Abstract
Forkhead transcription factor FoxO1 and the NAD+-dependent histone deacetylase SIRT1 are evolutionarily conserved regulators of the development of aging, oxidative stress resistance, insulin resistance, and metabolism in species ranging from invertebrates to mammals. SIRT1 deacetylates FoxO1 and enables activation of FoxO1 transcription in multiple systems. The functional consequences of the interactions between FoxO1 and SIRT1 remain incompletely understood. Here, we demonstrate that the 1.5-kb rat sirt1 promoter region contains a cluster of five putative FoxO1 core binding repeat motifs (5×IRS-1) and a forkhead-like consensus binding site (FKHD-L). Luciferase promoter assays demonstrate that FoxO1 directly activates SIRT1 promoter activity and that both the IRS-1 and FKHD-L enable FoxO1-dependent SIRT1 transcription. Electrophoretic mobility shift and chromatin immunoprecipitation assays show that FoxO1 binds to the IRS-1 and FKHD-L sites of the SIRT1 promoter. Consistently, FoxO1 overexpression increases SIRT1 expression, and FoxO1 depletion by siRNA reduces SIRT1 expression at both the messenger RNA and protein levels in vascular smooth muscle cells and HEK293 cells. Thus, endogenous FoxO1 is a positive transcriptional regulator of SIRT1. Conversely, SIRT1 promotes FoxO1-driven SIRT1 autotranscription through interacting with and deacetylating FoxO1. Moreover, resveratrol, a plant polyphenol activator of SIRT1, increases FoxO1-dependent SIRT1 transcription activity and thus induces its expression. These findings suggest that positive feedback mechanisms regulate FoxO1-dependent SIRT1 transcription and indicate a previously unappreciated function for FoxO1. This signaling network may coordinate multiple pathways acting upon immune, inflammatory, regenerative, and metabolic processes.
Keywords: Gene Regulation, Metabolic Diseases, Resveratrol, SIRT, Smooth Muscle, Feedback Loop, FoxO1, Resveratrol, VSMC
Introduction
The yeast Sir2 (silent information regulator 2) and its mammalian homologue SIRT1, class III histone deacetylases of the sirtuin family, modulate aging, oxidative stress resistance, cell metabolism, energy homeostasis, insulin resistance, and angiogenesis in several species (1, 2). In the yeast Saccharomyces cerevisiae and the nematode Caenorhabditis elegans, increased expression of Sir2 promotes longevity under calorie restriction conditions (3–5). In mammals, SIRT1 also promotes cell survival during calorie restriction (6–8). SIRT1 plays diverse roles in a number of cellular processes through deacetylation of histones, transcription factors, and transcriptional cofactors (9). SIRT1 mediates stress resistance in mammalian cells by deacetylating stress response mediators, such as FoxO (forkhead box O) transcription factors (10) and p53 (11). Reduced expression or activity of SIRT1 contributes to insulin resistance and its related diseases, such as type II diabetes mellitus (12, 13). In contrast, SIRT1 up-regulates adiponectin expression by deacetylating FoxO1 and thus protects against insulin resistance (14–16). SIRT1 transgenic mice display improved glucose tolerance and increased metabolic efficiency and have reduced aging-induced diabetes when fed a normal diet (17). Evidence from genetically engineered mouse models (18) shows a key role of SIRT1 in controlling endothelial angiogenic functions during vascular growth. These observations suggest that activation or increased expression of SIRT1 might have a broad spectrum of beneficial effects in metabolic diseases, including diabetes and obesity, as well as in aging-associated cardiovascular diseases, such as atherosclerosis.
Expression and activity of SIRT1 are tightly regulated at multiple levels. Apoptosis transcriptional regulator E2F1 induces SIRT1 transcriptional expression in response to the stress of DNA damage (19). Furthermore, deacetylation of E2F1 by SIRT1 inhibits its activity by a negative feedback mechanism (19). SIRT1 transcription is also negatively regulated by HIC1 (hypermethylated in cancer 1), a tumor suppressor gene, which binds the enhancer elements of the SIRT1 promoter and represses SIRT1 expression (20, 21). Interestingly, a concomitant induction of FoxO3a and SIRT1 expression was observed in response to acute nutritional stress (22). FoxO3a interacts with p53 and binds to the p53 response elements within the mouse Sirt1 promoter, thereby up-regulating SIRT1 transcription (22). HUR, an RNA-binding protein, associates with the 3′-untranslated region of SIRT1 mRNA and increases SIRT1 expression by stabilizing the SIRT1 mRNA (23). Oxidative stress reduces SIRT1 mRNA and protein levels, and a concomitant reduction of HUR and SIRT1 expression appears during the replicative senescence of human diploid fibroblasts (23). AROS (active regulator of SIRT1) (24) and DBC1 (deleted in breast cancer 1) (25) were recently identified as positive and negative regulators of SIRT1 enzymatic activity, respectively. SIRT1 activity is also regulated by post-translational modifications (2, 26). Thus, expression and activity of SIRT1 are subject to complex control patterns at multiple levels that might contribute to the highly multifunctional roles of SIRT1 in regulating cell homeostasis.
Transcription factor FoxO family members (and FoxO1 in particular) play important roles in aging, cell metabolism, insulin resistance, and oxidative stress resistance (27, 28). FoxO1 activity is tightly regulated. FoxO1 phosphorylation by phosphatidylinositol 3-kinase/Akt in response to insulin or growth factors leads to the export of FoxO1 from the nucleus to the cytoplasm, thereby inhibiting its transcription activity (27, 28). FoxO1 activity is also regulated by acetylation on specific lysine residues (29). CREB-binding protein acetylates FoxO1 and disrupts FoxO1-DNA interactions, thereby attenuating FoxO1-dependent gene expression (29). Conversely, SIRT1 increases FoxO1 DNA-binding ability by deacetylating FoxO1 and potentiates its transcription activity (16, 17, 29–31). Thus, FoxO1 and SIRT1 act synergistically to modulate diverse processes, such as aging, oxidative stress response, and insulin resistance, in a wide range of organisms. However, the precise mechanisms defining the interrelationship of FoxO1 and SIRT1 are incompletely characterized.
We posited that FoxO1 may regulate SIRT1 expression transcriptionally. Here we show that FoxO1 directly activates SIRT1 transcription through its binding to the IRS-1 and FKHD-like responsive elements within the rat sirt1 promoter region. Conversely, SIRT1 interacts with and deacetylates FoxO1 and thus potentiates FoxO1-dependent SIRT1 transcription, invoking a putative positive feedback loop mechanism. Finally, resveratrol, a known plant-derived polyphenol activator of SIRT1 that delays aging and protects against the metabolic consequences of high caloric intake in mice (32), increases FoxO1-mediated SIRT1 transcription and thus induces SIRT1 expression. These findings provide insights into a novel positive autofeedback mechanism by which this posited FoxO1-SIRT1 circuit may mediate beneficial effects on age-related immunoinflammatory diseases.
EXPERIMENTAL PROCEDURES
Materials
Antibodies to FKHR/FoxO1 (H-128), FKHRL1/FoxO3a (H-144), AFX1/FoxO4 (H-80), and SIRT2 (H-95) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibody to actin (20-33) was from Sigma. Anti-SIR2/SIRT1 antibody (07-131) was from Upstate. Anti-SIRT3 antibody (AP6242a) was from Abgent. Rabbit polyclonal GFP antibody (ab290) was from Abcam Inc. Antibodies to acetylated lysine (9681) and Myc tag (9B11) were from Cell Signaling. Anti-HA (12CA5) and anti-FLAG (F7425) antibodies were obtained from Roche Applied Science and Sigma, respectively. Expression plasmids for FLAG-FKHR/FoxO1 and HA-FoxO3a were purchased from Addgene. Resveratrol was purchased from Alexis Corp. Superscript® II reverse transcriptase was purchased from Invitrogen. SYBR Green JumpStart Taq ready mix was from Sigma. ECL Western blotting detection reagents and nitrocellulose membranes (Hybond-ECL) were obtained from Amersham Biosciences. The Basic Nucleofector® Amaxa transfection kit for primary smooth muscle cells was from Biosystems. All other chemicals and reagents, including DMEM with 25 mm Hepes and 4.5 g/liter glucose were from Sigma.
Cloning of Rat SIRT1 Promoter Constructs
Genomic DNA from rat vascular smooth muscle cells (VSMCs)2 was prepared using the PureLinkTM genomic DNA minikit (K1820-00, Invitrogen) following the manufacturer's instructions. Various fragments of the 5′-flanking sequence of the rat sirt1 gene (XM_001080493) were amplified directly from VSMC genomic DNA by PCR and subcloned into the pGL3-Basic vector (Promega) at XhoI and HindIII sites. The plasmids contain the regions from +54 or +84 to various sizes of the 5′-flanking region of the rat sirt1 gene with respect to the transcriptional start site. All PCRs were performed using the Expand High Fidelity PCR system (Roche Applied Science). Sequences of the constructs were determined by autosequencing to verify transcription in frame. The primer sequences are described in supplemental Table S1.
Site-directed Mutagenesis
Mutated constructs were generated with the QuikChange®-XL site-directed mutagenesis kit (Stratagene) following the manufacturer's directions. The FKHD-L binding site was disrupted by changing the TATGTAAATA into TATGTGCATA. The IRS-2 binding site was disrupted by changing the CAAAATA into CCGAATA. IRS-1a-M was obtained by changing the ACAAAAA into AGGAAAA. IRS-1b-M was obtained by changing the ACAAAAA into AGAAAAA. IRS-1cd-M was generated by changing the ACAAAAACAAACAAAAA into AGAAAAACAAAGAAAAA. IRS-1e-M was obtained by changing the ACAAAAA into AGCCAAA. Other combined mutants of IRS-1 binding sites, including IRS-1cde, IRS-1bcd, IRS-1abcd, IRS-1bcde, and IRS-1abcde mutants, were generated by multiple-round PCR step by step using the primers described in supplemental Table S2. All constructs generated by PCR were sequenced to verify mutation.
Luciferase Reporter Gene Assay
Cells in 24-well plates were cotransfected with luciferase constructs or empty pGL3 vector (400 ng), expression vector encoding FoxO1 or empty pcDNA3 vector (400 ng), and the internal control p-RL-TK vector (20 ng) using Lipofectamine 2000 for HEK293 or the Basic Nucleofector® Amaxa transfection kit for VSMCs for 24 h. Total amounts of DNA for each well were normalized by adding empty vector pcDNA3. Cells were harvested, and luciferase activity was measured using the Dual Luciferase reporter assay system (Promega) according to the manufacturer's instructions. The relative activity was normalized to the ratio of firefly luciferase activity to Renilla luciferase activity (internal control) and calculated as the -fold difference from control pGL3-basic or pcDNA3 empty vector. Data from 2–3 independent experiments (each performed in triplicate) are shown as mean ± S.E.
Cell Culture and Adenovirus Transduction
HEK293 cells were transfected using Lipofectamine 2000 (Invitrogen) with the indicated plasmids or siRNA duplexes (Applied Biosystems; Ambion). Sequences of custom siRNA duplexes are described in supplemental Table S4. VSMCs were growth-arrested for 24 h prior to treatment with various doses of resveratrol. Transfection of siRNA duplexes into VSMCs was performed following the instructions of the Basic Nucleofector® kit. For adenovirus-mediated protein overexpression, VSMCs were infected with various multiplicities of infection of either Ad.FoxO1 (33) (a gift from Dr. J. A. Hill) or control adenovirus (Ad.GFP) for 1 h in serum-free medium. Then the virus was removed, and cells were cultured for an additional 24–48 h before cell collection.
RT-PCR and Quantitative Real-time PCR
Total RNA from VSMCs or HEK293 cells was extracted as described previously (34). RT-PCR amplification was performed using the SuperScriptTM one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen). One microgram of total RNA was used as a template for subsequent one-step RT-PCRs. Rat sirt1 cDNA (residues 1021–1340) and human SIRT1 cDNA (residues 865–1188) were amplified with the PCR primers as described in supplemental Table S4. GAPDH served as an internal control of the reaction. One-step RT-PCR products were analyzed by electrophoresis on 1.8% agarose gels containing ethidium bromide. For quantitative real-time PCR, 1 μg of total RNA was reverse-transcribed with Superscript II (Invitrogen) using random primers. Synthesized cDNA samples were amplified in the LightCycler (Roche Applied Science) real-time thermocycler using SYBR Green JumpStart Taq ready mix (Sigma). The results of relative expression were normalized to GAPDH mRNA levels in each sample. Results are expressed as mean ± S.E.
Western Blotting and Immunoprecipitation
Cell lysates of VSMCs or HEK293 cells were prepared in lysis buffer (50 mm HEPES (pH 7.4), 50 mm NaCl, 1% Triton X-100, 5 mm EDTA, 1 mm DTT, 1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 mm sodium pyrophosphate, 50 mm sodium fluoride, and 1 mm sodium orthovanadate). For immunoprecipitation, VSMCs were infected with Ad.GFP-FoxO1 together with or without adenovirus encoding the Myc-tagged SIRT1 wild type or SIRT1 H363Y dominant negative mutant (35) for 1 h in serum-free medium. Then the virus was removed, and the cells were cultured for an additional 36 h before cell collection. 500 μg of total cell lysates were immunoprecipitated with GFP antibodies and protein A/G PLUS-agarose beads. Samples were separated by SDS-PAGE, transferred to nitrocellulose membranes, and incubated overnight with the indicated antibodies. After incubation with secondary antibodies, proteins were detected by enhanced chemiluminescence.
Electrophoretic Mobility Shift Assays (EMSAs)
Full-length FoxO1 protein was prepared using the T7 promoter of pcDNA3-FoxO1 expression plasmid by the TNT-coupled transcription/translation systems (Promega). In vitro translated FoxO1 protein was confirmed by Western blotting using anti-FoxO1 antibodies. EMSA was carried out using the LightShift® chemiluminescent EMSA kit (Pierce). Briefly, binding reactions were conducted in binding buffer for 20 min at room temperature using the in vitro translated FoxO1 proteins and biotin-labeled annealed double-stranded probes containing IRS-1bcd or FKHD-L binding sites or mutated probes as described in supplemental Table S3. For competition experiments, unlabeled wild-type probes were mixed with the labeled probes at 100- and 200-fold molar excess prior to the addition of in vitro translated proteins. The reaction mixtures were then loaded onto a 6% native polyacrylamide gel and electrophoresed at 4 °C for 150 min at 90 V in 0.5× TBE. Following electrophoresis, the gel was transferred to the positively charged nylon membrane (Biodyne®B, P/N 60209) and then analyzed following the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was done following the manufacturer's instructions (Upstate). VSMCs or HEK293 cells were fixed with 1% formaldehyde at 37 °C for 10 min, lysed, and sonicated. Soluble chromatin was coimmunoprecipitated with anti-FoxO1 or FoxO3a antiserum or an equal amount of rabbit IgG. Cells at ∼80% confluence grown in one 10-cm diameter dish were used per immunoprecipitation. After decross-linking of the DNAs, DNA purified from starting (1% input) and immunoprecipitated samples were subjected to PCR using the primers listed in supplemental Table S4. Those regions of amplification contain the IRS-1 or FKHD-L-binding element in the rat sirt1 promoter (XM_001080493) or two putative FoxO1 DNA-binding elements in the human SIRT1 promoter (NM_012238). Standard PCRs were performed.
Statistical Analysis
All values were expressed as mean ± S.E. Data were analyzed by Student's unpaired t test or one-way analysis of variance of the repeated experiments followed by Tukey's post hoc pairwise multiple comparisons when appropriate with Prism (GraphPad software). Statistical significance was accepted at p < 0.05.
RESULTS
Promoter Analysis of the Rat sirt1 Gene
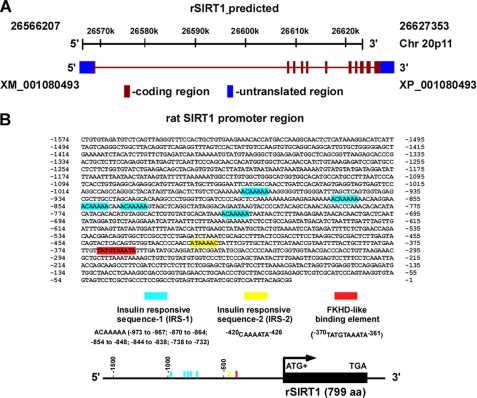
Aberrant growth, proliferation, differentiation, and migration of VSMCs are fundamental pathogenetic features of cardiovascular diseases, such as hypertension and atherosclerosis (36, 37). VSMC isolated from rat artery is a widely used in vitro model for studies of pathophysiological mechanisms of the development of vascular diseases (38–40). At present, little information about rat sirt1 gene regulation is available. To explore the rat homologue of yeast SIR2, we screened the National Center for Biotechnology Information expressed sequence tag and nonredundant databases using the conserved SIR2 domain with the tBlastn and Blastp algorithms. We found that the rat sirt1 gene (GenBankTM accession number XM_001080493) contains 11 exons, which encode a 799-amino acid protein with 94% (571 of 605) and 91% (551 of 604) sequence identities to mouse and human SIRT1 at the amino acid level, respectively. Mouse and human SIRT1 encode proteins with 737 and 747 amino acids, respectively (supplemental Fig. S1). Rat sirt1 was mapped on Rattus norvegicus chromosome 20p11 and spanned 61,147 base pairs according to the results of Blast analysis using the Rat Genome Celera Assembly database (Fig. 1A).
FIGURE 1.
Genomic location and promoter region of rat sirt1 (rSIRT1). A, the rat sirt1 gene (XM_001080493) was mapped on chromosome 20p11 with 11 exons and spanned 61,147 base pairs according to the computer analysis results using the Rat Genome Celera Assembly database. B, the 1.5-kb promoter region of the rat sirt1 gene contains a cluster of five putative FoxO1 core binding motifs (known as insulin-responsive sequence-1 (IRS-1)) (blue), insulin-responsive sequence-2 (IRS-2) (yellow), and a forkhead-like binding element (FKHD-L) (red). aa, amino acids.
We next analyzed the putative transcription factor binding sites within the 2.0-kb promoter region of the rat sirt1 gene using the MatInspector program of the Genomatix portal (available on the World Wide Web). We found that the 2.0-kb rat SIRT1 promoter region contains a cluster of five putative FoxO1 core binding repeat motifs (designated IRS-1abcde, ACAAAAA, from −973 to −732) (41–44), an insulin-responsive sequence (designated IRS-2, CAAAATA, from −420 to −426), and a putative forkhead-like binding site (designated FKHD-L, TATGTAAATA, from −370 to −361), which matches the FKHD consensus element (TTGTTTAC) in a reverse orientation at five of eight bases (44, 45) (Fig. 1B). This analysis suggests that SIRT1 expression could be regulated by FoxO family members. Interestingly, two putative consensus FoxO1 DNA-binding elements (−517ACAAAA−512 and −457TGTTTTAA−450, which is complementary at six of seven bases with the FoxO1 core consensus element (ACAAAAA)) are also found in the human SIRT1 promoter (supplemental Fig. S2), suggesting that these FoxO-dependent binding sequences are evolutionarily conserved within SIRT1 homologs.
FoxO1 Activates SIRT1 Transcription
To determine the roles of FoxO family members in rat SIRT1 transcriptional regulation, we cloned the proximal 5′-flanking segments of the sirt1 gene by PCR amplification using genomic DNA isolated from VSMCs. Sequences of cloned 5′-flanking fragments partially overlap and contain the 5′-transcriptional coding sequences within the first exon (supplemental Fig. S3). The sequencing results indicate that the isolated 5′-flanking fragments are indeed the rat sirt1 promoter regions containing the highly conserved FoxO core binding sites. To investigate whether FoxO family members might regulate the rat sirt1 gene transcription, we performed luciferase promoter assays in HEK293 cells using the 1.5-kb rat sirt1 reporter. We found that FoxO1 overexpression increased SIRT1 promoter activity 2–9-fold in a dose-dependent manner, whereas FoxO3a overexpression had a less stimulatory effect (Fig. 2A). These results indicate that FoxO1 robustly and selectively (relative to FoxO3a) activates the rat sirt1 promoter activity. In similar conditions, FoxO1 but not FoxO3a overexpression up-regulates endogenous SIRT1 expression in HEK293 cells (Fig. 2B), suggesting that FoxO1 is the main transcriptional regulator of SIRT1 expression. To investigate whether sirt1 mRNA abundance might be altered by FoxO1 overexpression, we conducted one-step RT-PCR and quantitative real-time PCR in VSMCs and HEK293 cells. We observed a robust dose-dependent induction in mRNA levels of SIRT1 when FoxO1 was overexpressed in both VSMCs and HEK293 cells (Fig. 2, C and D). In contrast, a dose-dependent reduction in SIRT1 mRNA levels was observed in FoxO1 siRNA-treated cells (Fig. 2, E and F), suggesting that endogenous FoxO1 up-regulates SIRT1 mRNA level basally. These results indicate that FoxO1 directly activates SIRT1 transcription.
FIGURE 2.
FoxO1 up-regulates sirt1 gene transcription. A, FoxO1 overexpression increases the rat sirt1 promoter transcriptional activity. HEK293 cells were transfected in 24-well cell culture plates with the 1.5-kb rat SIRT1 luciferase reporter, increasing amounts of expression vectors encoding FoxO1 or FoxO3a (10, 50, and 250 ng/well, respectively), and the internal control p-RL-TK vector for 24 h. Total amounts of DNA for each well were normalized by adding empty vector pcDNA3. Cells were harvested, and luciferase activity was determined as described under “Experimental Procedures.” Data from two independent experiments (each performed in triplicate) are shown as mean ± S.E. (error bars) (*, p < 0.05; **, p < 0.01 versus control vector). B, FoxO1 overexpression increases SIRT1 protein expression. HEK293 cells were transiently transfected in 100 × 20-mm tissue culture dishes with increasing amounts of FoxO1 or FoxO3a expression plasmids (1, 3, and 9 μg/dish, respectively) or control vector pcDNA3 for 24 h. Total equal amounts of DNA for each well were normalized by adding empty vector. Protein expression was analyzed by Western blotting using anti-SIRT1, FLAG, HA, and actin antibodies, respectively. One representative result is shown from three independent experiments. C and D, FoxO1 overexpression increases SIRT1 mRNA levels in VSMCs and HEK293 cells. VSMCs were infected with increasing amounts of adeno-FoxO1 or control adeno-GFP for 24 h. HEK293 cells in 100 × 20-mm tissue culture dishes were transiently transfected with increasing amounts of FoxO1 expression plasmid (1, 3, and 9 μg/dish, respectively) or control vector pcDNA3 for 24 h. Total equal amounts of DNA for each well were normalized by adding empty vector. Total RNA was prepared. One-step RT-PCR products were analyzed by electrophoresis on 1.8% agarose gels containing ethidium bromide (C). D, expression of rat SIRT1 and human SIRT1 mRNA levels was quantified by a two-step quantitative real-time RT-PCR using the same RNA samples from C. E and F, FoxO1 depletion by siRNA reduces SIRT1 mRNA levels in VSMCs and HEK293 cells. VSMCs or HEK293 cells were transfected with increasing concentrations of FoxO1 siRNA (10, 25, and 50 nm, respectively) or scrambled siRNA for 48 h. SIRT1 and GAPDH mRNA levels were analyzed by one-step RT-PCR (E) or quantified by a real-time RT-PCR (F). The results for relative expression were normalized by measuring GAPDH mRNA levels in each sample (n = 3). The expression level of SIRT1 mRNA from scrambled siRNA was assigned the value of 100%. Results are expressed as mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01 versus control vector or scrambled siRNA. rSIRT1, rat SIRT1; hSIRT1, human SIRT1.
Identification of Necessary FoxO1 Binding Sites in the sirt1 Promoter
To investigate which of the putative FoxO1-dependent DNA-binding elements in the rat sirt1 promoter might be required for SIRT1 transcription, we generated 5′-sequential deletions, which were cloned into the reporter gene vector pGL3-basic. The serial deletion reporter constructs of the SIRT1 promoter and the FoxO1 expression vector were cotransfected transiently into HEK293 cells. The reporter assay (Fig. 3A) shows that transfection of the reporter constructs (−1360/+84) or (−1027/+54), containing the putative 5×IRS-1-binding elements, together with the FoxO1 expression plasmid resulted in a robust increase (7.7 ± 0.6- and 7.6 ± 0.7-fold, respectively) in luciferase expression, compared with pGL3 control vector. The deletion construct (−652/+54), which lacks the putative 5×IRS-1-binding elements, has a significantly decreased luciferase activity (3.9 ± 0.3-fold) as compared with the construct (−1027/+54) (7.6 ± 0.7-fold). Transfection of the reporter construct (−1027/−628) containing only the cluster of 5×IRS-1-binding elements led to the highest increase in luciferase activity (9.1 ± 1.1-fold versus control). These results suggest that the 5×IRS-1-binding elements within the SIRT1 promoter are sufficient and necessary for FoxO1-dependent SIRT1 transcription. No significant difference of luciferase activity was observed between the deletion constructs (−652/+54) (3.9 ± 0.3-fold) and (−415/+54) (3.8 ± 0.4-fold), which lack the putative IRS-2 binding site, suggesting that this binding element is not required for FoxO1-dependent SIRT1 transcription. The deletion construct (−325/+54), which lacks the putative FKHD-L-binding element, caused a substantial reduction in luciferase expression as compared with construct (−415/+54) (1.0 ± 0.2-fold versus 3.8 ± 0.4-fold, respectively), suggesting that the FKHD-L element also contributes to SIRT1 transcription. Conversely, transfection of the proximal 5′-flanking construct (−325/+54), which lacks any putative FoxO1-binding elements, failed to activate SIRT1 transcription. These results imply that the putative 5×IRS-1- and FKHD-L-binding elements are required for FoxO1-dependent SIRT1 transcription.
FIGURE 3.
Identification of essential FoxO1 DNA-binding elements in the rat sirt1 (rSIRT1) promoter. A, HEK293 cells were transiently transfected with the indicated deletion luciferase constructs or pGL3.0 basic control vector together with FoxO1 expression vector, and the internal control p-RL-TK vector for 24 h. B and C, HEK293 cells were transiently transfected with the indicated single and combined mutations or the 1.5-kb reporter (wild type) together with expression vector encoding FoxO1 or control pcDNA3 and the internal control p-RL-TK vector for 24 h. Luciferase activity was determined. Data from 2–4 independent experiments (each performed in triplicate) are shown as mean ± S.E. (error bars). ns, no significant difference; **, p < 0.01 versus control or wild type or as indicated.
To identify the functional significance of the potential FoxO1-dependent binding elements, we disrupted each core sequence by site-directed mutagenesis and assayed their effects on luciferase activity when FoxO1 was overexpressed in HEK293 cells (Fig. 3B). Disruption of FKHD-L, IRS-1b, IRS-1cd, or IRS-1e, but not IRS-2 or IRS-1a, led to a significant reduction in luciferase activity compared with the wild type of the SIRT1 promoter, suggesting that IRS-1b, IRS-1cd, IRS-1e, and FKHD-L binding sites are potential FoxO1-dependent binding sites that act as essential positive regulatory elements within the SIRT1 promoter.
To explore this possibility further, we constructed combined mutations of IRS-1-binding elements and assayed their roles in luciferase expression (Fig. 3C). We found that the combined mutations IRS-1bcde and IRS-1abcde failed to activate SIRT1 transcription, although the FKHD-L site was intact (1.2 ± 0.1- and 0.9 ± 0.1-fold, respectively, versus control). One possible explanation is that the combined mutations IRS-1bcde and IRS-1abcde may affect the normal secondary binding conformation of the FKHD-L site for FoxO1. Furthermore, no significant differences in luciferase activity were observed between the IRS-1abcd and IRS-1bcd mutations (2.5 ± 0.5- versus 3.1 ± 0.5-fold, respectively). Similar results were obtained by comparing the effects of the IRS-1abcde versus IRS-1bcde mutations on luciferase expression, suggesting that the putative IRS-1a-binding element is not required for SIRT1 transcription. These data demonstrate that the potential DNA-binding elements, including IRS-1b, IRS-1c, IRS-1d, IRS-1e, and FKHD-L within the rat sirt1 promoter regulate FoxO1-dependent SIRT1 transcription.
FoxO1 Binds to IRS-1 and FKHD-L Elements in the SIRT1 Promoter
To evaluate whether FoxO1 directly binds to the putative IRS-1 and FKHD-L elements in the SIRT1 promoter, we performed EMSAs using the wild type and mutant oligomers corresponding to the adjacent IRS-1bcd and FKHD-L sites. We found that FoxO1 is indeed able to bind to both the IRS-1bcd (Fig. 4A) and FKHD-L (Fig. 4B) target sequences in vitro. The binding specificity of the DNA-protein complex was verified by using the mutated probes (Fig. 4, A (compare lane 5 with lane 4) and B (compare lane 5 with lane 4)) and the addition of an excess of unlabeled oligonucleotide competitors (Fig. 4A, compare lane 5 with lanes 6 and 7; Fig. 4B, compare lane 5 with lanes 6 and 7). To provide further insights as to whether FoxO1 interacts with IRS-1 and FKHD-L-binding elements in situ, we performed ChIP assays using FoxO1 and FoxO3a antibodies. The data revealed that endogenous FoxO1 was indeed recruited to both the IRS-1 and FKHD-L sites of the rat sirt1 promoter in VSMCs (Fig. 4, C and D). FoxO3a appeared to interact slightly with the IRS-1 site but not with the FKHD-L site. Together, these results provide strong evidence indicating that FoxO1 binds to the IRS-1 and FKHD-L elements of the rat sirt1 promoter both in vitro and in situ. Similarly, we also found that FoxO1 but not FoxO3a interacted with the putative FoxO1 DNA-binding elements of the human SIRT1 promoter in HEK293 cells (Fig. 4E).
FIGURE 4.
Binding of FoxO1 to the sirt1 promoter. A and B, FoxO1 binds directly to the IRS-1bcd and FKHD-L elements in the sirt1 promoter in vitro. EMSA was conducted using in vitro translated FoxO1 proteins as described under “Experimental Procedures.” The partial sequences of the biotin-labeled IRS-1bcd (A) and FKHD-L (B) probes (wild type, underlined) and mutated probes (framed) are shown above the gel shift. The running orientation of the gels and the mobility-shifted bands are indicated by arrows. C and D, FoxO1 associates with the rat sirt1 promoter in VSMCs by ChIP assays. Soluble chromatin from VSMCs was coimmunoprecipitated with anti-FoxO1 or FoxO3a antibodies or an equal amount of rabbit IgG. DNA purified from starting (1% input) and immunoprecipitated samples was subjected to PCR. The amplified region surrounds the IRS-1-binding element (C) (−1035 to −686) or the FKHD-L-binding element (D) (−415 to −89) within the rat sirt1 promoter. One representative result from three independent experiments is shown. E, FoxO1 associates with the human SIRT1 promoter in HEK293 cells by ChIP assays. Soluble chromatin from HEK293 cells was coimmunoprecipitated with anti-FoxO1 or FoxO3a antibodies or an equal amount of rabbit IgG. DNA purified from starting (1% input) and immunoprecipitated samples was subjected to PCR. The amplified region surrounds two putative FoxO1 DNA-binding elements (−587 to −278) within the human SIRT1 promoter. One representative result from three independent experiments is shown. rSIRT1, rat sirt1; hSIRT1, human SIRT1.
FoxO1 Up-regulates SIRT1 Protein Expression
We determined the effects of FoxO1 and FoxO3a siRNA on SIRT1 expression in VSMCs and HEK293 cells. As shown in Fig. 5, A and B, knockdown of FoxO1 but not FoxO3a by siRNA resulted in a reduction in SIRT1 protein expression, indicating that endogenous FoxO1 up-regulates SIRT1 expression basally and specifically. Consistently, FoxO1 overexpression increased SIRT1 protein levels in both VSMCs (Fig. 5C) and HEK293 cells (Fig. 5D). FoxO1 overexpression had no effect on expression of other sirtuin members, including SIRT2 and -3 in VSMCs (Fig. 5E). Together, these results indicate that endogenous FoxO1 but not FoxO3a is a positive regulator of SIRT1 expression.
FIGURE 5.
Induction of SIRT1 protein expression by FoxO1. A and B, FoxO1 depletion by siRNA decreases SIRT1 expression. VSMCs (A) or HEK293 cells (B) were transfected with 30 nm FoxO1 or FoxO3a siRNA or scrambled siRNA for 48 h. C and D, FoxO1 overexpression increases SIRT1 protein level. C, VSMCs were infected with adeno-FoxO1 or control adeno-GFP for 24 h. D, HEK293 cells in 100 × 20-mm tissue culture dishes were transiently transfected with 10 μg/dish of FoxO1 expression plasmid or control vector pcDNA3 for 24 h. E, SIRT2 and SIRT3 expression were unaffected by FoxO1 overexpression in VSMCs. Protein expression of the samples from two independent experiments was analyzed by Western blotting. Data shown are from one of 3–4 independent experiments.
FoxO1 Mediates a Positive Autofeedback Regulation of SIRT1 Expression
SIRT1 interacts with and deacetylates FoxO1 and thus affects FoxO1-mediated transcription of specific target genes in other systems (10, 17, 29, 30). We thus performed the luciferase assays to evaluate whether SIRT1 overexpression could affect FoxO1-dependent SIRT1 transcription in VSMCs. As shown in Fig. 6A, coexpression of SIRT1 wild type significantly augmented FoxO1-mediated SIRT1 transcription (4.0 ± 0.3- versus 2.6 ± 0.2-fold, p < 0.01), as compared with FoxO1 alone. Coexpression of a catalytically inactive dominant negative mutant of SIRT1 (H363Y) had no significant effect (2.7 ± 0.5- versus 2.6 ± 0.2-fold, p > 0.05 as indicated), suggesting that the deacetylation of FoxO1 by SIRT1 might impact FoxO1-mediated SIRT1 transcription. To test this possibility, we performed coimmunoprecipitation assays to investigate whether SIRT1 interacts with and deacetylates FoxO1 in VSMCs. Both SIRT1 wild type and its dominant negative mutant interacted with FoxO1 (Fig. 6B, middle), but only SIRT1 wild type deacetylated FoxO1 (Fig. 6B, top). These data imply that SIRT1 potentiates FoxO1-dependent SIRT1 transcription through its binding to and deacetylation of FoxO1.
FIGURE 6.
A positive feedback regulation of SIRT1 expression via FoxO1. A, SIRT1 promotes FoxO1-mediated SIRT1 promoter activity. VSMCs were transfected with the 1.5-kb SIRT1 reporter and p-RL-TK vector for 24 h and then infected with the indicated Ad.GFP-FoxO1 with or without Ad.SIRT1 (WT) or its dominant negative mutant (H363Y) for 24 h before luciferase activity was determined. B, SIRT1 interacts with and deacetylates FoxO1. VSMCs were infected with the adenovirus encoding the indicated Myc-tagged SIRT1 (WT), SIRT1 (H363Y), GFP-tagged FoxO1, or Ad.GFP (control) for 36 h. Total cell lysates were immunoprecipitated (IP) using anti-GFP polyclonal antibody and blotted (IB) with anti-acetylated lysine (top panel) or anti-Myc monoclonal antibody (second panel). Equal amounts of cell lysates (5%) were loaded as input. Total levels of FoxO1 were analyzed by Western blotting using anti-GFP antibody (bottom panel). C, resveratrol (RSV) increases FoxO1-mediated SIRT1 transcription. VSMCs were transfected with the 1.5-kb SIRT1 reporter, FoxO1 expression plasmids, and p-RL-TK vector for 24 h. Cells were treated with either vehicle (DMSO) or increasing doses of resveratrol for 16 h before luciferase activity was determined. *, p < 0.05; **, p < 0.01 versus vehicle. D and E, resveratrol induces endogenous expression of SIRT1 but not SIRT2 and -3. Growth-arrested VSMCs were treated with 10, 30, and 60 μm resveratrol or vehicle (DMSO) for 24 h. Protein expression was analyzed by Western blotting with the indicated antibodies. F, FoxO1 is required for resveratrol-induced SIRT1 expression. VSMCs transfected with 50 nm FoxO1 siRNA or control scrambled siRNA were treated with 50 μm resveratrol or vehicle (DMSO) for 24 h. Protein expression of the samples from two independent experiments was analyzed by Western blotting.
Resveratrol is a potent activator of SIRT1. However, whether resveratrol activates SIRT1 directly or indirectly is being debated currently (46–48). We evaluated the impact of resveratrol on FoxO1-mediated SIRT1 transcription in VSMCs. As shown in Fig. 6C, resveratrol increased FoxO1-dependent SIRT1 transcriptional activity in a dose-dependent manner. Consistently, resveratrol treatment caused a dose-dependent induction in endogenous expression of SIRT1 despite the fact that FoxO1 expression was not affected (Fig. 6D). No effect of resveratrol on protein expression of other sirtuin members, including SIRT2 and -3, was observed in VSMCs (Fig. 6E). These results support the notion that resveratrol induces SIRT1 expression possibly through the activation of FoxO1-dependent SIRT1 transcription. To confirm this possibility, we determined the impact of FoxO1 depletion on resveratrol-induced SIRT1 expression (Fig. 6F). Resveratrol treatment increased SIRT1 expression, whereas FoxO1 depletion by siRNA decreased SIRT1 expression basally. Strikingly, the induction in SIRT1 expression by resveratrol was completely prevented by FoxO1 depletion, indicating that FoxO1 is required for resveratrol-induced SIRT1 expression in VSMCs.
DISCUSSION
SIRT1 and FoxO1 are evolutionarily conserved longevity genes involved in a broad spectrum of biological processes, many of which are salutary for health and development. In other systems, SIRT1 has been shown to interact physically with FoxO1 and to regulate its transcriptional activity through SIRT1-mediated deacetylation (16, 17, 29, 30). Here we show that FoxO1 binds to two conserved FoxO1-dependent DNA regulatory elements (IRS-1 and FKHD-L) in the rat sirt1 promoter. Further, endogenous FoxO1 is a positive regulator of SIRT1 transcription and expression in VSMCs and HEK293 cells. Importantly, we show that FoxO1-dependent SIRT1 transcription is augmented, probably by SIRT1 deacetylation of FoxO1, implying that SIRT1 modulates its own transcription through a positive feedback mechanism (Fig. 7). Finally, we demonstrate that resveratrol potentiates FoxO1-dependent SIRT1 transcription and thus up-regulates SIRT1 expression. These findings provide new insights into functional, self-amplifying consequences of interactions between FoxO1 and SIRT1. Further, these new data may be generally important in informing the centrality of these pathways in controlling processes as diverse as metabolism, senescence, and longevity.
FIGURE 7.

Schematic model of an autofeedback regulation of SIRT1 expression via FoxO1. FoxO1 binds to the IRS-1 and FKHD-L sites of the SIRT1 promoter and activates SIRT1 transcription. Newly synthesized SIRT1 interacts with and deacetylates FoxO1 and promotes FoxO1-dependent SIRT1 transcription. Resveratrol (RSV) promotes FoxO1-dependent SIRT1 transcription and thus induces SIRT1 expression.
FoxO transcription factors are important regulators of diverse cellular functions, including proliferation, differentiation, apoptosis, and defense against oxidative stress and aging in response to growth factors, such as insulin and insulin-like growth factor (27, 28). FoxO proteins regulate expression of their downstream target genes through binding to the highly conserved FoxO-binding element (TTGTTTAC) (44, 45) or the insulin-responsive core sequences (TGTTTT) (41, 42, 44). We found that the 1.5-kb rat SIRT1 promoter region contains a putative forkhead-like binding site (FKHD-L), which matches the FKHD consensus element (TTGTTTAC) in a reverse orientation at five of eight bases, and a cluster of five insulin-responsive core repeat motifs (IRS-1), which are required for FoxO1 binding and SIRT1 transcription. A main finding of this study is that FoxO1 directly activates SIRT1 transcription via binding to those binding elements. Nemoto et al. (22) previously showed that FoxO3a interacts with p53 and binds to the p53 response elements within the mouse Sirt1 promoter, thereby up-regulating SIRT1 transcription. We find that the putative p53 DNA-binding elements are also present in the promoter regions of the rat and human SIRT1 genes. Therefore, other FoxO proteins may influence SIRT1 expression via this indirect mechanism.
SIRT1 plays a broad range of physiological roles, including stimulating antioxidant stress response enzymes through targeting substrates, such as FoxO transcription factors and PGC-1α (peroxisome proliferator-activated receptor-γ coactivator 1α) (10, 49, 50). SIRT1 interacts with and deacetylates FoxO1 and PGC-1α at multiple lysine sites, consequently increasing PGC-1α coactivation of FoxO1-mediated transcriptional activity (50, 51). We showed previously that PGC-1α coactivation of FoxO1-mediated catalase transcription is inhibited by PGC-1α acetylation in VSMCs (34). Here we show that SIRT1 interacts with and deacetylates FoxO1 and potentiates FoxO1-driven SIRT1 transcription. SIRT1 probably binds to and deacetylates the FoxO1·PGC-1α elements of a transcriptional complex at the SIRT1 promoter, thus self-regulating SIRT1 transcription.
Resveratrol has been shown to delay aging in many organisms from yeast to mammals (32). Initially, it was generally believed that resveratrol, as a direct activator of SIRT1, was involved in longevity, improvement of insulin sensitivity, and increase of mitochondrial biogenesis. However, controversy has surrounded the direct effect of resveratrol on SIRT1 activation. Recent evidence in vitro (47, 48) indicated that resveratrol exerts indirect effects on SIRT1 activation. Evidence in vivo (52, 53) also supported a notion that resveratrol may increase SIRT1 enzymatic activity through the sustained elevation of the cellular NAD+ levels, which depends on AMP-activated protein kinase activation. Here we show that SIRT1 wild type, but not a catalytically inactive dominant negative mutant, increases FoxO1-dependent promoter activity, suggesting that activation of SIRT1 enzymatic activity by resveratrol is the likely mechanism for resveratrol-induced SIRT1 expression. Additionally, we show that resveratrol promotes FoxO1-dependent SIRT1 transcription and induces SIRT1 protein expression. Interestingly, depletion of FoxO1 by siRNA prevents resveratrol-induced SIRT1 expression, indicating that FoxO1 is required for SIRT1 up-regulation in response to resveratrol treatment. Therefore, it is reasonable to anticipate that the effects of resveratrol on SIRT1 are possibly mediated by the increase of NAD+-dependent SIRT1 enzymatic activity via AMP-activated protein kinase leading to deacetylation of FoxO1. However, the extent to which FoxO1 deacetylation by SIRT1 contributes to resveratrol-activated SIRT1 expression remains to be investigated (Fig. 7). The FoxO1/resveratrol-dependent regulation of SIRT1 is not a general mechanism controlling all sirtuin family members because SIRT2 and SIRT3 expression was unaffected by FoxO1 (Fig. 5E) and resveratrol (Fig. 6E). Finally, we cannot exclude the possibility that elevated stability of SIRT1 mRNA (23) or protein (54) is involved in resveratrol-induced SIRT1 expression.
In summary, we demonstrate that FoxO1 binds to two cis-acting elements, IRS-1 and FKHD-L, in the rat sirt1 promoter and directly activates SIRT1 transcription. Further, SIRT1 interacts with and deacetylates FoxO1, enhancing its own transcription. These observations indicate a possible forward autofeedback loop mechanism that may sustain SIRT1 expression. Moreover, the SIRT1 activator resveratrol augments FoxO1-mediated SIRT1 transcription, which may increase the gain of the feedback system. These findings provide novel insights into the role of FoxO1 in controlling SIRT1 expression and into its self-amplifying features, which are enhanced by resveratrol. Further, the increased understanding of the transcriptional mechanisms controlling SIRT1 expression provided here may enable new therapeutic approaches to a broad spectrum of aging-related, oxidative stress-dependent diseases.
Supplementary Material
Acknowledgments
We are grateful to J. A. Hill (University of Texas Southwestern Medical Center, Dallas, TX) for AdFoxO1-WT and K. Irani (University of Pittsburgh Medical Center, Pittsburgh, PA) for AdSIRT1-WT and SIRT1-H363Y constructs. We especially thank Drs. Didier Merlin and Yutao Yan (Emory University, Atlanta, GA) for assistance with luciferase assays. We also thank the members of the Alexander laboratory for helpful discussion and support.
This work was supported, in whole or in part, by National Institutes of Health Grants UO1 HL80711 and HL60728.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S3.
- VSMC
- vascular smooth muscle cell.
REFERENCES
- 1. Haigis M. C., Guarente L. P. (2006) Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 2. Brooks C. L., Gu W. (2009) Nat. Rev. Cancer 9, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaeberlein M., McVey M., Guarente L. (1999) Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin S. J., Defossez P. A., Guarente L. (2000) Science 289, 2126–2128 [DOI] [PubMed] [Google Scholar]
- 5. Tissenbaum H. A., Guarente L. (2001) Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 6. Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 7. Boily G., Seifert E. L., Bevilacqua L., He X. H., Sabourin G., Estey C., Moffat C., Crawford S., Saliba S., Jardine K., Xuan J., Evans M., Harper M. E., McBurney M. W. (2008) PLoS One 3, e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Y., Xu W., McBurney M. W., Longo V. D. (2008) Cell Metab. 8, 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donmez G., Guarente L. (2010) Aging Cell 9, 285–290 [DOI] [PubMed] [Google Scholar]
- 10. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 11. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 12. Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. (2007) Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 14. Kadowaki T., Yamauchi T., Kubota N., Hara K., Ueki K., Tobe K. (2006) J. Clin. Invest. 116, 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamauchi T., Kamon J., Waki H., Terauchi Y., Kubota N., Hara K., Mori Y., Ide T., Murakami K., Tsuboyama-Kasaoka N., Ezaki O., Akanuma Y., Gavrilova O., Vinson C., Reitman M. L., Kagechika H., Shudo K., Yoda M., Nakano Y., Tobe K., Nagai R., Kimura S., Tomita M., Froguel P., Kadowaki T. (2001) Nat. Med. 7, 941–946 [DOI] [PubMed] [Google Scholar]
- 16. Qiao L., Shao J. (2006) J. Biol. Chem. 281, 39915–39924 [DOI] [PubMed] [Google Scholar]
- 17. Banks A. S., Kon N., Knight C., Matsumoto M., Gutiérrez-Juárez R., Rossetti L., Gu W., Accili D. (2008) Cell Metab. 8, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Potente M., Ghaeni L., Baldessari D., Mostoslavsky R., Rossig L., Dequiedt F., Haendeler J., Mione M., Dejana E., Alt F. W., Zeiher A. M., Dimmeler S. (2007) Genes Dev. 21, 2644–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 20. Chen W. Y., Wang D. H., Yen R. C., Luo J., Gu W., Baylin S. B. (2005) Cell 123, 437–448 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Q., Wang S. Y., Fleuriel C., Leprince D., Rocheleau J. V., Piston D. W., Goodman R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Nemoto S., Fergusson M. M., Finkel T. (2004) Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 23. Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. (2007) Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim E. J., Kho J. H., Kang M. R., Um S. J. (2007) Mol. Cell 28, 277–290 [DOI] [PubMed] [Google Scholar]
- 25. Kim J. E., Chen J., Lou Z. (2008) Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 26. Haigis M. C., Sinclair D. A. (2010) Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ronnebaum S. M., Patterson C. (2010) Annu. Rev. Physiol. 72, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gross D. N., van den Heuvel A. P., Birnbaum M. J. (2008) Oncogene 27, 2320–2336 [DOI] [PubMed] [Google Scholar]
- 29. Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10042–10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakae J., Cao Y., Daitoku H., Fukamizu A., Ogawa W., Yano Y., Hayashi Y. (2006) J. Clin. Invest. 116, 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jing E., Gesta S., Kahn C. R. (2007) Cell Metab. 6, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ni Y. G., Berenji K., Wang N., Oh M., Sachan N., Dey A., Cheng J., Lu G., Morris D. J., Castrillon D. H., Gerard R. D., Rothermel B. A., Hill J. A. (2006) Circulation 114, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiong S., Salazar G., San Martin A., Ahmad M., Patrushev N., Hilenski L., Nazarewicz R. R., Ma M., Ushio-Fukai M., Alexander R. W. (2010) J. Biol. Chem. 285, 2474–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattagajasingh I., Kim C. S., Naqvi A., Yamamori T., Hoffman T. A., Jung S. B., DeRicco J., Kasuno K., Irani K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berk B. C., Vekshtein V., Gordon H. M., Tsuda T. (1989) Hypertension 13, 305–314 [DOI] [PubMed] [Google Scholar]
- 37. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 38. Gunther S., Alexander R. W., Atkinson W. J., Gimbrone M. A., Jr. (1982) J. Cell Biol. 92, 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Griendling K. K., Berk B. C., Socorro L., Tsuda T., Delafontaine P., Alexander R. W. (1988) Clin. Exp. Pharmacol. Physiol. 15, 105–112 [DOI] [PubMed] [Google Scholar]
- 40. Griendling K. K., Tsuda T., Berk B. C., Alexander R. W. (1989) J. Cardiovasc. Pharmacol. 14, Suppl. 6, S27–S33 [PubMed] [Google Scholar]
- 41. Biggs W. H., 3rd, Cavenee W. K., Arden K. C. (2001) Mamm. Genome 12, 416–425 [DOI] [PubMed] [Google Scholar]
- 42. Vander Kooi B. T., Streeper R. S., Svitek C. A., Oeser J. K., Powell D. R., O'Brien R. M. (2003) J. Biol. Chem. 278, 11782–11793 [DOI] [PubMed] [Google Scholar]
- 43. Schilling M. M., Oeser J. K., Boustead J. N., Flemming B. P., O'Brien R. M. (2006) Nature 443, E10–E11 [DOI] [PubMed] [Google Scholar]
- 44. Becker T., Loch G., Beyer M., Zinke I., Aschenbrenner A. C., Carrera P., Inhester T., Schultze J. L., Hoch M. (2010) Nature 463, 369–373 [DOI] [PubMed] [Google Scholar]
- 45. Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. (2003) Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 47. Beher D., Wu J., Cumine S., Kim K. W., Lu S. C., Atangan L., Wang M. (2009) Chem. Biol. Drug Des. 74, 619–624 [DOI] [PubMed] [Google Scholar]
- 48. Pacholec M., Bleasdale J. E., Chrunyk B., Cunningham D., Flynn D., Garofalo R. S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., Withka J., Ahn K. (2010) J. Biol. Chem. 285, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sedding D. G. (2008) Biol. Chem. 389, 279–283 [DOI] [PubMed] [Google Scholar]
- 50. Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 51. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 52. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Um J. H., Park S. J., Kang H., Yang S., Foretz M., McBurney M. W., Kim M. K., Viollet B., Chung J. H. Diabetes 59, 554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ford J., Ahmed S., Allison S., Jiang M., Milner J. (2008) Cell Cycle 7, 3091–3097 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.