacco suspension-cultured cells. We think that it is important to increase the amount of image data from other plant species, including crops, to contribute to applied science (Shinozaki and Sakakibara 2009) as well as basic science (International Arabidopsis Information Consortium 2010) as one of bioinformatics infrastructures. One of the unique features of the organelles movie database is to deal with movie data that are derived from various tissue types and culture conditions. For example, there are movies for peroxisome movement in leaf and root cells (Mano et al. 2002), and actin dynamics at various stages during cell division (Higaki et al. 2008). Besides these, there are movies that were taken in various tissues and under various conditions, such as mitochondrial movement in pollen cells (Miura et al. 2007), movement of Rubisco-containing bodies in dark-adapted leaves (Ishida et al. 2008) and vacuolar membrane dynamics under high salt conditions (Hamaji et al. 2009). Users can compare organelle dynamics in different conditions, even for the same organelle.
Plant Organelles World
The image and movie data in PODB2 are visually engaging, and contain educationally valuable information for members of the non-scientific community. It was therefore our belief that the data in PODB2 should be used to promote the awareness of plant organelles and the importance of plant biology, and this led to the construction of Plant Organelles World. Some databases provide image data information on individual organisms, including plants. For example, ImageBank (http://bio.ltsn.ac.uk/imagebank/), the Plants Database (http://plants.usda.gov/) and Plant Image Database (http://www4.albion.edu/plants/) contain image data from various organisms, including plant species. In addition, the Science and Plants for Schools website (SAPS; http://www-saps.plantsci.cam.ac.uk/) provides support to educators who teach plant science and molecular biology in schools and colleges. However, there were no databases for the general public that were dedicated to plant organelles and contained movie data. Therefore, Plant Organelles World is a unique and useful platform that can provide information about plant organelles to members of the non-scientific community via a visually engaging interface.
The image and movie data in Plant Organelles World are freely downloadable and may be used for educational purposes, such as learning and teaching. In such cases, we would like users to acknowledge the contributor and the copyright holder of the images and movies every time these data are used, because accurate and appropriate credit leads to improvement of the status and better management of the database.
Use of image and movie data for quantitative analysis
Image and movie data contain quantitative information, such as the size, length, morphology and velocity of objects. Therefore, PODB2 also represents a potentially rich data source for computational analyses. Some users may want to handle a variety of data deposited in PODB2, to calculate the size, length, number, etc. of organelles. At present, PODB2 is unable to download a large data set that contains image and movie data automatically. However, we can generate extra links for downloading large data sets in response to user requests.
Currently, there are no general protocols for image-based quantitative analysis that cover all plant organelles. Various aspects of imaging analysis, such as image processing, pattern recognition algorithms, machine learning and statistical modeling, have recently advanced (Coelho et al. 2010, Hu et al. 2010, Langhammer et al. 2010). Indeed, endoplasmic reticulum body mutants in Arabidopsis were isolated using image-based quantitative analysis (Nagano et al. 2009). In addition, high-throughput live cell imaging was developed to screen for human cells defective in cell division in a library generated by RNA interference (Held et al. 2010, Schmitz et al. 2010). We are currently collaborating with mathematical morphologists to establish organelle-specific algorithm(s) that can be used to extract organelle-specific information from image data for use in statistical analyses (Kimori et al. 2010). The information extracted from image data, together with other information such as expression data, can be incorporated into simulation models of plants in systems biology studies.
Future perspectives
As part of our maintenance of PODB2, we continuously update records of movie data, static images and protocols, as well as links to other websites. To facilitate the ongoing expansion of information, the organization and layout of PODB2 have been modified. We will, therefore, continue to improve the search function so that it can handle both the organellome database and the organelle movie database simultaneously, and we will modify the layout of pages to integrate gene expression data and metabolomics data. We are also working to develop the next version of this database, which will incorporate electron microscopy images, in order to compile detailed information of suborganellar structures. In addition, we plan to construct a system for image-based data mining to identify organelles in cases where users encounter organelles that have not been identified.
As for PODB2, we continuously update records of movie data, static images and electron micrographs in Plant Organelles World. To ensure that Plant Organelles World is readily accessible to members of the non-scientific community, we will improve the organization and layout of the pages therein, and add data and descriptions. We plan to add new content that shows 3D computer graphics that were generated from bioimaging data. These images will help users understand the structure and functioning of plant organelles via a visually engaging interface.
Finally, we aim to continue working with other biological databases and colleagues in the plant research and bioinformatics fields to distribute our data and provide pertinent links to external websites. Currently, high-throughput live cell imaging techniques are being developed to profile and identify mutants in mammalian cells (Held et al. 2010, Neumann et al. 2010, Schmitz et al. 2010). It is important for an ‘omics’ approach to combine the large data sets derived from high-throughput imaging experiments with other databases (Neumann et al. 2010). Since the high-throughput imaging techniques will have a profound impact on future plant organelle research, PODB2 and later versions of our database will be a valuable platform for imaging analysis in the field of plant biology. We believe that PODB2 and Plant Organelles World are easily accessible platforms for those who wish to explore the basics of plant biology, and who are interested in the field of plant organelle.
Design and implementation
The PODB2 engine is a Mac OS X server that runs FileMaker Server 8 Advanced. The web interface was also designed in the PHP server-side scripting language (version 4.4.9) and in the web-based JavaScript and XML languages. Movies in the organelles movie database are in QuickTime format (Apple Inc.). As in the previous version, each image in the organellome database in PODB2 is hyperlinked to a datum that displays more information and provides links to NCBI (Sayers et al. 2009).
Availability
PODB2 and Plant Organelles World can be accessed at the websites http://podb.nibb.ac.jp/Organellome and http://podb.nibb.ac.jp/Organellome/PODBworld/en/index.html, respectively, at the National Institute for Basic Biology. All data in PODB2 and Plant Organelles World are free for educational and academic use. We request that the submitter of data/protocols and PODB2 and Plant Organelles World be given proper credit in scientific publications and classroom presentations that use these data/tools. All technical questions and concerns, including comments, suggestions, revisions and updates of data, should be directed to podb@nibb.ac.jp.
PODB2 and Plant Organelles World provide biological information exclusively, and do not collect, generate or distribute genetic resources, such as the DNA, seeds and transgenic plants that were used to produce the data. To request these genetic resources, users should contact the submitter directly.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japanese Ministry of Education, Sports, Culture, Science, and Technology [Grant-in-Aid for Scientific Research of Priority Areas ‘Organelle Differentiation as the Strategy for Environmental Adaptation in Plants (No. 16085101)’]; the Japan Society for the Promotion of Science [Grant-in-Aid for Publication of Scientific Research Results (No. 228052)].
Acknowledgments
We would like to thank Ms. Yuko Kuboki and Chizuru Ueda at the Division of Cell Mechanisms, Department of Cell Biology, and Dr. Tomoko Kurata and Mr. Colin Kawaguchi at the Office of Public Relations and International Cooperation of the National Institute for Basic Biology for their assistance in proofreading sections written in English and in maintaining the website.
Glossary
Abbreviations
- GFP
green fluorescent protein
- PODB
Plant Organelles Database.
References
- Brown J.W.S., Shaw P.J., Shaw P., Marshall D.F. Arabidopsis nucleolar protein database (AtNoPDB) Nucleic Acids Res. 2005;33:D633–D636. doi: 10.1093/nar/gki052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho L.P., Peng T., Murphy R.F. Quantifying the distribution of probes between subcellular locations using unsupervised pattern unmixing. Bioinformatics. 2010;26:i7–i12. doi: 10.1093/bioinformatics/btq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl Acad. Sci. USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damme D.V., Bouget F.Y., Poucke K.V., Inzé D., Geelen D. Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 2004;40:386–398. doi: 10.1111/j.1365-313X.2004.02222.x. [DOI] [PubMed] [Google Scholar]
- Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Hamaji K., Nagira M., Yoshida K., Ohnishi M., Oda Y., Uemura T., et al. Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 2009;50:2023–2033. doi: 10.1093/pcp/pcp143. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Nishimura M. Frontiers of research on organelle differentiation. Plant Cell Physiol. 2009;50:1995–1999. doi: 10.1093/pcp/pcp161. [DOI] [PubMed] [Google Scholar]
- Held M., Schmitz M.H.A., Fischer B., Walter T., Neumann B., Olma M.H., et al. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods. 2010;7:747–754. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Hasezawa S. Quantitative analysis of changes in actin microfilament contribution to cell plate development in plant cytokinesis. BMC Plant Biol. 2008;8:80. doi: 10.1186/1471-2229-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Sano T., Hasezawa S. Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 2007;10:549–556. doi: 10.1016/j.pbi.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Hu Y., Osuna-Highley E., Hua J., Nowicki T.S., Stolz R., McKayle C., et al. Automated analysis of protein subcellular location in time series images. Bioinformatics. 2010;26:1630–1636. doi: 10.1093/bioinformatics/btq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Arabidopsis Information Consortium. An international bioinformatics infrastructure to underpin the Arabidopsis community. Plant Cell. 2010;22:2530–2536. doi: 10.1105/tpc.110.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., et al. Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008;148:142–155. doi: 10.1104/pp.108.122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J., Noel B., Policriti A., Clepet C., Casagrande A., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Kadota A., Yamada N., Suetsugu N., Hirose M., Saito C., Shoda K., et al. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:13106–13111. doi: 10.1073/pnas.0906250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimori Y., Baba N., Morone N. Extended morphological processing: a practical method for automatic spot detection of biological markers from microscopic images. BMC Bioinformatics. 2010;11:373. doi: 10.1186/1471-2105-11-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva O.A., Tomlinson M.L., Leader D., Shaw P., Doonan J.H. High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP–ORF fusions. Plant J. 2005;41:162–174. doi: 10.1111/j.1365-313X.2004.02281.x. [DOI] [PubMed] [Google Scholar]
- Langhammer C.G., Zahn J.D., Firestein B.L. Identification and quantification of skeletal myotube contraction and association in vitro by video microscopy. Cytoskeleton. 2010;67:413–424. doi: 10.1002/cm.20457. [DOI] [PubMed] [Google Scholar]
- Li S., Ehrhardt D.W., Rhee S.Y. Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol. 2006;141:527–539. doi: 10.1104/pp.106.078881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Hayashi M., Nishimura M. Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 1999;17:309–320. doi: 10.1046/j.1365-313x.1999.00378.x. [DOI] [PubMed] [Google Scholar]
- Mano S., Miwa T., Nishikawa S.-i., Mimura T., Nishimura M. The plant organelles database (PODB): a collection of visualized plant organelles and protocols for plant organelle research. Nucleic Acids Res. 2008;36:D929–D937. doi: 10.1093/nar/gkm789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Miwa T., Nishikawa S., Mimura T., Nishimura M. Seeing is believing: on the use of image databases for visually exploring plant organelle dynamics. Plant Cell Physiol. 2009;50:2000–2014. doi: 10.1093/pcp/pcp128. [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., Nishimura M. Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol. 2002;43:331–341. doi: 10.1093/pcp/pcf037. [DOI] [PubMed] [Google Scholar]
- Mathur J. The illuminated plant cell. Trends Plant Sci. 2007;12:506–513. doi: 10.1016/j.tplants.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Miura E., Kato Y., Matsushima R., Albrecht V., Laalami S., Sakamoto W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell. 2007;19:1313–1328. doi: 10.1105/tpc.106.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K., Shinozaki K. Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 2010;51:497–523. doi: 10.1093/pcp/pcq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano A.J., Maekawa A., Nakano R.T., Miyahara M., Higaki T., Kutsuna N., et al. Quantitative analysis of ER body morphology in an Arabidopsis mutant. Plant Cell Physiol. 2009;50:2015–2022. doi: 10.1093/pcp/pcp157. [DOI] [PubMed] [Google Scholar]
- Neumann B., Walter T., Hériché J.K., Bulkescher J., Erfle H., Conrad C., et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M.H.A., Held M., Janssens V., Hutchins J.R.A., Hudecz O., Ivanova E., et al. Live-cell imaging RNAi screen identifies PP2A-B55α and importin-β1 as key mitotic exit regulators in human cells. Nature Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Sakakibara H. Omics and bioinformatics: an essential toolbox for systems analyses of plant functions beyond 2010. Plant Cell Physiol. 2009;50:1177–1180. doi: 10.1093/pcp/pcp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidosis Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. The genome of black cottonwood, Populus trichocarpa. Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G.K.S., Li S., Liu B., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.