Abstract
Molecular breeding approaches are of growing importance to crop improvement. However, closely related cultivars generally used for crossing material lack sufficient known DNA polymorphisms due to their genetic relatedness. Next-generation sequencing allows the identification of a massive number of DNA polymorphisms such as single nucleotide polymorphisms (SNPs) and insertions–deletions (InDels) between highly homologous genomes. Using this technology, we performed whole-genome sequencing of a landrace of japonica rice, Omachi, which is used for sake brewing and is an important source for modern cultivars. A total of 229 million reads, each comprising 75 nucleotides of the Omachi genome, was generated with 45-fold coverage and uniquely mapped to 89.7% of the Nipponbare genome, a closely related cultivar. We identified 132,462 SNPs, 16,448 insertions and 19,318 deletions between the Omachi and Nipponbare genomes. An SNP array was designed to validate 731 selected SNPs, resulting in validation rates of 95 and 88% for the Omachi and Nipponbare genomes, respectively. Among the 577 SNPs validated in both genomes, 532 are entirely new SNP markers not previously reported between related rice cultivars. We also validated InDels on a part of chromosome 2 as DNA markers and successfully genotyped five japonica rice cultivars. Our results present the methodology and extensive data on SNPs and InDels available for whole-genome genotyping and marker-assisted breeding. The polymorphism information between Omachi and Nipponbare is available at NGRC_Rice_Omachi (http://www.nodai-genome.org/oryza_sativa_en.html).
Keywords: Deletion, DNA marker, Insertion, Next-generation sequencer, SNP, SNP array
Introduction
Progress in next-generation sequencing technologies allows entire genomes to be sequenced more efficiently and economically than ever before and provides the opportunity to discover numerous DNA polymorphisms throughout a genome. Although this approach requires resequencing and bioinformatic tools, millions of DNA polymorphisms such as single nucleotide polymorphisms (SNPs) and insertion–deletion polymorphisms (InDels) can be obtained by high-throughput methods. The extremely large volume of SNPs makes whole-genome genotyping and genome-wide association studies possible in animals and higher plants (Lee et al. 2008, Atwell et al. 2010, Huang et al. 2010, Yamamoto et al. 2010). It also provides us with a powerful tool for marker-assisted breeding and quantitative trait locus (QTL) analysis. Genes of scientific and agronomic interest can be isolated with molecular markers and subsequently identified as key genes.
Numerous types of DNA polymorphisms have been developed for use as DNA markers in genetic analysis (Jena and Mackill 2008). SNPs are based on direct detection of sequence-level polymorphisms and represent the most abundant DNA sequence variation present in most organisms, and their detection is expected to be easier in most single-copy regions of the genome than that of other DNA markers (Rafalski 2002). In some self-fertilizing crops such as rice (Oryza sativa L.) and soybean (Glycine max), only low levels of DNA polymorphism have been detected between cultivars due to their genetic relatedness. Generally, modern cultivars have been bred from the progeny of crossed hybrids between closely related cultivars, resulting in decreased genetic diversity (Yamamoto et al. 2010). To detect sequence polymorphisms within cultivars or individuals and improve them by molecular breeding approaches, more comprehensive technologies must be developed.
SNPs are of growing importance as DNA markers in crop genetic research (Ganal et al. 2009, McCouch et al. 2010). In the Oryza SNP project, genetic variations have been discovered within 20 rice cultivars and landraces using 160,000 SNPs distributed over 100 Mb of the rice genome, which were determined by resequencing microarrays (McNally et al. 2009). Introgression patterns of shared SNPs revealed breeding history and some regions associated with agronomic traits. Recently, whole-genome resequencing of the japonica rice cultivar Koshihikari, which is closely related to Nipponbare, was completed using high-throughput sequencers (Yamamoto et al. 2010). In total, 67,051 SNPs have been identified by comparisons between these two genomes. SNP array analysis of 151 japonica cultivars defined the genome-wide pedigree haplotypes of these cultivars. However, those SNPs were distributed unevenly on each chromosome, suggesting that some regions with low SNP frequencies might be conserved regions that share a common ancestral cultivar between Koshihikari and Nipponbare (Yamamoto et al. 2010). Since the uneven distribution of SNPs hinders the advanced analysis of rice QTLs and marker-assisted selection, the construction of extremely high-density maps of DNA markers is needed. To accomplish this, resequencing of additional japonica cultivars and identification of new DNA markers have been carried out (Nagasaki et al. 2010).
A next-generation sequencer can generate a massive amount of InDels as well as SNPs. Recently, the importance of InDels has increased as a DNA polymorphism contributing to genetic divergence. The utility of an InDel array for accurate mapping of recessive mutations has been demonstrated in Arabidopsis (Salathia et al. 2007). In addition to the use of InDel polymorphisms as a DNA marker, Liu et al. (2008) have developed a new strategy to identify 215 rice genes with alternative expression isoforms related to InDels between indica and japonica rice. Thus, the use of InDel polymorphisms may reinforce the value of massive sequences to obtain millions of DNA polymorphisms.
Although massive sequences can provide fine haplotype information among genetically related cultivars (Yamamoto et al. 2010), a landrace, an ancestral cultivar of current improved cultivars, seems valuable to obtain an increased number of DNA polymorphisms between landrace and improved cultivars. One rice landrace, Omachi, is also used for the production of Japanese rice wine, sake, and appears to differ from cooking rice cultivars such as Koshihikari and Nipponbare. For example, rice cultivars used strictly for sake brewing are characterized by a larger grain size than ordinary cooking rice and the presence of an opaque structure called the white core located in the center of rice grains (Yoshida et al. 2002).
In this study, we obtained a large number of SNPs and InDels between two rice cultivars, Omachi and Nipponbare, by using a next-generation sequencer. In addition to validation of SNPs by an SNP array, InDels were also partially validated by actual use as DNA markers, indicating that, like SNPs, genome-wide InDel polymorphisms were promising DNA markers. The information described here can be exploited in future studies to provide novel observations for rice genetics and breeding.
Results
In silico mapping of genome analyzer reads to the reference rice genome
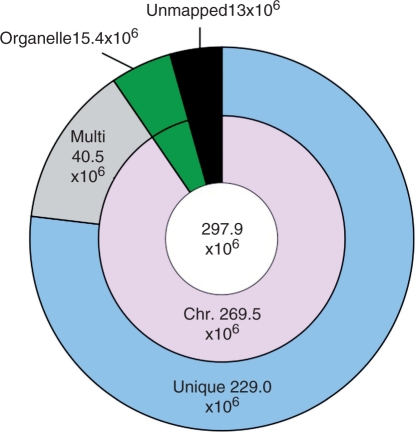
Whole-genome sequencing was performed on the genomic DNA of O. sativa L. cv. Omachi using a Genome Analyzer (GA), and 297,891,092 short reads of 75 nucleotides (297.9 × 106 reads henceforth) were generated by eight paired-end lanes. We mapped the short reads of the Omachi genome onto the Nipponbare genome (IRGSP Build 4) using Burrows–Wheeler alignment tool (BWA) software (Li and Durbin 2009), and 269.5 × 106 and 15.4 × 106 of the obtained reads were successfully mapped to chromosome and organelle genomes, respectively (Fig. 1). A total of 229.0 × 106 reads were uniquely mapped to chromosomes, corresponding to 17.2 Gb of the Nipponbare genome, and the remaining 40.5 × 106 reads were mapped to multiple locations (Fig. 1). The sequencing depth of the 229.0 × 106 reads varied from 40.7× the genome on chromosome 11 to 50.4× on chromosome 2, and averaged 45.0× across the entire genome (Supplementary Figs. S1, S2). The uniquely mapped reads were almost evenly distributed across the chromosomes of Nipponbare, as shown in Supplementary Fig. S1, and covered 342,646,566 bp (approximately 89.7%) of 382,150,945 bp of the Nipponbare genome.
Fig. 1.
Classification of Omachi reads mapped onto the Nipponbare genome. The total number of mapped reads (297.9 × 106) is in the center circle. The numbers of reads mapped onto chromosome and organelle genomes and unmapped reads are shown in the middle circle. The outer circle represents unique or multiple mapping on chromosomes.
The remaining regions of the Nipponbare genome not covered by any of the unique reads comprised two types of sequences: 23.7 Mb of sequences covered by reads mapped to multiple locations such as repeat sequences and 15.8 Mb of unmapped sequences. The unmapped regions included the following sequences: repetitive regions; undefined bases, dominant in the Nipponbare reference genome; sequences homologous to the chloroplast and mitochondrial genomes; and sequences containing multiple SNPs and long InDels between the Omachi and Nipponbare genomes.
Detection and distribution of SNPs and InDels between Omachi and Nipponbare
We detected a total of 84,473,707 SNPs and 2,143,594 InDels between the Omachi and Nipponbare genome sequences using BWA software with default parameters. Three filters were then applied to decrease the rate of false-positive SNPs and InDels: target depth of ≥5; target minimum mapping quality of 30, which implies that on average one per 1,000 reads was incorrectly identified; and a call rate of a polymorphism ≥90% for SNPs and 30% for InDels (Supplementary Fig. S2C). After filtering, we detected a total of 132,462 SNPs and 35,766 InDels (16,448 insertions and 19,318 deletions) between the Omachi and Nipponbare genomes (Table 1 and Supplementary Fig. S2B; http://www.nodai-genome.org/oryza_sativa_en.html).
Table 1.
Densities of SNPs and InDels on individual chromosomes detected between Omachi and Nipponbare genomes
| No. of SNPs | No. of insertions | No. of deletions | |
|---|---|---|---|
| Chromosome 1 | 14,436 (320.3) | 1,883 (41.8) | 1,926 (42.7) |
| Chromosome 2 | 10,103 (274.4) | 1,468 (39.9) | 1,621 (44.0) |
| Chromosome 3 | 5,877 (157.7) | 877 (23.5) | 992 (26.6) |
| Chromosome 4 | 27,576 (768.9) | 3,452 (96.3) | 4,371 (121.9) |
| Chromosome 5 | 5,715 (190.2) | 933 (31.1) | 1,091 (36.3) |
| Chromosome 6 | 15,107 (470.3) | 1,489 (46.3) | 1,690 (52.6) |
| Chromosome 7 | 12,806 (421.8) | 1,452 (47.8) | 1,686 (55.5) |
| Chromosome 8 | 10,682 (374.4) | 1,222 (42.8) | 1,447 (50.7) |
| Chromosome 9 | 2,902 (121.7) | 411 (17.2) | 490 (20.6) |
| Chromosome 10 | 4,405 (186.2) | 659 (27.9) | 756 (31.9) |
| Chromosome 11 | 17,258 (559.8) | 1,759 (57.1) | 2,102 (68.2) |
| Chromosome 12 | 5,595 (201.6) | 843 (30.4) | 1,146 (41.3) |
| Total | 132,462 (346.6) | 16,448 (43.0) | 19,318 (50.5) |
The numbers in parentheses show the average numbers of SNPs and InDels described as the number per 1 Mb genome sequence.
To confirm whether the sequencing depth of the output GA reads was sufficient for genome-wide detection of inherent SNPs and InDels, we analyzed the relationship between sequencing depth and the number of SNPs and InDels detected. The number of mapped reads increased in a linear fashion across eight lanes in which sequencing depth increased from 5.0× in the first lane to 10.8× in the second lane, 16.6× in the third lane, 22.3× in the fourth lane, 28.2× in the fifth lane, 34.0× in the sixth lane, 39.7× in the seventh lane and 45.0× in the eighth lane. In contrast, genome coverage only slightly increased across lanes (Supplementary Fig. S2A). The number of SNPs detected by BWA was calculated in every lane. The number of SNPs and InDels increased markedly up to a sequencing depth of 22.3× the genome (lane 4) and then at a slower rate (Supplementary Fig. S2B). This finding suggested that a 45.0× sequencing depth was sufficient to detect SNPs and InDels distributed throughout the genome and that nearly all SNPs and InDels were detected by this method.
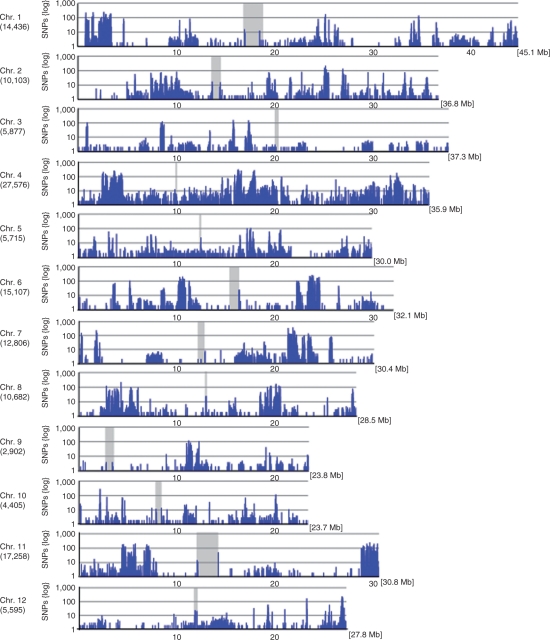
The average densities of detected polymorphisms between the Omachi and Nipponbare genomes were 346.6 Mb−1 (SNPs), 43.0 Mb−1 (insertions) and 50.5 Mb−1 (deletions) in the Omachi genome (Table 1). The number of SNPs per 1 Mb varied across individual chromosomes. Chromosome 4 had the highest density of SNPs, 768.9 Mb−1, and chromosome 9 had the lowest SNP density, 121.7 Mb−1 (Table 1). Fig. 2 shows the chromosomal distribution of the SNPs per 0.1 Mb. The distribution of SNPs was uneven within chromosomes. Sixty-six high-density regions with >500 SNPs per Mb and 27 low-density regions with <10 SNPs per Mb were identified. All chromosomes except chromosome 4 were composed of a mixture of dense and sparse SNP regions (Fig. 2). For example, on chromosome 1, SNPs were dense from the region of 2.5–3.0 Mb (2,874 SNPs) but sparse from 4.7 to 6.5 Mb (26 SNPs) and from 27 to 33 Mb (92 SNPs). On chromosome 9, the region from 11.1 to 12.4 Mb contained 1,841 SNPs, but the region from 1 to 10 Mb contained only 122 SNPs. Similarly, on chromosome 12, the region from 27 to 27.5 Mb contained 2,753 SNPs, but the region from 2.9 to 9.8 Mb had only 18 SNPs.
Fig. 2.
Distribution of SNPs between Omachi and Nipponbare in the 12 rice chromosomes. The x-axis represents the physical distance along each chromosome, split into 100 kb windows. The total genome size of each chromosome is shown in brackets. The y-axis indicates the number of SNPs. The total SNP number in each chromosome is shown in parentheses. The gray areas show centromere regions.
The above high SNP density regions on chromosome 1 and 12 have also been detected between Koshihikari and Nipponbare rice (Yamamoto et al. 2010). However, unlike our present results, the region from 11.1 to 12.4 Mb on chromosome 9, which we found to have a high SNP density, contained only a few SNPs between Koshihikari and Nipponbare, and the regions from 27 to 33 Mb of chromosomes 1, in which SNPs were sparse in our study, had a high SNP density between Koshihikari and Nipponbare. These results suggested that different SNP sets, such as those between Omachi and Nipponbare and between Nipponbare and Koshihikari, may be useful to identify the SNPs distributed over all chromosomes. The distribution patterns of InDel densities were similar to those of SNPs (Supplementary Fig. S3).
Annotation of SNPs and InDels
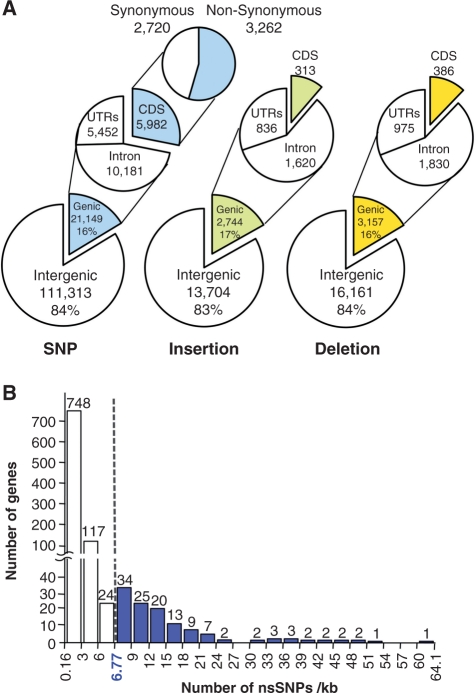
A large number of SNPs and InDels were annotated with The Rice Annotation Project Database release 2 (RAP-DB2) (see Materials and Methods) to assign the 132,462 SNPs and 35,766 InDels detected with a gene. Although 21,149 SNPs (16% of the total), 2,744 insertions (17% of the total) and 3,157 deletions (16% of the total) occurred in a gene region, only 5,982 SNPs, 313 insertions and 386 deletions were located in coding sequences (Fig. 3A). Among these 5,982 SNPs, 3,262 SNPs (55%) were non-synonymous in 1,017 genes (Fig. 3A). The number of non-synonymous SNPs (nsSNPs) per 1,000 bp of each gene had a large distribution (Fig. 3B), varying widely from a minimum of 0.16 to a maximum of 64.1, with a mean of 3.6. Using the five-number summary of the box-and-whisker plot to calculate outlier values, 128 genes were classified as outliers, having >6.77 SNPs per kb (Fig. 3B). They deviated markedly from the majority of genes, demonstrating the biased occurrence of SNPs in genes.
Fig. 3.
Annotation of SNPs and InDels and distribution of SNPs. (A) SNPs, insertions and deletions on the IRGSP rice pseudomolecules were classified as genic and intergenic, and locations within the gene models were annotated. The number of SNPs, insertions and deletions in each class is shown. (B) The degree of distribution and skewness of the nsSNP number per 1 kb of the 1,017 genes that were annotated by the 3,262 nsSNPs. The outlier value calculation indicated that 128 genes (blue bars) had an nsSNP number >6.77 per 1 kb in a gene.
Utilization of SNPs as DNA markers on an array
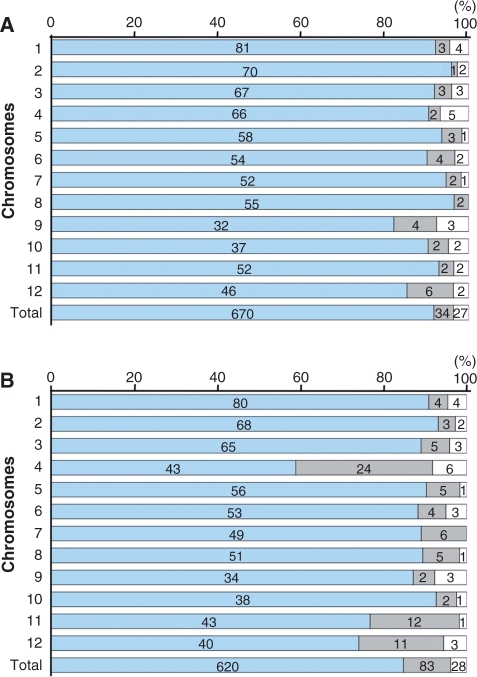
The SNPs identified by GA analysis were expected to be useful for genome-wide genetic analysis. To evaluate the quality of the detected SNPs for a genotyping system, we selected 731 SNPs at a spacing of approximately 500 kb (Supplementary Table S1), and designed oligonucleotides for an array containing the selected SNPs, which were widely distributed across the Omachi genome; we analyzed this array on an Illumina Bead Station 500G system. Genomic DNA from Omachi and Nipponbare produced fluorescence signals for 704 and 703 of the 731 selected SNPs, whereas no signals were detected for the remaining 27 and 28 SNPs, respectively (Fig. 4). Of the 704 SNPs, 670 SNPs matched the Omachi sequence obtained by GA analysis, and 34 SNPs mismatched (Fig. 4A), indicating that the SNP validation rate was approximately 95%. In contrast, for the Nipponbare genome, the SNP validation rate was 88%, with 620 SNPs matched to the Nipponbare reference sequence and 83 SNPs mismatched (Fig. 4B).
Fig. 4.
Validation of genome-wide SNPs on an array. The genomic DNA of Omachi (A) and Nipponbare (B) was analyzed with an array consisting of 731 selected SNPs using the Illumina Bead Station 500G system. The numbers of SNPs in blue and gray boxes represent matched and mismatched sequences of Omachi (A) and Nipponbare (B), respectively. White boxes represent the number of SNPs without signals.
The number of mismatched SNPs varied across individual Nipponbare chromosomes, and their highest concentration was observed on chromosomes 4, 11 and 12 (Fig. 4B). Among the above SNPs matched to Omachi and/or Nipponbare sequences, 577 SNPs produced signals in analyses of both genomes, indicating that they were promising as DNA markers. They were distributed over the genome with an average of 48 SNPs per chromosome, ranging from a maximum of 74 at chromosome 1 to a minimum of 30 at chromosome 9 (Fig. 5). Thus, the selected SNPs provided quality oligonucleotides for an SNP genotyping system.
Fig. 5.
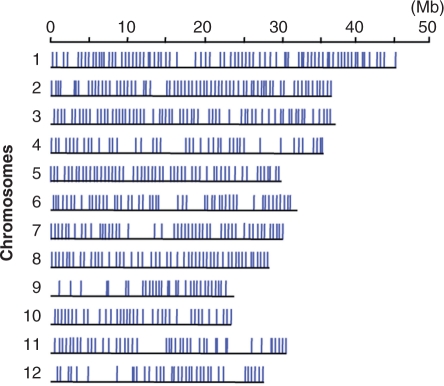
Distribution of SNPs validated on an SNP array. The positions of 577 SNPs that produced signals with both Omachi and Nipponbare genomic DNA are represented by blue lines. The x-axis represents the physical distance along each chromosome.
The possible use of InDels as DNA markers
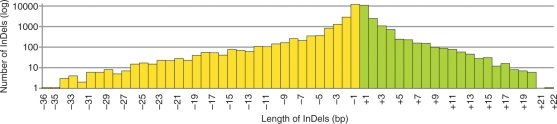
Whole-genome sequencing by GA allowed us to detect InDel polymorphisms as well as SNPs (Table 1). A total of 35,766 InDels including 16,448 insertions and 19,318 deletions were detected between Omachi and Nipponbare genomes by GA analysis. As shown in Fig. 6, the length of InDels was distributed from 1 to 36 bp. The majority of InDels (64%) were 1 bp mononucleotide insertion–deletions, 26% were 2–4 bp and 10% were ≥5 bp.
Fig. 6.
Distribution of the length of InDels. The x-axis shows the number of nucleotides of insertion (green) and deletion (yellow). The y-axis shows the number of InDels at each length.
To verify the availability of InDels as a new DNA marker, we focused on the region from 30 to 36.8 Mb on chromosome 2 as an example. We selected 20 InDels ≥5 bp in length from this region and sequenced them using a capillary sequencer. Although the presence of all these InDels in the genome was confirmed as expected, nine InDel sequences were mismatched to the sequence obtained by GA analysis (Supplementary Table S2). A detailed analysis of these nine InDels revealed that two showed base substitutions, three showed long insertions or deletions and four contained both mismatch types (Supplementary Table S2). However, because no InDels were shorter than their predicted GA sequences, all tested InDels were successfully amplified by PCR.
To investigate whether these InDels were specific to the Omachi genome, we sequenced the amplified fragments from the genomes of three other japonica cultivars: Koshihikari, Norin 8 and Kameji. Fragments containing 18 of the 20 InDels were successfully amplified by all three genomes (Supplementary Fig. S4). One InDel-containing fragment showed multiple bands in the Kameji genome, and another did not stably produce an amplified band in any of the three cultivar genomes. The diversity of the 18 InDels across the three cultivar genomes was highest between Omachi and Norin 8, where 17 InDels were polymorphic compared with Omachi. Koshihikari, one of the most popular japonica rice cultivars, and Kameji, a landrace cultivar, showed mixed patterns with Omachi and Nipponbare, with eight of 19 InDels and nine of 18 InDels, respectively, the same as Omachi (Supplementary Fig. S4). This result provided strong support that the InDels detected by GA analyses may be widely applicable as DNA markers among rice cultivars.
Discussion
In this study, we generated comprehensive SNP and InDel data from the closely related rice cultivars Omachi and Nipponbare. The discovery of DNA markers between two major groups of rice in Asia, japonica and indica, is fairly easy because their genetic diversity is quite large. However, the discovery of DNA markers within japonica rice cultivars is more difficult, due to their significantly lower diversity than indica rice cultivars (Yang et al. 1994). Moreover, modern japonica rice cultivars such as Nipponbare and Koshihikari have been bred through selection among the progeny of crossed hybrids between related cultivars, leading to an observed decrease in their genetic diversity (Yamamoto et al. 2010). Recurrent selection and crossing between related cultivars were common in self-fertilizing crops. This fact accrued limitation of resources for breeding in rice and other self-fertilizing crops. Accumulation and saturation of available genetic markers between related cultivars or individuals are essential for the efficient breeding and genetic research of crops. The increased availability of DNA markers provides a powerful tool to advance marker-assisted selection and QTL analysis and to identify novel genes for important functions of rice cultivars.
Whole-genome sequencing of Omachi, a japonica rice landrace, was accomplished, and in silico mapping of the reads to the reference Nipponbare genome was performed using BWA software. Uniquely mapped reads covered almost 90% of the Nipponbare genome sequence; 6.2% was covered by multimapped reads due to conserved domains and repeat sequences. The remaining 4.1% was not covered by any reads, possibly due to large deletions. We detected 132,462 SNPs between Omachi and Nipponbare, with an average density of 346.6 SNPs per Mb (Table 1), which is twice the SNP density between Nipponbare and Koshihikari cultivars. This high SNP density between Omachi and Nipponbare resulted in increased coverage for regions of extremely low SNP density between Koshihikari and Nipponbare. For example, we identified 76 SNPs in the region from 6.5 to 9.5 Mb on chromosome 1, 102 SNPs in the region from 12 to 14 Mb on chromosome 2 and 514 SNPs in the region from 2.5 to 3 Mb on chromosome 8, where no SNPs were observed between Koshihikari and Nipponbare (Yamamoto et al. 2010). However, some low SNP density regions shared between Koshihikari and Nipponbare and between Omachi and Nipponbare remain, for example the region from 1 to 4.5 Mb of chromosome 9 (Fig. 2). These common low SNP density regions seem to include highly conserved sequences among the three cultivars, Omachi, Koshihikari and Nipponbare, or the unmapped Nipponbare sequence.
SNP arrays allow thousands of markers to be genotyped in parallel and are also applicable to other fields such as marker-assisted breeding, positional cloning and genomic selection (Meuwissen et al. 2001, Zhong et al. 2009). To evaluate the quality of SNPs identified between Omachi and Nipponbare for genotyping, we prepared an array consisting of 731 SNP sites across the Omachi genome, which indicated that 577 SNPs were potential DNA markers (Fig. 5). A high-throughput typing array consisting of 1,917 SNP sites between Nipponbare and Koshihikari was used to genotype 151 japonica cultivars and reveal their pedigree information (Yamamoto et al. 2010). Because only 45 of our 577 SNPs overlapped with the 1,917 SNPs investigated previously, our SNP array may provide additional resolution of rice genotyping. For example, 16 and 18 SNPs were identified on the regions of 9.2–17.6 Mb of chromosome 5 and 11.3–21.0 Mb of chromosome 9, respectively, where they contained no SNP markers in the previous report (Supplementary Table S1). Our results also highlight the usefulness of landraces for the identification of more genetic polymorphisms with GA analysis.
Recently, InDels have become increasingly important sources of genetic variation. InDel polymorphisms were cost-efficiently applied to QTL mapping in salmon (Vasemägi et al. 2010). Our genome-wide analyses with BWA software also allowed us to identify a large amount of InDels in addition to SNPs. To validate InDels as new DNA markers, we focused on 20 InDels selected from the 6.8 Mb region of chromosome 2 and sequenced them with Sanger sequencing. Mismatches between Sanger and GA sequences were found in nine InDels, four insertions and five deletions (Supplementary Table S2). Krawitz et al. (2010) used the short-read mapping tools of BWA and Novoalign (http://www.novocraft.com/) to demonstrate that a short sequence read containing an InDel might be aligned with mismatched bases instead of gaps, resulting in a higher rate of variant bases at InDel positions. The mismatched deletions in our study might also have resulted from alignment with mismatched bases instead of gaps, leading to deleted nucleotide lengths shorter than those observed by Sanger sequencing and successful PCR amplification of InDel-containing fragments. The locations of tested InDel markers were also confirmed in other japonica rice cultivars, Nipponbare, Koshihikari, Norin 8 and Kameji. Thus, InDels with short sequences (5–18 bp) appear promising as DNA markers similar in form to microsatellite length polymorphisms.
A greater number of SNPs and InDel markers by GA could prove to be a powerful tool for narrowing the range of QTLs. Yoshida et al. (2002) identified the QTL for grain characters by using a doubled haploid population derived from a cross between rice cultivars, Yamada-nishiki and Reiho, with 145 markers that consisted of random amplified polymorphic DNA, amplified fragment length polymorphism and simple sequence repeats. One Yamada-nishiki allele at this QTL that was detected by the marker B01482 on chromosome 11 was found to have the greatest effect on grain length. Yamada-nishiki, as well as Omachi and one of its progeny, are sake brewing cultivars with large grain size. The Omachi region corresponding to the above-mentioned QTL has 55 genes, which contains a total of 308 non-synonymous SNPs and 21 InDels. These genes seem to include a promising candidate gene for the grain length QTL. This kind of information obtained by GA will be inevitable to define a candidate gene for a QTL. Furthermore, these SNPs and InDels themselves will be an additional source of genetic markers in fine-scale linkage mapping. Thus, information on polymorphisms between Omachi and Nipponbare sequences would be valuable for narrowing the QTL region and identifying functional genes.
In our analyses, some results remain unexplainable. Although we detected 3,262 SNPs and 699 InDels as mutations that would affect gene functions, 267 InDels (38.2%) were located on chromosome 4 (data not shown). The densities of detected SNPs and InDels on chromosome 4 were 2.3-fold higher than the total genome densities (Table 1). Moreover, in the SNP array analysis of the Nipponbare genome, 24 of 67 SNPs (35.8%) located on chromosome 4 were mismatched to the Nipponbare reference sequence (Fig. 4). These results induce a sense of uncertainty regarding the reference sequences of chromosome 4. In fact, sequence misassemblies have been reported in all chromosomes of the IRGSP and TIGR (The Institute for Genomic Research) (Zhou et al. 2007). Possible false-positive SNPs and InDels may in part be due to misassembled Nipponbare sequences. An increase in quality reference sequences is needed to make more efficient use of next-generation sequencing technology.
We obtained very large numbers of SNPs and InDels between related rice cultivars. Because related cultivars are commonly used for crossing in crop breeding, DNA polymorphisms between them provide tools to advance the study of genetic diversity and molecular breeding. The obtained SNPs and InDels will be applied to QTL analysis and marker-assisted breeding in rice.
Materials and Methods
Library construction and sequencing
Genomic DNA was extracted from 10 O. sativa L. cv. Omachi plants (Rice Genome Resource Center, NIAS, ID: JRC 32, http://www.gene.affrc.go.jp/databases-core_collections_jr.php) with Nucleon PhytoPure (GE Healthcare BioSciences) and used for the preparation of a GA sequencing library according to the manufacturer's protocols (Illumina). Fragments of the library were paired-end sequenced using Genome Analyzer II (Illumina). The length of all sequences generated was 75 nucleotides. The GA reads of the Omachi genome have been submitted to DDBJ (http://www.ddbj.nig.ac.jp/index-e.html) under accession No. DDBJ: DRA000307.
Sequence mapping and SNP/InDel identification
Filtering rules were applied to select reads that were mapped to the rice chromosomal genome (O. sativa L. cv. Nipponbare, Pseudomolecules Build 4.0, http://rgp.dna.affrc.go.jp/E/IRGSP/Build4/build4.htm, International Rice Genome Sequencing Project 2005) using ELAND (optional software for the Illumina GA pipeline system ver. 1.4). Mapping to the chromosomal and organelle genomes (O. sativa ssp. japonica group chloroplasts and Nipponbare mitochondria, with DDBJ accession Nos. X15901 and DQ167400) was performed with a BWA software (ver. 0.5.1) algorithm allowing two mismatches from the first to 35th bases of the read and three mismatches in total in the sequence (Li and Durbin 2009). BWA outputs were analyzed by SAMtools software (Li et al. 2009). For SNP and InDel discovery, the algorithm implemented in SAMtools considered only reads that aligned to a unique location of the Nipponbare genome. We integrated the physical positions of Omachi sequences, including SNP and InDel information, into the annotated RAP-DB2 (Ohyanagi et al. 2006, Itho et al. 2007; Rice Annotation Project 2008; http://rapdb.dna.affrc.go.jp/) using the Generic Genome Browser (Stein et al. 2002). These data are available at a GBrowse, NGRC_Rice_Omachi (http://www.nodai-genome.org/oryza_sativa_en.html).
Annotation of SNPs and InDels
GFF files including positional information about the SNPs and InDels were constructed and annotated with a data set of RAP-DB2 transcripts (representative cDNA) downloaded from the GBrowse component of RAP-DB2. The gene region, which consisted of rep-UTR5′, rep-CDS and rep-UTR3′, was listed in the data set along with data including positions, directions and descriptions, which we used for annotations. SNPs and InDels in the gene region and other genome regions were annotated as genic and intergenic, respectively. The genic SNPs and InDels were classified as rep-CDS (coding sequences), rep-UTR5′ and rep-UTR3′ (untranslated regions), or other (introns) according to those positions. SNPs in the coding sequences were separated into synonymous and non-synonymous amino acid substitutions. The data are available at http://www.nodai-genome.org/oryza_sativa_en.html.
Validation of SNPs and InDels
Genomic DNAs were extracted from leaves of O. sativa L. cv. Omachi, Nipponbare, Koshihikari, Norin8 and Kameji with Nucleon PhytoPure (GE Healthcare BioSciences). Validation of detected SNPs was performed with the Illumina Golden Gate detection system (Illumina). The adaptability of detected SNPs to the system was scored using the Illumina online scoring system (https://icom.illumina.com). After scoring the SNPs and their neighboring sequences, SNPs with a score >0.4 were selected to design an SNP array for the Illumina GoldenGate BeadArray technology platform. We used 5 μl of genomic DNA (50 ng μl−1) in the SNP analysis. These SNPs were detected using the Illumina Bead Station 500G system. All experimental procedures for SNP typing followed the manufacturer's instructions (Illumina). Validation of detected InDels was performed with an ABI 3730xl sequencer (Applied Biosystems) according to the manufacturer's protocols.
Supplementary data
Supplementary data are available at PCP online.
Funding
This study was supported by the Ministry of Education, Culture, Sports, Science and Technology [Grants-in-Aid for Scientific Research (S0801025)].
Acknowledgments
We thank S. Sano for helping with informatic analyses.
Glossary
Abbreviations
- BWA
Burrows–Wheeler alignment tool
- GA
Genome Analyzer
- InDel
insertion–deletion
- nsSNP
non-synonymous single nucleotide polymorphism
- QTL
quantitative trait locus
- RAP-DB2
The Rice Annotation Project Database release 2
- SNP
single nucleotide polymorphism.
References
- Atwell S., Huang Y.S., Vilhjálmsson B.J., Willems G., Horton M., Li Y., et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal M.W., Altmann T., Röder M.S. SNP identification in crop plants. Curr. Opin. Plant Biol. 2009;12:211–217. doi: 10.1016/j.pbi.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Itho T., Tanaka T., Barrero R.A., Yamasaki C., Fujii Y., Hilton P.B., et al. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena K.K., Mackill D.J. Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 2008;48:1266–1276. [Google Scholar]
- Krawitz P., Rödelsperger C., Jäger M., Jostins L., Bauer S., Robinson P.N. Microindel detection in short-read sequence data. Bioinformatics. 2010;26:722–729. doi: 10.1093/bioinformatics/btq027. [DOI] [PubMed] [Google Scholar]
- Lee S.H., van der Werf J.H., Hayes B.J., Goddard M.E., Visscher P.M. Predicting unobserved phenotypes for complex traits from whole-genome SNP data. PLoS Genet. 2008;4:e1000231. doi: 10.1371/journal.pgen.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Xu W., Tan L., Xue Y., Sun C., Su Z. Case study for identification of potentially indel-caused alternative expression isoforms in the rice subspecies japonica and indica by integrative genome analysis. Genomics. 2008;91:186–194. doi: 10.1016/j.ygeno.2007.10.001. [DOI] [PubMed] [Google Scholar]
- McCouch S.R., Zhao K., Wright M., Tung C., Ebana K., Thomson M., et al. Development of genome-wide SNP assays for rice. Breed. Sci. 2010;60:524–535. [Google Scholar]
- McNally K.L., Childs K.L., Bohnert R., Davidson R.M., Zhao K., Ulat V.J., et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl Acad. Sci. USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T.H., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H., Ebana K., Shibaya T., Yonemaru J., Yano M. Core single-nucleotide polymorphisms—a tool for genetic analysis of the Japanese rice population. Breed. Sci. 2010;60:648–655. [Google Scholar]
- Ohyanagi H., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:741–744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002;5:94–100. doi: 10.1016/s1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- Rice Annotation Project. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathia N., Lee H.N., Sangster T.A., Morneau K., Landry C.R., Schellenberg K., et al. Indel arrays: an affordable alternative for genotyping. Plant J. 2007;51:727–737. doi: 10.1111/j.1365-313X.2007.03194.x. [DOI] [PubMed] [Google Scholar]
- Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasemägi A., Gross R., Palm D., Paaver T., Primmer C.R. Discovery and application of insertion–deletion (INDEL) polymorphisms for QTL mapping of early life-history traits in Atlantic salmon. BMC Genomics. 2010;11:156. doi: 10.1186/1471-2164-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Nagasaki H., Yonemaru J., Ebana K., Nakajima M., Shibaya T., et al. Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics. 2010;11:267. doi: 10.1186/1471-2164-11-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G.P., Maroof M.A., Xu C.G., Zhang Q., Biyashev R.M. Comparative analysis of microsatellite DNA polymorphism in landraces and cultivars of rice. Mol. Gen. Genet. 1994;245:187–194. doi: 10.1007/BF00283266. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Ikegami M., Kuze J., Sawada K., Hashimoto Z., Ishii T., et al. QTL analysis for plant and grain characters of sake-brewing rice using a doubled haploid population. Breed. Sci. 2002;52:309–317. [Google Scholar]
- Zhong S., Dekkers J.C., Fernando R.L., Jannink J.L. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a barley case study. Genetics. 2009;182:355–364. doi: 10.1534/genetics.108.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Bechner M.C., Place M., Churas C.P., Pape L., Leong S.A., et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8:278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.