Abstract
The developmental potential of stem cells and progenitor cells must be functionally distinguished to ensure the generation of diverse cell types while maintaining the stem cell pool throughout the lifetime of an organism. In contrast to stem cells, progenitor cells possess restricted developmental potential, allowing them to give rise to only a limited number of post-mitotic progeny. Failure to establish or maintain restricted progenitor cell potential can perturb tissue development and homeostasis, and likely contributes to tumor initiation. Recent studies using the developing fruit fly Drosophila larval brain have provided molecular insight into how the developmental potential is restricted in neural progenitor cells.
Introduction
Restricted developmental potential allows progenitor cells to generate a limited number of terminally-differentiated progeny, amplifying the output of stem cells while safeguarding the stem cell pool throughout the natural lifespan of an organism. Expanded progenitor cell potential might result in the formation of aberrant stem-like cells, contributing to developmental defects and possibly tumor initiation. In contrast to stem cells, how progenitor cell potential is restricted remains largely unknown due to their short-lived nature. The fruit fly Drosophila larval brain, which consists of the central brain and optic lobe, possesses well-defined lineages of neural stem cells that generate progenitor cells in a highly reproducible pattern (Figure 1). These lineages provide an excellent in vivo system for studying regulation of the progenitor cell potential at a single-cell resolution. Conservation in gene function between flies and mammals suggests that molecular mechanisms that regulate progenitor cell potential in Drosophila neural stem cell lineages might be similarly employed during vertebrate neurogenesis.
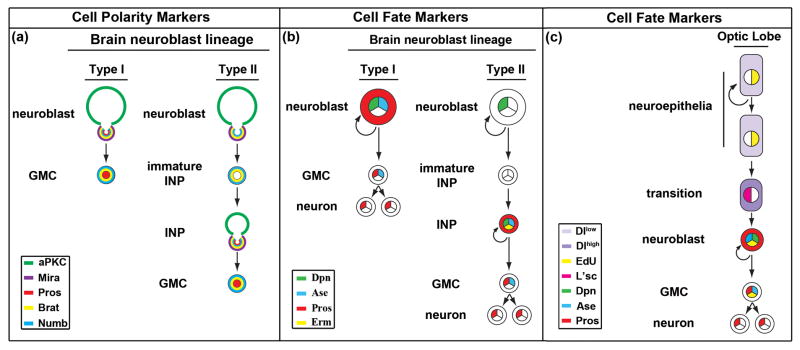
Figure 1.
Neural stem cell lineages in the developing Drosophila larval brain. (a) The apical and basal protein complexes unequally segregate during asymmetric divisions of neural stem/progenitor cells in the type I and type II neuroblast lineage in the larval brain. Abbreviation: aPKC: atypical Protein Kinase C; Mira: Miranda; Pros: Prospero; Brat: Brain tumor. (b) The cell fate markers allow unambiguous identification of neural stem/progenitor cells in the type I and type II neuroblast lineage in the larval brain. Abbreviation: Dpn: Deadpan; Ase: Asense; Pros: Prospero; Erm: Earmuff. (c) The cell fate markers allow unambiguous identification of neuroepithelial stem cells and progenitor cells in the optic lobe. Abbreviation: Dl: Delta; EdU: 5-ethynyl-2′-deoxyuridine; L’sc: Lethal of scute; Dpn: Deadpan; Ase: Asense; Pros: Prospero.
Central brain neuroblasts generate neural progenitor cells with distinct developmental potential
All neural stem cells in the central brain (called neuroblasts) undergo repetitive asymmetric divisions to self-renew and to generate a neural progenitor cell with limited developmental potential. The cortex of a mitotic central brain neuroblast is highly polarized, and the role of this polarity in neuroblast asymmetric division has been extensively reviewed [1–4]. Discrete protein complexes are assembled in the apical and basal cortical domains. In telophase, the apical protein complexes segregate into the self-renewing neuroblast, whereas the basal protein complexes segregate into the neural progenitor cell. Both genetic and correlative live imaging studies indicate that the apical protein complexes have dual functions: promoting neuroblast identity and targeting the basal protein complexes into the neural progenitor cell. The basal protein complexes function specifically in restricting the neural progenitor cell potential [5]. Two classes of central brain neuroblast lineages (types I and II) can be unambiguously identified based on the progenitor progeny generated and the combination of cell fate markers expressed [6• •,7• •,8• •] (Figure 1). Below, we discuss the functional properties of neural progenitor cells generated in the type I and type II neuroblast lineages and the molecular mechanisms that restrict their developmental potential.
Neuroblasts and neural progenitor cells in the type I lineage
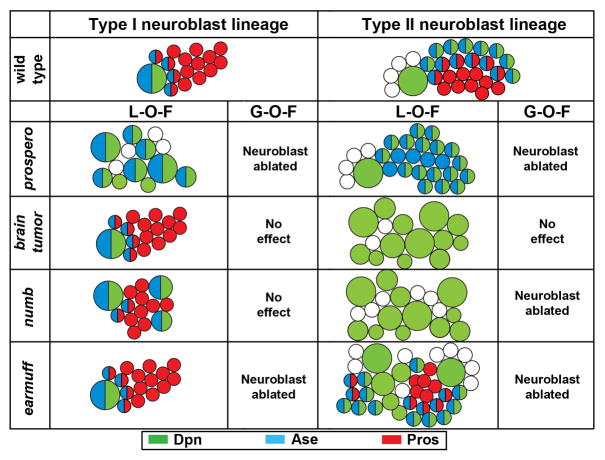
A type I neuroblast divides asymmetrically to generate a self-renewing daughter neuroblast and a neural progenitor cell called a ganglion mother cell (GMC) which divides once to produce two post-mitotic neurons [6• •,7• •,8• •]. During this asymmetric division, the basal proteins Brain tumor and Prospero exclusively segregate into the GMC by binding to the scaffolding protein Miranda, while Numb partitions into the GMC independently of Miranda. The basal proteins remain asymmetrically segregated into GMCs in a telophase brain tumor mutant neuroblast, and genetic clones derived from single brain tumor mutant neuroblasts always contain one neuroblast and many neurons per clone (Figure 2). Thus, Brain tumor is either dispensable or functionally redundant with other proteins in restricting the GMC potential.
Figure 2.
A summary of the identity of cells derived from type I and II neuroblasts lacking or over-expressing key proteins required to restrict the progenitor cell potential. Type I neuroblasts are Dpn+Ase+ whereas type II neuroblasts are Dpn+Ase-. Abbreviation: L-O-F: loss-of-function; G-O-F: gain-of-function; Dpn: Deadpan; Ase: Asense; Pros: Prospero.
prospero encodes a homeodomain transcription factor, and plays a key role in specifying neuronal and glial cell types in the developing nervous system [9–12]. Although Prospero is expressed in neuroblasts, it is kept out of neuroblast nuclei by the combination of nuclear exclusion and binding to the scaffolding protein Miranda [13–16]. The Miranda-Prospero complex localizes to the basal cortex of a mitotic neuroblast in metaphase and asymmetrically segregates into the GMC in telophase. Upon completion of cell division, Miranda becomes proteolytically degraded, and Prospero is released from the cortex and localizes to the GMC nuclei [17]. Nuclear Prospero restricts the GMC potential by suppressing genes that promote the neuroblast identity and activating genes that promote differentiation and cell cycle exit [12,18]. While mitotic prospero mutant type I neuroblasts exhibit normal apical-basal cortical polarity, prospero mutant neuroblast lineage clones contain almost exclusively neuroblasts at the expense of neurons [12,19–22•] (Figure 2). Over-expression of Prospero leads to constitutive accumulation of Prospero in neuroblast nuclei, triggering premature loss of neuroblasts. These data indicate that Prospero is necessary and sufficient to restrict the GMC potential.
numb encodes an evolutionarily conserved protein essential for proper neuronal fate specification in the developing nervous system [23–27]. Eighty-five percent of numb mutant type I neuroblast lineage clones contain more than one neuroblast per clone despite asymmetric segregation of Miranda into GMCs [8• •,28] (Figure 2). Furthermore, mutations that perturb asymmetric segregation of Numb into GMCs lead to formation of ectopic neuroblasts, a phenotype that can be suppressed by over-expression of Numb in neuroblasts [28,29]. Thus, Numb likely restricts the GMC potential independent of Prospero. Fly and mouse studies have shown that Numb suppresses Notch signaling in the developing nervous system, raising the possibility that Numb might restrict the GMC potential by antagonizing Notch signaling. Expression of multiple Notch reporters is detectable in neuroblasts but is undetectable in GMCs in the wild type brain [8• •,30]. Additionally, ectopic expression of a constitutively active form of Notch (Notchintra) perturbs neuroblast asymmetric divisions, leading to a massive increase in neuroblasts at the expense of neurons [8• •,30]. Unlike Prospero, ectopic expression of Numb or knock-down of the Notch function by RNA interference is insufficient to trigger premature loss of type I neuroblasts [8• •]. Thus, inhibition of the Notch signaling by Numb is necessary but not sufficient to limit the GMC potential.
Neuroblasts and neural progenitors in the type II lineage
A type II neuroblast divides asymmetrically to self-renew and generate an intermediate neural progenitor cell (INP), previously referred to as a transit amplifying GMC, a secondary neuroblast or an intermediate progenitor [6• •,7• •,8• •] (Figure 1). A newly born INP is immature, and is arrested in the G2 phase of the cell cycle and must undergo maturation, during which it acquires restricted developmental potential prior to resuming proliferation [8• •]. A mature INP divides asymmetrically several times, each time self-renewing by producing a daughter INP and a GMC. The basal proteins Brain tumor and Numb, inherited from the asymmetrically dividing parental neuroblasts, establish the restricted developmental potential in an immature INP [8• •]. Following completion of maturation, the transcription factor Earmuff maintains the INP potential [22• •]. These sequential mechanisms play key roles in restricting the INPs potential.
Establishment of the restricted developmental potential in INPs
While a wild-type type II neuroblast clone always contains one neuroblast, 3–5 immature INPs and 20–30 INPs, a brain tumor mutant type II neuroblast clone contains almost exclusively neuroblasts [8• •] (Figure 2). Interestingly, a mitotic brain tumor mutant type II neuroblast shows normal apical-basal cortical polarity and asymmetric segregation of Numb into immature INPs. Thus, ectopic type II neuroblasts in the brain tumor mutant brain likely arise from de-differentiation of immature INPs that fail to acquire restricted developmental potential despite inheriting Numb. These data suggest that Brain tumor likely functions parallel to Numb to promote restriction of the INP potential. Over-expression of Brain tumor does not effect the expression of a Notch reporter in neuroblasts, and removal of brain tumor does not alter binary cell fate determination in the sensory organ precursor lineage, a system highly sensitive to the loss of Notch function [8• •]. Together, these data strongly suggest that Brain tumor is necessary but not sufficient to restrict the INP potential.
Despite showing normal apical-basal cortical polarity and asymmetric segregation of Brain tumor into immature INPs, numb mutant type II neuroblast clones also consist of mostly neuroblasts [8• •,28] (Figure 2). Thus, ectopic type II neuroblasts in the numb mutant brain might also arise from de-differentiation of immature INPs due to aberrant activation of the Notch signaling mechanism. Indeed, ectopic expression of Notchintra leads to ectopic type II neuroblasts at the expense of immature INPs, whereas over-expression of Numb or knock-down of the Notch function by RNA interference results in the premature loss of type II neuroblasts [8• •]. Thus, by antagonizing Notch, Numb is necessary and sufficient to establish the restricted developmental potential in immature INPs. Taken together, Brain tumor and Numb function non-redundantly to establish the INP potential during maturation.
Maintenance of the restricted developmental potential in INPs
Following maturation, the INP potential requires an active mechanism mediated by the earmuff gene for maintenance during limited rounds of asymmetric divisions [22• •]. While the number of type I neuroblasts remain unchanged in the earmuff mutant brain, the population of type II neuroblasts becomes drastically expanded (Figure 2). Surprisingly, earmuff mutant mitotic type II neuroblasts exhibit normal apical-basal cortical polarity and undergo repeated asymmetric divisions to self-renew and to generate immature INPs that mature into INPs. Furthermore, earmuff mutant mitotic INPs also exhibit normal cortical polarity and asymmetric segregation of the basal proteins Brain tumor, Prospero and Numb into GMCs that produce differentiated neurons. Thus, it is unlikely that ectopic type II neuroblasts in the earmuff mutant brain arise from de-differentiation of immature INPs due to failure to acquire restricted developmental potential. Analyses of the cell fate markers in lineage clones derived from earmuff mutant type II neuroblasts indicate that following maturation, INPs fail to maintain restricted developmental potential and de-differentiate back into type II neuroblasts. Analyses of its promoter expression pattern reveal that earmuff is undetectable in type II neuroblasts and immature INPs and instead, is detected in INPs. Additionally, ectopic type II neuroblasts in the earmuff mutant brain can be suppressed by restoring the expression of Earmuff in INPs under the control of its own promoter. Thus, Earmuff specifically maintains the INP potential.
One way to maintain the restricted potential of INPs is to limit their proliferation capacity. In the wild-type brain, an INP shows a limited proliferation capacity prior to exit from cell cycle and terminal differentiation, processes likely regulated by nuclear localization of Prospero [12,31]. While nuclear Prospero is rarely detected in INPs in the wild-type brain, over-expression of Earmuff in neuroblasts or INPs can induce almost a ten-fold increase in the frequency of nuclear Prospero and premature loss of these cells [22• •]. Furthermore, INP-specific expression of Prospero can partially suppress the ectopic neuroblast phenotype in the earmuff mutant brain. Moreover, prospero mutant INPs generate ectopic INPs at the expense of neurons, but do not de-differentiate back into type II neuroblasts [22• •]. Thus, a Prospero-dependent mechanism limits INP proliferation and promotes INP differentiation, whereas a Prospero-independent mechanism prevents INPs from acquiring the type II neuroblast identity.
Neuroblast-specific expression of Notchintra leads to ectopic neuroblasts at the expense of GMCs and immature INPs, suggesting that down-regulation of Notch might be a general mechanism to restrict the developmental potential in neural progenitor cells. Similarly, ectopic expression of Notchintra in INPs is sufficient to trigger formation of ectopic type II neuroblasts, raising the possibility that earmuff might restrict the developmental potential of INPs by antagonizing Notch signaling [22• •]. In agreement with this hypothesis, knock-down of Notch function by RNA interference partially suppresses the ectopic type II neuroblast phenotype in the earmuff mutant brain. Furthermore, over-expression of Earmuff in INPs can suppress the formation of ectopic type II neuroblasts induced by over-expression of Notchintra. A recent study demonstrates that the vertebrate homologs of Earmuff can suppress Notch signaling by directly binding to the promoter of a Notch target gene Hes5 during mouse cortical neurogenesis [32•]. Notch signaling plays a critical role in distinguishing neural stem cell from intermediate progenitors during both embryonic and adult brain neurogenesis [32•,33]. Thus, Earmuff and its vertebrate homologs likely regulate the progenitor cell potential during neurogenesis through antagonizing the Notch signaling.
Optic lobe neuroepithelial stem cells generate two types of neural progenitor cells
Neuroepithelial stem cells in the developing optic lobe initially undergo symmetric divisions to expand the stem cell population, then differentiate into neural progenitors that generate terminally differentiated neurons through limited rounds of asymmetric divisions [34• •] (Figure 1). This dynamic mechanism allows rapid generation of a large number of post-mitotic progeny from a relatively small population of stem cells, and is widely used in the context of development and regeneration [35,36]. Failure to properly restrict the developmental potential in neuroepithelial stem cells and their progenitor progeny might contribute to childhood tumors of epithelial origin [37,38]. Thus, understanding how developmental potential is precisely specified in neuroepithelial stem cells and neural progenitor cells will likely provide novel insight into development and tumorigenesis.
The functional property of neuroepithelial stem cells changes dynamically in the outer proliferation center of the developing optic lobe. Prior to the third larval instar, most neuroepithelial stem cells predominantly undergo symmetric divisions to expand the stem cell population, forming a C-shaped swath flanked with few neuroblasts at the medial edge bordering the central brain. In the third larval instar, neuroepithelial stem cells progressively transition into neuroblasts from the medial edge toward the lateral edge of the optic lobe, leading to narrowing of the neuroepithelia and widening of the neuroblast swath [34• •,39,40• •]. Neuroblasts in the optic lobe share many parallels with INPs in the central brain, including expression of similar cell fate markers and asymmetric segregation of similar cell polarity proteins. A neuroblast in the optic lobe also undergoes limited rounds of asymmetric divisions to regenerate and to produce a GMC that gives rise to two terminally differentiated progeny [21,34• •]. However, the molecular mechanism that restricts the neuroblast potential in the optic lobe has yet to be investigated and will not be discussed further. Below, we will focus on the molecular mechanism that regulates the neuroepithelial stem cells.
Comparative expression profiling of micro-dissected neuroepithelia and neuroblasts from the optic lobe suggests that the Notch signaling mechanism likely plays a key role in maintaining the neuroepithelial stem cell identity [41• •]. Removal of the Notch function triggers premature transition from neuroepithelia to neuroblasts, whereas constitutive activation of Notch signaling prevents the transition. Thus, down-regulation of Notch signaling is necessary and sufficient for the transition from neuroepithelia to neuroblasts in the larval optic lobe.
How is the Notch signaling spatially and temporally regulated in the developing optic lobe allowing synchronous transition from neuroepithelial stem cells to neuroblasts in a medial-to- lateral manner? Neuroepithelial stem cells become transiently arrested in cell cycle prior to reaching the transition zone where they lose their epithelial characteristics and assume the stereotypical round neuroblast morphology [42• •]. The expression of delta, encoding a Notch ligand, is detected at a high level in 1-2 rows of cells that are among those transiently arrested in cell cycle [40• •,42• •]. Since Delta activates Notch signaling cell non-autonomously and suppresses Notch signaling cell autonomously, over-expression or removal of delta leads to both inhibition and acceleration of neuroblast formation. This result suggests that the coordinated change between the level of Delta and the Notch signaling provides the cue that times the transition from neuroepithelia to neuroblasts. Interestingly, the proneural gene lethal of scute is also highly expressed in 1–2 rows of cells that are among those transiently arrested in cell cycle [40• •,42• •]. While removal of the lethal of scute function mildly delays the transition of neuroepithelial stem cells to neuroblasts, over-expression of lethal of scute suppresses Notch signaling and promotes premature transition. The dynamic integration of Delta and Lethal of scute specifies the transition from neuroepithelia to neuroblasts spatially in the optic lobe by repressing the Notch signaling.
The swath of neuroblasts widens synchronously from the medial edge toward the lateral edge of the developing optic lobe, suggesting that the transition from neuroepithelia to neuroblasts might also be temporally coordinated. Intriguingly, the output of the Janus kinase (Jak/Stat) signaling mechanism coincides with the timing of neuroepithelia transitioning into neuroblasts: Jak/Stat signaling is the highest at the lateral edge and the lowest at the medial edge. Removal of the components in the Jak/Stat signaling mechanism leads to precocious transition of neuroepithelia into neuroblasts, while constitutive activation of the Jak/Stat signaling delays the transition [40• •]. In addition, inactivation of the Fat-Hippo signaling mechanism delays the transition from neuroepithelia to neuroblasts, whereas constitutive activation of the Fat-Hippo signaling accelerates the transition at the medial edge of neuroepithelia [42• •]. Taken together, the Jak/Stat and the Fat-Hippo signaling mechanisms provide temporal control of the transition from neuroepithelia to neuroblasts. More experiments will be necessary to elucidate whether these two signaling pathways promote the transition through Notch or independent of Notch.
Discussion
The developmental potential in stem cells and progenitor cells must be precisely defined to ensure normal development and prevent accumulation of aberrant stem-like cells. Studies of the neural stem cell lineages in the developing Drosophila larval brain have begun to unravel the molecular mechanisms underlying how neural stem cells and neural progenitor cells are functionally distinguished at a single-cell resolution. Accumulating data point to down-regulation of the Notch signaling by various mechanisms as a critical step in establishing the restricted developmental potential in neural progenitor cells. However, additional mechanisms mediated by Brain tumor or Prospero function non-redundantly to the Notch signaling, and play important roles in restricting the developmental potential of neural progenitor cells. Notch also distinguishes neural stem cells from neural progenitor cells in the developing mouse brain [43•,44]. It will be interesting to test whether Brain tumor and Prospero indeed function in parallel of the Notch signaling in restricting the developmental potential in neural progenitor cells, and whether the vertebrate homologs of Brain tumor or Prospero might also play similar roles in regulating neural progenitor cells during mouse cortical neurogenesis. Emerging evidence strongly suggests that the Jak-Stat and Fat-Hippo signaling mechanisms regulate the timing of restricting the developmental potential in neuroepithelial stem cells. It will be important to determine whether these two signaling mechanisms might promote the transition from neuroepithelia to neuroblasts in the developing optic lobe via a Notch-dependent or -independent mechanism.
Acknowledgments
We would like to thank members of the Lee lab for helpful discussion and comments. We would like to acknowledge the editorial support of Jon Kilner, MS, MA (Pittsburgh, PA). M.W and C.-Y. L are supported by the Sontag Foundation and NIH (GM092818).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 2.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 3.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Wu P-S, Egger B, Brand AH. Asymmetric stem cell division: Lessons from Drosophila. Seminars in Cell & Developmental Biology. 2008;19:283–293. doi: 10.1016/j.semcdb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 6••.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. One of the three studies identifies and characterizes the type II neuroblast lineage. Bello et al., demonstrates the existence of intermediate progenitor cells in the developing Drosophila larval brain, which serve as a novel model to study the mechanism restricting the developmental potential in neural progenitor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68 :1185–1195. doi: 10.1002/dneu.20648. One of the three studies identifies and characterizes the type II neuroblast lineage. Boone and Doe demonstrate the existence of transit amplifying GMCs in the developing Drosophila larval brain, which serve as a novel model to study the mechanism restricting the developmental potential in neural progenitor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. One of the three studies identifies and characterizes the type II neuroblast lineage. Bowman et al., demonstrate the existence of secondary neuroblasts in the developing Drosophila larval brain, which serve as a novel model to study the mechanism restricting the developmental potential in neural progenitor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu-Lagraff Q, Wright DM, McNeil LK, Doe CQ. The prospero gene encodes a divergent homeodomain protein that controls neuronal identity in Drosophila. Development. 1991;113:79–85. [PubMed] [Google Scholar]
- 10.Manning L, Doe CQ. Prospero distinguishes sibling cell fate without asymmetric localization in the Drosophila adult external sense organ lineage. Development. 1999;126:2063–2071. doi: 10.1242/dev.126.10.2063. [DOI] [PubMed] [Google Scholar]
- 11.Freeman MR, Doe CQ. Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development. 2001;128:4103–4112. doi: 10.1242/dev.128.20.4103. [DOI] [PubMed] [Google Scholar]
- 12.Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-Renewal and differentiation in Drosophila neural stem Cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 14.Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. Miranda localizes Staufen and Prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 16.Schuldt AJ, Adams JHJ, Davidson CM, Micklem DR, Haseloff J, Johnston DS, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Cai Y, Chia W, Yang X. Drosophila homologs of mammalian TNF/TNFR-related molecules regulate segregation of Miranda/Prospero in neuroblasts. EMBO J. 2006;25:5783–5793. doi: 10.1038/sj.emboj.7601461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- 19.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 20.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 22••.Weng M, Golden KL, Lee CY. dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev Cell. 2010;18:126–135. doi: 10.1016/j.devcel.2009.12.007. Weng et al., shows that following specification of the intermediate neural progenitor cell (INP) identity, their developmental potential requires an active mechanism mediated by Earmuff for maintenance. Earmuff maintains the INP potential by limiting proliferation by promoting nuclear localization of Prospero and suppressing de-differentiation by antagonizing Notch signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 24.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 25.Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- 26.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhong W, Feder JN, Jiang M-M, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17 :43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 32•.Shimizu T, Nakazawa M, Kani S, Bae Y-K, Shimizu T, Kageyama R, Hibi M. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–1885. doi: 10.1242/dev.047167. Shimizu et al., shows the mouse homologs of Earmuff antagonize Notch signaling by directly binding to the promoter of the Hes5 gene during cortex neurogenesis. [DOI] [PubMed] [Google Scholar]
- 33.Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–22. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- 34••.Egger B, Boone J, Stevens N, Brand A, Doe C. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural. Development. 2007;2:1. doi: 10.1186/1749-8104-2-1. Egger et al., shows that neuroepithelia in the developing larval optic lobe generate neuroblasts, which in turn generate neurons. This study characterizes symmetric and asymmetric divisions of neuroepithelial cells and neuroblasts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Hadjipanayis CG, Van Meir EG. Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol Med. 2009;15:519–530. doi: 10.1016/j.molmed.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubuc A, Northcott P, Mack S, Witt H, Pfister S, Taylor M. The genetics of pediatric brain tumors. Curr Neurol and Neurosci Rep. 2010;10:215–223. doi: 10.1007/s11910-010-0103-9. [DOI] [PubMed] [Google Scholar]
- 39.Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- 40••.Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. Yasugi et al., shows that conversion of neuroepithelia into neuroblasts in the developing larval optic lobe occurs in a medial-to-lateral manner. These authors demonstrate that the expression of Lethal of scute coincides with the transition from neuroepithelia to neuroblasts, and that the JAK/STAT pathway modulates this transition. [DOI] [PubMed] [Google Scholar]
- 41••.Egger B, Gold K, Brand A. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010 doi: 10.1242/dev.051250. in press. Egger et al., shows that Notch signaling plays a key role in maintaining the identity of neuroepithelia in the developing larval optic lobe. The authors demonstrate that Notch loss of function leads to premature conversion of neuroepithelia into neuroblasts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Reddy BVVG, Rauskolb C, Irvine KD. Influence of fat-hippo and Notch signaling on the proliferation and differentiation of Drosophla optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. Reddy et al., shows that the Notch signaling is required for the maintenance of optic lobe neuroepithelia, and that fat-hippo signaling pathway regulates the neuroepithelia-neuroblast transition by promoting transient cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Mizutani K-i, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. Mizutani et al., identifies a population of intermediate neural progenitors that co-exist with neural stem cells in the ventricular zone and shows that Notch signaling functionally distinguishes intermediate progenitors from neural stem cells. [DOI] [PubMed] [Google Scholar]
- 44.Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF. Regulation of neural progenitor cell development in the nervous system. J Neurochem. 2008;106:2272–2287. doi: 10.1111/j.1471-4159.2008.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]