Abstract
The multiple transferable resistance (MTR) pump, from Neisseria gonorrhoeae, is typical of the specialized machinery used to translocate drugs across the inner and outer membranes of Gram-negative bacteria. It consists of a tripartite complex composed of an inner-membrane transporter, MtrD, a periplasmic membrane fusion protein, MtrC, and an outer-membrane channel, MtrE. We have expressed the components of the pump in Escherichia coli and used the antibiotic vancomycin, which is too large to cross the outer-membrane by passive diffusion, to test for opening of the MtrE channel. Cells expressing MtrCDE are not susceptible to vancomycin, indicating that the channel is closed; but become susceptible to vancomycin in the presence of transported substrates, consistent with drug-induced opening of the MtrE channel. A mutational analysis identified residues Asn-198, Glu-434, and Gln-441, lining an intraprotomer groove on the surface of MtrE, to be important for pump function; mutation of these residues yielded cells that were sensitive to vancomycin. Pull-down assays and micro-calorimetry measurements indicated that this functional impairment is not due to the inability of MtrC to interact with the MtrE mutants; nor was it due to the MtrE mutants adopting an open conformation, because cells expressing these MtrE mutants alone are relatively insensitive to vancomycin. However, cells expressing the MtrE mutants with MtrC are sensitive to vancomycin, indicating that residues lining the intra-protomer groove control opening of the MtrE channel in response to binding of MtrC.
Keywords: Antibiotics, Bacteria, Drug Resistance, Membrane, Multidrug Transporters
Introduction
Gram-negative bacteria frequently utilize transport systems composed of a tripartite assembly of proteins that span both the inner and outer membranes to pump antibiotics from the cell. These assemblies consist of an inner-membrane protein (IMP),3 which transduces electrochemical energy, and an outer-membrane protein (OMP) that are connected by a periplasmic membrane fusion protein (MFP), which is anchored to the inner membrane (1–3). The systems largely utilize proton-antiporters but there are some that utilize ABC transporters (4–5). Analogous tripartite pumps exist that export proteins such as toxins and adhesion factors, and for such pumps the IMP is typically an ABC transporter (6). Indeed, the importance of such systems is emphasized by the fact that the same OMP can be utilized for both drug and toxin extrusion: for example, TolC functions with antibiotic H+-antiporters, such as AcrB that belongs to the RND family (7–8), with ABC transporters, such as the macrolide transporter MacB (4), and with HlyB that extrudes the protein-toxin α-hemolysin (6).
Although the structures of representatives of all the individual components have been elucidated, the structure of an assembled tripartite complex has not yet been determined. A well studied example for which the structures are known for all the constituent components is the AcrABTolC multidrug-pump from Escherichia coli; for which the structure of the RND transporter AcrB (9–10) and of its cognate MFP, AcrA (11), and OMP, TolC (12), have been determined. Cross-linking studies (13) indicate that the six β-hairpins from the upper headpiece of the AcrB-trimer contact the tips of the six helical pairs of the TolC trimer (14), forming a continuous path across the periplasm. Because of this internal pseudo 6-fold symmetry in both OMP and IMP, the exact stoichiometry of the MFP binding is still strongly debated with 3:3:3 (15) and 3:6:3 (16) being the most popular candidates. The latter suggests that there are both intra-protomer and inter-protomer grooves on the surface of the OMP that can accommodate the MFP. Modeling studies, supported by cross-linking, suggest that the coiled-coil domain of AcrA can fit into the intra-protomer grooves of the open state of TolC, to make contacts that resemble helical bundles, with corresponding contacts between the β-/lipoyl-domains of AcrA and the periplasmic domain of AcrB (15, 17, 18). There are several lines of evidence to suggest that assembly of the tripartite complex is a dynamic process in which the protein components undergo conformational changes. In most of the available structures of OMPs the channel is closed at one or both sides, so that conformational changes are required to allow passage of the substrate (14, 15, 17, 19). It is conceivable that a small number of key interactions between adjacent coiled-coils of the OMP are broken to allow the inner coils to untwist and realign with the outer coils, increasing the aperture of the OMP entrance (14, 20–22). Interaction with AcrA may enable the realignment of the coiled-coils, resulting in stabilization of TolC in the open state (11, 14, 20). Recent crystallographic studies of AcrB indicate that it can adopt at least three different conformations, lending support to a mechanistic model in which there is a functional rotation in the periplasmic domains driving transfer of drugs into the OMP (23–26). These conformational changes in AcrB could be transmitted to TolC via AcrA (14).
We have sought to further investigate the assembly of a tripartite multidrug efflux pump using as a model the MtrCDE-system from Neisseria gonorrhoeae, which is responsible for the extrusion of antibiotics, including β-lactams and macrolides, and hydrophobic agents, such as detergents and antimicrobial peptides (27–29). This efflux pump is an important contributor in conferring resistance to antibiotics and, because of its ability to extrude antimicrobial oligopeptides, such as PC-8 and LL-37, produced by mammalian cells as part of their innate defense, contributes to the virulence of the human pathogens N. gonorrhoeae and N. menigitidis (30–35). This system has the advantage that all the components of the pump are encoded by the same operon and, consequently, the interactions between the components are likely to be specific, facilitating analyses of pump assembly. We present evidence for a model for the transport mechanism in which drug binding to the IMP MtrD, an RND transporter, induces a tight interaction of the coiled-coil hairpin domain of the MFP MtrC with an intra-protomer groove on the surface of the OMP MtrE, to induce the open state necessary for drug efflux.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
The E. coli strains and plasmids used are described in supplemental Table S1, and the primers used for construction of plasmid vectors in supplemental Table S2. The mtrC, mtrD, and mtrE genes were amplified from the genomic DNA of N. gonorrhoeae FA19 (36).
Site-directed Mutagenesis of mtrE
Site-directed mutagenesis was performed with the QuickChange XL kit (Stratagene). For in vivo functional analyses of the pump the pACYCDuet-mtrC/mtrD/mtrE plasmid was used as the template; while for the purification of MtrE derivatives, the pET-mtrE plasmid was used as the template; with the primers given in supplemental Table S3 used to introduce the required mutations into mtrE. PCR reactions were performed using the following cycling parameters: initial activation 95 °C-1 min, denaturation 95 °C-50 s, annealing 55 °C-50 s, and extension 68 °C-10 min for 17 cycles, with a final extension at 68 °C for 10 min. After PCR, the template was digested with Dpn1 for 3 h and 5 μl of the PCR mix was transformed into Novoblue E. coli and colonies were selected for chloramphenicol resistance. Plasmids were extracted from the colonies and their DNA sequenced.
Protein Overexpression and Purification
All the proteins used in this study were purified as fusion proteins with a six-histidine tag. The E. coli cells harboring plasmids were grown at 37 °C in 2xYT media, with shaking at 220 rpm. When the cell density reached an A600 of 0.6, 0.5 mm IPTG was added, the temperature was adjusted to 25 °C and cells left to overexpress proteins for 6 h for membrane proteins and 3 h for soluble proteins. Cells were harvested by centrifugation at 4 °C and resuspended in 20 mm Tris-HCl buffer, 300 mm NaCl, pH 7.5, 10% v/v glycerol. Cell paste was supplemented with complete protease inhibitor (Roche) and DNase I (Sigma) prior to lysis using three passes through a cell disrupter (1 × 10 kpsi and 2 × 25 kpsi, model Z-plus 1.1kW, Constant Systems). Cell debris were removed by centrifugation at 37,000 × g (45 min at 4 °C), and cell membranes isolated by ultracentrifugation at 125,000 × g (90 min at 4 °C) for membrane proteins, while the supernatant was collected for soluble proteins. Membranes pellets were then solubilized in 20 mm Tris-HCl buffer, pH 7.5, 100 mm NaCl, 10% (v/v) glycerol, 2% w/v βDDM.
MtrD Purification
Membranes pellets were dissolved in 2% w/v DDM and MtrD protein was purified under 0.1% w/v βDDM by affinity chromatography using a Ni2+-charged HiTrap chelating column (GE Healthcare). After desalting, MtrD samples were subjected to ion exchange chromatography, using a HiTrap Q column (GE Healthcare) and eluted using a NaCl gradient.
Δ1–34-MtrC Purification
NT-MtrC supernatant was purified by affinity chromatography using a HiTrap Ni2+-chelating column, followed by gel filtration chromatography with buffer 20 mm Tris-HCl buffer, pH 7.5, 300 mm NaCl, 10% v/v glycerol.
MtrE Purification
MtrE was purified as described for the purification of MtrD but was subject to cation exchange chromatography using a HiTrap SP column (GE Healthcare) after the chelating column step.
MtrC Hairpin Purification
MtrC α-helical hairpin was purified as described for the purification of NT-MtrC.
Growth Curve Analyses
E. coli cells, of strain (ΔacrB) KAM3(DE3)(37) or (ΔtolC) TG1(DE3) (38), harboring plasmids were grown at 37 °C (200 rpm) until the cell density reached an A600 of 0.5, and then induced with1 mm IPTG. The cells were grown for a further 3 h, diluted with 2xYT media containing antibiotic and the growth curve recorded; or the cells monitored for growth after 24 h for MIC measurements.
Microcalorimetry
ITC measurements were carried out at 25 °C using a VP-ITC Micro-Calorimeter (MicroCal). MtrD and MtrE were prepared in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10% (v/v) glycerol, 0.1% w/v βDDM (buffer 1). After adding βDDM to NT-MtrC and MtrC hairpin to a final concentration of 0.1% (w/v), they were dialyzed in different dialysis cassettes at the same time with MtrD or MtrE against buffer 1. An ITC experiment involved making a single 2 μl and a series of 8 μl injections of protein into the instrument mixing chamber, which contained the other protein. All data were corrected for the heat changes arising from injection of proteins into buffer before data analysis with ORIGIN software (MicroCal). Experiments were carried out in triplicate and a statistical average taken.
Pulldown Assays
For pulldown assays, proteins were overexpressed with a C-terminal S-tag or GST-tag for use with the cognate protein that was His-tagged. Membrane pellets, containing the S-tagged or GST-tagged prey protein, were solubilized in 2% w/v βDDM and mixed with purified His-tagged bait-protein (∼2 mg in 1 ml). The mixture was loaded onto a Ni2+-charged Hitrap chelating column (GE Healthcare), so that the bait-protein could be immobilized on the column along with the prey protein if they interact. The column was then washed with 20 column volumes of Tris buffer containing 25 mm imidazole and 0.1% w/v βDDM to eliminate any nonspecific interactions. The bait-prey protein complex was eluted with 500 mm imidazole Tris buffer, containing 0.1% w/v βDDM, and SDS-PAGE used to visualize the bait and prey proteins. A Western blot was performed with anti-S tag or anti-GST tag antibodies to confirm the presence of the S-tagged and GST-tagged prey proteins. For the MtrE E434K pulldown assay, the purified NT MtrC GST-tagged protein was used as bait, immobilized on GST column beads (GE Healthcare), to which was added purified His-tagged MtrE E434K prey protein (∼5 mg protein in 0.5 ml). The beads were washed with Tris buffer, containing 0.1% w/v βDDM, and finally the beads were boiled at 90 °C for 5 min to elute the complex. Western blotting was performed with both anti-GST and anti-His tag antibodies to detect bait and prey proteins, respectively.
Disc Diffusion Assays
E. coli cells were grown to an A600 of about 0.5, induced with 0.5 mm IPTG, the cells were diluted to 0.1 and then 200 μl of cell culture were spread on an MHA plate previously incorporated with 0.5 mm IPTG; antibiotic discs were placed on the plate medium and incubated at 37 °C overnight. The zone of culture growth inhibition was determined. The discs used were impregnated with 1 μg/ml nafcillin, 30 μg/ml tetracycline, 10 μg/ml erythromycin, 5 μg/ml novobiocin, and 30 μg/ml vancomycin.
Mass Spectrometry
Analyses were performed in a nanoflow electro-spray mass spectrometer (nES-MS) QSTAR and Q-ToF-2 (Waters, Milford, MA). The following experimental parameters were used on QSTAR to record mass spectra of 16 mg/ml MtrE: capillary voltage up to 1.2 kV, declustering potential 150 V, focusing potential 150 V, de-clustering potential-2 15 V, and collision energy up to 70 V. In MS mode, the following QToF-2 parameters were used for analysis of MtrE: capillary voltage 1668 V, sample cone 150 V, extractor cone 0 V, accelerating voltage into the collision cell (termed “collision energy” as per the manufacturer's terminology) 150 V, ion transfer stage pressure 6.1 × 10−3 mbar and ToF analyzer pressure 1.0 × 10−4 mbar.
Molecular Modeling
Because of the low sequence identity between MtrE and OMPs of known structure, combined sequence and structural alignments were created using T-coffee (39) and Expresso3D (40), respectively, and manually analyzed, followed by homology model building with Modeler (41), using the TolC (1ek9.pdb, 2vde.pdb) and OprM (1wp1.pdb) structures as templates. Because of the higher level of sequence identity (43%) and homology (75%) as calculated using the Smith-Waterman sequence similarity algorithm (42), we have used OprM as a guidance for the manual refinement of the MtrE model using geometrical (Ramachandran and torsion) restraints in Coot (43). The helical regions of OprM share sufficient homology with MtrE to allow the unambiguous assignment of the periplasmic part of the protein (see structural alignment of MtrE and OprM provided in supplemental Fig. S1). The optimized model was stereo-chemically consistent as witnessed by a Ramachandran plot analysis (performed with RAMPAGE, Ref. 44) with 96.6% of the residues in favored regions, 2.7% in allowed and only 0.7% outliers, as well as a ProSa model quality Z-score of −7.01 (45).
RESULTS
MtrC Interacts with MtrD and MtrE
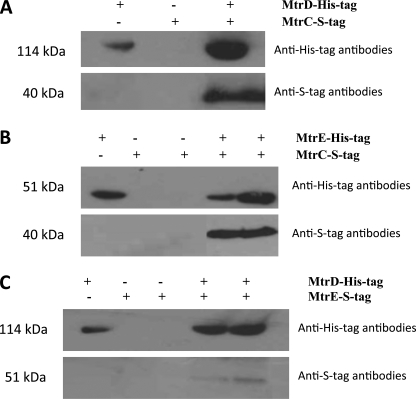
Interactions between N. gonorrhoeae MtrC, MtrD, and MtrE were tested for using detergent-solubilized proteins for pulldown assays (Fig. 1). One protein was expressed as a fusion protein with a His tag, which was used to purify the protein on an IMAC column, over which was passed a complex mixture of detergent-solubilized membrane proteins that included the cognate protein with an S tag. Interacting proteins were tested for by Western blotting using an antibody to the S tag. Using this approach MtrC was found to interact with MtrD (Fig. 1A) and MtrE (Fig. 1B). Although we detected an interaction between MtrD and MtrE (Fig. 1C), the intensity of the band suggested a weak interaction. The fact that in each case the cognate pump protein could be pulled-out of a complex mixture of detergent-solubilized proteins from membranes or cells indicated that the interactions are specific.
FIGURE 1.
The interaction of MtrC, MtrD, and MtrE. The pulldown of MtrC by MtrD and MtrE: purified His-tagged MtrD (A) and MtrE (B) were used as bait, immobilized on a Ni2+-agarose column, over which a slurry of detergent-solubilized membranes, from strains overexpressing S-tagged MtrC, was passed to test whether the MtrC protein could be pulled-out of this complex mixture of proteins. The MtrC prey protein was detected by Western blotting, using antibodies to the S tag. The pulldown of MtrE by MtrD: purified His-tagged MtrD (C) was used as bait, immobilized on a Ni2+-agarose column, over which a slurry of detergent-solubilized membranes, from strains overexpressing S-tagged MtrE, was passed to test whether this protein could be pulled-out of this complex mixture of proteins. The MtrE prey protein was detected by Western blotting, using antibodies to the S tag. As negative controls, either the bait protein or the prey protein was omitted from the assay (the presence and absence of proteins is indicated by + and −). As shown in each assay when either of these proteins was omitted from the assay, the corresponding prey protein could not be detected. As a positive control for the presence of the bait protein in each assay, the His-tagged protein was detected by Western blotting, using antibodies to the His-tag. In pulldown assays both the His-tagged bait and S-tagged prey proteins were detected. These assays indicate that MtrC interacts with MtrD and MtrE and that there is a weak interaction between MtrD and MtrE.
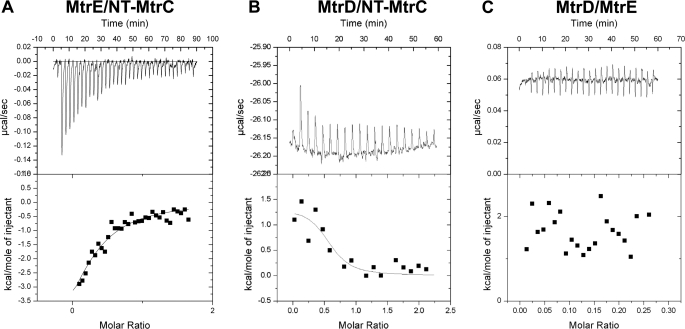
Microcalorimetry was used to assess the affinities of these interactions. When NT-MtrC was titrated into MtrE a weak exothermic interaction, with a Kd of 9.8 (±4.9) μm, was detected (Fig. 2A), which was characterized by a change in enthalpy (ΔH) of −6.8 (±3.58) kcal·mol−1 and entropy (ΔS) of −0.01 cal·mol−1·K−1. In contrast, when NT-MtrC was titrated into MtrD, a strong endothermic reaction, with a Kd of 0.8 (±0.70) μm, was detected (Fig. 2B), which was characterized by a ΔH of 1.4 (±0.32) kcal·mol−1 and ΔS of 32.5 cal·mol−1·K−1. In accord with our pulldown experiments that suggested that MtrD interacts weakly with MtrE, no appreciable interaction was detected by ITC (Fig. 2C). Our ITC measurements indicated that MtrE and MtrD have similar affinities for MtrC to that of TolC and AcrB for AcrA (7). In each case it is notable that the OMP binds the MFP with an affinity that is about an order-of-magnitude less than the IMP binds the MFP. In the case of TolC and AcrA this might have arisen because TolC needs to interact with a number of different transport systems. However, our studies established MtrC binds MtrE with a lower affinity than MtrD, suggesting that this is an intrinsic feature of such pumps, which might reflect a need to load and unload the OMP.
FIGURE 2.
ITC analyses of the interaction of MtrC, MtrD, and MtrE. 200 μm NT-MtrC was titrated into (A) 15 μm MtrE and (B) 12 μm MtrD; and in (C) 15 μm MtrE was titrated into 12 μm MtrD in a VP-ITC microcalorimeter and the heat exchange determined at 25 °C. In each case, the upper panel shows the raw energy changes during the titration, while the lower panel represents the derived integrated total energy change as a function of the molar ratio (based on the molecular weight of the monomeric protein) of the interactants. Non-linear regression fitting of the data (shown as a solid line through the data points in the lower panel) to a monophasic one-site model yielded the following thermodynamic parameters for the interaction: the interaction of Nt-MtrC with MtrE was characterized by a Ka, ΔH and ΔS of 1.0 (±0.50) × 105 m−1, −6836 (±3575) cal·mol−1 and −0.01 cal·mol−1·K−1; while the interaction of Nt-MtrC with MtrD was characterized by a Ka, ΔH, and ΔS of 1.2 (±1.0) × 106 m−1, 1412 (±318) cal·mol−1 and 32.5 cal·mol−1·K−1.
MtrC Interacts with MtrD and Mtr Eto Form a Functional Multidrug Pump
To establish that the MtrCDE proteins formed a functional multidrug pump we expressed them in the E. coli ΔacrB strain KAM3 (37) and a ΔtolC TG1 strain (38) that are hypersensitive to antibiotics; revealing that while MtrD alone did not confer resistance, it was able to confer resistance when expressed with MtrC and E (Table 1).
TABLE 1.
MICs for (ΔacrB)KAM3(DE3) and (ΔtolC)TG1(DE3) strains expressing mtr genes
| Strain/constructs | Penicillin G | Nafcillin | Tetracycline | Erythromycin |
|---|---|---|---|---|
| μg/ml | μg/ml | μg/ml | μg/ml | |
| KAM3/pACYC | 32 | 32 | 4 | 0.5 |
| KAM3/pACYC MtrD | 32 | 32 | 16 | 2 |
| KAM3/pACYC MtrCDE | 64 | 1024 | 64 | 128 |
| TG1/pACYC vector | 16 | 1.5625 | <0.5 | 2 |
| TG1/pACYC MtrD | 16 | 1.5625 | <0.5 | 2 |
| TG1/pACYC MtrCDE | 64 | 32 | 12.5 | 64 |
We used the antibiotic vancomycin, which is too large (1450 Da) to cross the outer-membrane by passive diffusion (14), to test for opening of MtrE, which would allow its passage into the cells to disrupt peptidoglycan synthesis. In comparison to control ΔtolC TG1 cells, transformed with pACYC, cells transformed with pACYC-mtrCDE were equally insensitive to vancomycin; but only the latter became highly susceptible to vancomycin in the presence of nafcillin (Table 2). For example, while both ΔtolC TG1 control cells, transformed with pACYC, and cells expressing MtrCDE, transformed with pACYC-mtrCDE, both had an MIC for vancomycin of 512 μg/ml; in the presence of (1 μg/ml) nafcillin this had reduced to 128 μg/ml and 8 μg/ml for control and MtrCDE-expressing cells, respectively. Although the data for the control cells suggest nafcillin and vancomycin can act synergistically, presumably to do so the vancomycin must gain entry to the cells. Our data for cells expressing MtrCDE suggest that there is a more substantive uptake of vancomycin by these cells than control cells. Similarly, KAM3 cells expressing MtrCDE were more susceptible to vancomycin in the presence, than the absence, of nafcillin (supplemental Fig. S2). We also tested the effect of tetracycline on the susceptibility of the ΔtolC TG1 cells expressing MtrCDE to vancomycin: revealing an apparent sensitivity of the cells to vancomycin in the presence of tetracycline (supplemental Fig. S3). That this effect is small might be due to the ability of the pump to remove vancomycin from the cells, so that while some vancomycin leaks into the cells, some is pumped out again. It is notable that cells expressing MtrCDE tend to grow better than control cells, lacking the pump, in the presence of vancomycin. In any event, our data clearly indicate that the MtrCDE channel is effectively closed (to vancomycin) in the absence of transported drugs and suggests that MtrD, upon drug binding, induces conformational changes in MtrC that enable it to interact with MtrE to induce opening of its channel (allowing vancomycin entry).
TABLE 2.
MICs determined by broth-dilution method for mtrE mutants expressed with mtrCD in E.coli (ΔtolC) TG1(DE3)
MtrE intra-protomer groove mutants (N198L, E434K and Q441E) are shown in bold; underline indicates faint growth; antibiotic abbreviations are Pen, penicillin G; Naf, nafcillin; Tet, tetracycline; Ery, erythromycin; Nov, novobiocin; and Van, vancomycin; and for studies of the simultaneous effects of nafcillin and vancomycin, the MIC for vancomycin was determined in the presence of 1 μg/ml nafcillin.
| Constructs | Pen | Naf | Tet | Ery | Nov | Van | Van (+ 1 μg/ml Naf) |
|---|---|---|---|---|---|---|---|
| μg/ml | μg/ml | μg/ml | μg/ml | μg/ml | μg/ml | μg/ml | |
| Vector pACYC | 16 | 1.5625 | <0.5 | 2 | <0.5 | 512 | 128 |
| MtrE WT-CD | 64 | 32 | 12.5 | 64 | 2 | 512 | 8 |
| MtrE N198L-CD | 32 | 8 | <0.5 | 2 | 1 | 256 | 8 |
| MtrE Q441E-CD | 16 | 1 | <0.5 | 64 | <0.5 | 64 | 8 |
| MtrE E434K-CD | 16 | 1 | <0.5 | 64 | <0.5 | 64 | 8 |
| MtrE R239K-CD | 64 | 32 | 12.5 | <0.5 | 2 | 256 | 8 |
| MtrE K397E-CD | 64 | 32 | 12.5 | <0.5 | 2 | 256 | 8 |
Residues Within the Intra-protomer Groove of the MtrE trimer Are Important for the Function of the Assembled Tripartite Pump
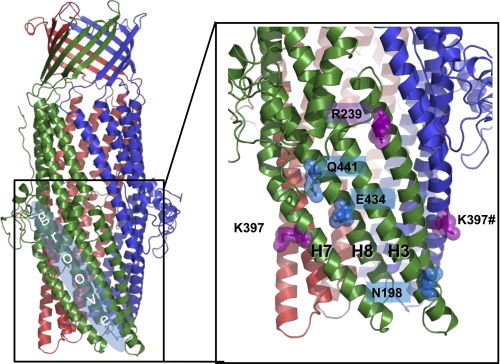
Previous studies of the interaction of AcrA with TolC indicate that the hairpin domain of the MFP binds within an intra-protomer groove, bounded by helices 3, 7, and 8, of the OMP (15). To pinpoint the possible MtrC-interacting residues on the surface of MtrE, we have created a homology model of the MtrE (Fig. 3 and supplemental Fig. S4B) based upon the available three-dimensional structures of OMPs. To experimentally test that MtrE has the same oligomeric state as that of other OMPs already crystallized, we used mass spectrometry (46) to determine the molecular weight of the intact assembly; which indicated a trimeric stoichiometry with a molecular mass of 148,890 (+32) Da (supplemental Fig. S4A).
FIGURE 3.
A molecular homology model of the structure of MtrE showing the intra-protomer grooves and the position of the mutations introduced into the intra-protomer groove and the flanking region.
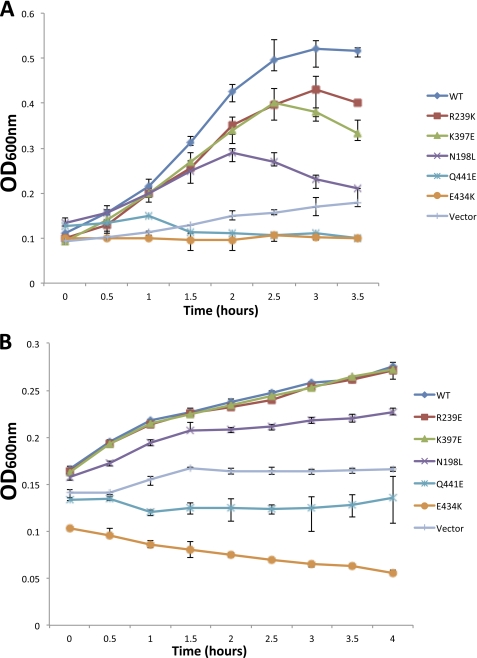
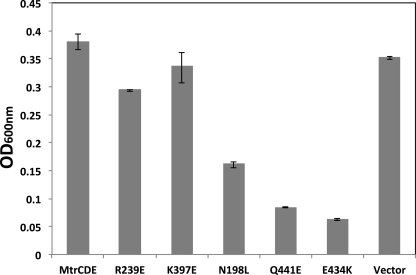
A comparative analysis of the modeled MtrE and TolC molecules was performed with a view to identifying conserved solvent accessible residues, and taking into account previous mutagenesis data (14, 15, 21). Based on this analysis, three point mutants (e.g. E434K, Q441E, and N198L), positioned toward the proximal, middle, and distal end of the intra-protomer groove, were constructed, along with mutants (e.g. R239E and K397E) located outside the intra-protomer groove as a control, and the effects of these in respect to their antibiotic sensitivity were tested. In contrast to the cells expressing MtrCD with the R239E and K397E derivatives of MtrE, those expressing the N198L, E434K, and Q441E derivatives clearly had an increased sensitivity (as indicated by lower MICs) to a range of antibiotics (Table 2). Surprisingly, when we compared the growth curves for these MtrE derivatives, we found that mutations in the intra-protomer groove had a marked effect, with these mutants being even more susceptible to antibiotics than control cells lacking a drug pump (Fig. 4). This behavior was apparent for both nafcillin, which targets penicillin-binding proteins in the periplasm, and tetracycline, which targets the ribosome in the cytoplasm. Consequently, this data would argue against these mutations simply disrupting the interaction of MtrCD with MtrE but suggests that the pump assembles and that this is detrimental to the growth of the cells.
FIGURE 4.
Mutations in the intra-protomer groove of MtrE impair the MtrCDE pump. Growth curves for a ΔtolC TG1(DE3) strain expressing the mtrCDE genes, in the presence of (A) 16 μg/ml nafcillin and (B) 3 μg/ml tetracycline, for wild-type mtrE and a series of mtrE mutants, and the same strain transformed with the empty vector as control. The growth curve corresponding to each mutant, the WT and control are indicated to the right of each curve.
We also analyzed the effects of antibiotics, including vancomycin, on the growth of these mutants on plates, confirming that the MtrE N198L, E434K, and Q441E mutants were susceptible to antibiotics and more so than the R239E and K397E mutants (Table 3). Importantly, these studies revealed more pronounced differences between the mutants: cells expressing MtrCD, with wild-type MtrE or its R239E and K397E derivatives, were insensitive to vancomycin; while the growth of the E434K and Q441E mutants was completely inhibited by vancomycin (Table 3). These data strongly suggest that the impaired function of the N198L, E434K and Q441E mutants is due to the pump becoming leaky, which would also explain why its assembly is detrimental to the cells.
TABLE 3.
The inhibitory zone determined by the disc diffusion method for mtrE mutants expressed with or without mtrC or mtrCD in E.coli (ΔtolC) TG1(DE3) and (ΔacrB) KAM3(DE3) cells
The inhibitory zone diameter is given in mm; abbreviations are NI, no inhibition of growth; NG, no growth due to complete growth inhibition; Naf, nafcillin; Tet, tetracycline; Ery, erythromycin; Nov, novobiocin; and Van, vancomycin. The MtrE derivatives with intra-protomer groove mutations are shown in bold.
| Strain/constructs | Naf (1 μg/ml) | Tet (30 μg/ml) | Ery (10 μg/ml) | Nov (5 μg/ml) | Van (30 μg/ml) |
|---|---|---|---|---|---|
| TG1/pACYC | 20 | 35 | 31 | 27 | NI |
| TG1/MtrE WT-CD | 12 | 25 | 20 | 19 | NI |
| TG1/MtrE N198L-CD | 20 | 33 | 31 | 27 | 7 |
| TG1/MtrE E434K-CD | 25 | 40 | 33 | 28 | NG |
| TG1/MtrE Q441E-CD | 19 | 40 | 33 | 27 | NG |
| TG1/MtrE R239E-CD | 14 | 29 | 27 | 25 | NI |
| TG1/MtrE K397E-CD | 14 | 29 | 29 | 21 | NI |
| KAM3/pET21a | 7 | 30 | 26 | 20 | NI |
| KAM3/MtrE WT | NI | 26 | 21 | 15 | NI |
| KAM3/MtrE N198L | NI | 26 | 21 | 15 | NI |
| KAM3/MtrE E434K | NI | 26 | 21 | 15 | NI |
| KAM3/MtrE Q441E | NI | 26 | 21 | 15 | NI |
| KAM3/MtrE R239E | NI | 26 | 21 | 15 | NI |
| KAM3/MtrE K397E | NI | 26 | 21 | 15 | NI |
| KAM3 pACYC | 7 | 36 | 31 | 17 | NI |
| KAM3/MtrE WT-C | 24 | 36 | 37 | 21 | 9 |
| KAM3/MtrE N198L-C | NI | 26 | 21 | 15 | NI |
| KAM3/MtrE E434K-C | NG | 50 | 40 | NG | NG |
| KAM3/MtrE Q441E-C | NG | 36 | 37 | 21 | 27 |
| KAM3/MtrE E434K-CD | 20 | 46 | 40 | 19 | 20 |
| KAM3/MtrE WT-CD | NI | 27 | 21 | 10 | NI |
Residues in the Intra-protomer Groove of MtrE Influence Opening of the Channel
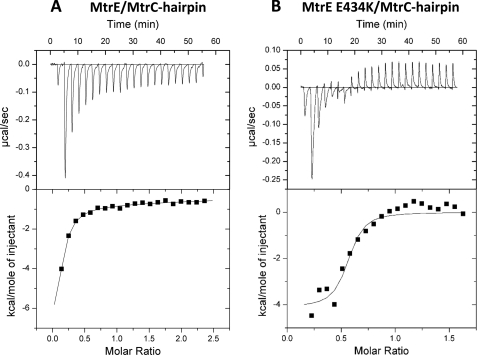
Because mutation of the intra-protomer groove impaired pump function, we tested whether this arose due to a reduced interaction of MtrC with the MtrE derivatives. A pulldown experiment was undertaken using GST-tagged NT MtrC as the bait and His-tagged MtrE E434K as the prey; revealing that the interaction was not compromised and that MtrC was capable of pulling-down MtrE E434K (Fig. 5). In comparison to the wild-type MtrE control, we did not see any reduction in the intensity of the band corresponding to His-tagged MtrE E434K on a Western blot, suggesting that MtrE and its E434K derivative have at least similar affinities for MtrC. Furthermore, we sought to test if the MtrE E434K derivative interacted with the coiled-coil hairpin domain of MtrC. For this experiment, we made a construct to express the hairpin domain of MtrC, consisting of residues 103–183; the purified protein had a CD spectrum that indicated that it retained α-helical structure (data not shown). Using ITC, we established that the hairpin binds to both wild-type MtrE and its E434K derivative (Fig. 6). Analysis of the binding isotherms indicated that the hairpin binds to a site on MtrE that is characterized by a Kd, ΔH and ΔS of 2.2(±0.29) μm, −11 (±13.1) kcal·mol−1 and −11.6 cal·mol−1·K−1; while its binding to the E434K derivative was characterized by a Kd, ΔH and ΔS of 0.13 (±0.078)μm, −4.1 (±0.31) kcal·mol−1 and 17.8 cal·mol−1·K−1. Interestingly, these data indicate that MtrE E434K binds the hairpin of MtrC with an affinity that is one-to-two orders of magnitude greater than the wild-type protein binds the hairpin or NT MtrC (see 0.13 μm versus 2.2/9.8 μm), suggesting that this derivative forms a tight assembly with MtrCD.
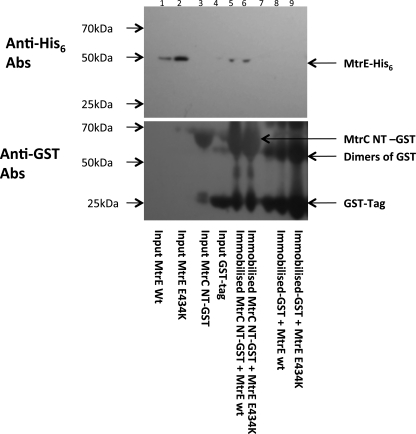
FIGURE 5.
Analysis of the interaction of the MtrC with MtrE E434K. Purified GST-tagged NT-MtrC protein was used as bait, immobilized on GST column beads, to which was added purified His-tagged MtrE E434K prey protein. The beads were washed with Tris buffer, containing 0.1% w/v βDDM, and finally the beads were boiled at 90 °C for 5 min to elute the complex. This experiment was repeated using purified His-tagged wild-type MtrE as a positive control. The following samples were run on an SDS-PAGE gel: wild-type MtrE (lane 1), MtrE E434K input control (lane 2), NT-MtrC GST input control (lane 3), GST tag input control (lane 4), NT MtrC-GST + wild-type MtrE pulldown (lane 5), NT-MtrC-GST + MtrE E434K pulldown (lane 6), MW marker (lane 7), GST tag + wild-type MtrE pulldown control (lane 8), and GST tag + MtrE E434K pulldown control (lane 9). The MtrC bait protein and the MtrE prey protein were detected by Western blotting using antibodies to the GST tag and the His tag (lower and upper panel), respectively. In pulldown assays both the His-tagged prey- and GST-tagged bait proteins were detected, indicating that NT-MtrC interacts with the MtrE E434K derivative.
FIGURE 6.
ITC analysis of the interaction of the MtrC hairpin with MtrE and its E434K. 500 μm MtrC hairpin was titrated into (A) 15 μm MtrE and (B) 15 μm) MtrE E434K in a VP-ITC micro-calorimeter and the heat exchange determined at 25 °C. The upper panel shows the raw energy changes during the titration, while the lower panel represents the derived integrated total energy change as a function of the molar ratio (based on the molecular weight of the monomeric protein) of the interactant. In the case of wild-type MtrE, for which the titration was extended to a molar ratio of 2.5, the data were best-fitted to a two site model yielded the following thermodynamic parameters for the interaction: Ka1, Ka2, ΔH1, ΔH2, ΔS1, and ΔS2 of 4.6 (±0.61) × 105 m−1, 3.2 (±0.96) × 103 m−1, −1.2 (±1.31) × 104 cal·mol−1,−6.2 (±0.01) × 104 cal·mol−1, −11.4 cal·mol−1·K−1, and −190 cal·mol−1·K−1, respectively. In the case of the MtrE E434K derivative, the titration was restricted to a molar ratio of 1.5, and consequently it was only appropriate to fit the data to a one site model; the high-affinity site was characterized by a Ka, ΔH and ΔS of 8.2 (±4.8) × 106 m−1, −4115 (±311) cal·mol−1 and 17.8 cal·mol−1·K−1.
One possibility is that MtrC traps MtrE E434K in the open conformation, so that it becomes leaky. Consequently, we sought to determine if the mutations in the intra-protomer groove of MtrE had caused the channel to become leaky, by testing the susceptibility of cells expressing these MtrE mutants with MtrCD to vancomycin. These studies revealed that all of the intra-protomer groove mutants were susceptible to vancomycin, while the R239E and K397E derivatives of MtrE were relatively insensitive to vancomycin (Fig. 7 and Table 3). This behavior cannot be attributed to a decrease in the expression of the MtrE derivatives because this would be expected to decrease, not increase, the susceptibility of the cells to vancomycin. Furthermore, purification of the MtrE E434K derivative indicated that it was expressed and targeted to the membrane at similar levels to wild-type MtrE, while our pulldown studies confirmed that it was equally capable of forming a complex with MtrC as wild-type MtrE (Fig. 5).
FIGURE 7.
Mutation of the intra-protomer groove induces opening of the MtrE channel in the MtrCDE pump. A bar chart showing the 4-h growth for a ΔtolC TG1(DE3) strain expressing the mtrCDE genes in the presence of 160 μg/ml vancomycin, for wild-type mtrE and a series of mtrE mutants (as indicated), and the same strain transformed with the empty vector as control. Cells expressing MtrCDE, MtrCD, and the R239E and K397E derivatives of MtrE, as well as the control cells, were insensitive to vancomycin; while cells expressing MtrCD and the N198L, E434K, and Q441E derivatives of MtrE, which are located in the intra-protomer groove, were clearly susceptible to vancomycin.
Although the above data suggest that the MtrE N198L, E434K, and Q441E mutants, when expressed with MtrCD, become leaky to vancomycin, presumably because of opening of the MtrE channel, it was not clear if this arose due to an intrinsic leakiness of the MtrE mutants. To determine if these MtrE mutants were intrinsically leaky, and to avoid any potential interaction with AcrAB, they were expressed alone in KAM3 cells, showing that none of them were as sensitive to vancomycin as when expressed with MtrCD (Table 3). This suggests that the interaction of the MtrE derivatives with MtrC, and possibly MtrD, is needed to stabilize the open conformation of MtrE. However, expression of MtrC with these MtrE mutants conferred sensitivity to vancomycin to the cells, indicating that they only required an interaction with MtrC to become leaky (Table 3). This contrasts with cells expressing wild-type MtrCE and MtrCDE, which are relatively insensitive to vancomycin (Table 3), suggesting that the interaction with drug-loaded MtrD is required to fully unlock the channel for the wild-type MtrCDE assembly.
DISCUSSION
Recent studies suggest that the MFP plays more than a passive role in coupling the IMP and OMP: in that it interacts with and induces opening of the OMP channel. For example, in one study, the TolC K383E and AcrA D149K mutants were shown to be individually hypersensitive to novobiocin but, when expressed together, complemented one another to confer resistance to novobiocin (14). This suggests that there is a direct interaction of TolC and AcrA involving these residues, possibly via the formation of a salt bridge. Furthermore, in contrast to mutations in the TolC channel, which conferred vancomycin sensitivity on the mutant cells, the TolC K383E mutant was relatively insensitive to vancomycin, and it was concluded that the K383E derivative was probably impaired in its ability to interact with AcrA. In another study, four residues, 147GLVA150, located in the turn region between the outer helices H3 and H4 of TolC, were simultaneously altered to 147AGSG150; the resulting mutant was hypersensitive to several antibiotics, but insensitive to vancomycin (47). Because this mutant interacted with AcrA, its impaired function was attributed to closure of the TolC channel. The characterization of antibiotic-resistant revertants of cells expressing TolC 147AGSG150 with AcrAB revealed compensatory changes in AcrA; suggesting that these induced or stabilized an opening of the TolC channel. Although these studies indicate that AcrA can support the open state of TolC, its exact role in regulating the transition of TolC from its closed to open state remains unclear as in these studies TolC was mutated so that the channel was either open (14) or closed (47). For example, in the first study a salt bridge in the channel of TolC was disrupted, by introducing a Y362F/R367E double mutation, to make it leaky (14); while in the other study the periplasmic end of TolC, comprising of residues 147–150, was mutated from GLVA to AGSG, causing it to close (47). In addition to these studies, two mutants of TolC, R367H, and R390C, have been reported that cause TolC to adopt a constitutively open state (14, 20, 48).
We have sought to investigate the potential role of the MFP in inducing opening of the OMP channel using the components of the MtrCDE multidrug pump from N. gonorrhoeae. The introduction of N198L, E434K, and Q441E mutations, positioned toward the proximal, middle, and distal end of the intra-protomer groove of MtrE, all impaired pump function, so that the cells expressing the mtrE mutants with mtrCD became hypersensitive to a range of antibiotics (Fig. 4 and Table 2). Surprisingly, an analysis of the growth curves of the E434K and Q441E mutants revealed that they were more susceptible to nafcillin and tetracycline than control cells (Fig. 4), suggesting that for these mutants assembly of the pump was detrimental for the cells. Consistent with this suggestion, the E434K and Q441E mutants grew poorly in the presence of vancomycin (Fig. 7), indicating that they form a leaky pump. However, although the E434K and Q441E mutants were sensitive to vancomycin, this did not prevent their growth (Fig. 7). Furthermore, MIC measurements indicated that neither mutant was hypersensitive to vancomycin but that they became highly susceptible to it in the presence of nafcillin (Table 2). These data indicate that, although leaky, these mutants are able to undergo the transition from the closed to open state in response to transported drugs. It is worth noting that Glu-434 occupies a position corresponding to Lys-383 in TolC, the exact residue, which has been shown to be directly involved in an interaction with AcrA (14) and thus it is plausible that it is also directly involved in the interaction with the MFP MtrC in the MtrCDE system, as it appears to be exposed on the surface as well.
Taken together, these data suggest that the intra-protomer groove of MtrE has a role in controlling the closed to open transition and mutations in the groove can induce the open state. However, it is possible that this behavior could arise because the MtrE mutant is unable to interact with MtrC. Consequently, we performed a pulldown assay, which revealed that MtrC interacts with MtrE E434K as strongly as with wild-type MtrE (Fig. 5). Considering this, we tested the vancomycin sensitivity of cells expressing the E434K and Q441E derivatives in the absence and presence of MtrC. These studies revealed that while the cells expressing E434K and Q441E were insenstive to vancomycin, those co-expressing these mutants in conjunction with MtrC were hypersensitive to vancomycin (Table 3). These data suggest that although mutation of the intra-protomer groove may facilitate opening of the channel, the interaction with MtrC is required for this to occur. Interestingly, while cells expressing MtrE E434K with MtrC did not grow at all on vancomycin plates, those expressing MtrE E434K with MtrCD grew, albeit poorly (Table 3). In comparison, cells expressing wild-type MtrCE had a slight sensitivity to vancomycin, again suggesting that the MFP unlocks the channel. In contrast, cells expressing MtrCDE appeared to be sensitive to vancomycin only in the presence of transported substrates (Tables 2 and 3). These findings suggest that in the fully assembled transporter, MtrC is blocked from opening the MtrE channel, which is otherwise locked, until induced to do so by MtrD as it binds drugs.
Previous studies have been interpreted in terms of a transport mechanism in which there are two energetically distinct steps (14). Firstly, the helical protrusions of the OMP engage in the exposed periplasmic crown of the IMP, regardless of the drug-bound state, resulting in a partial opening of the OMP channel. Consistent with this proposal, our studies confirm that there is a weak interaction of MtrD and MtrE in the absence of drugs (Fig. 1C). Upon opening of the periplasmic gates of the OMP, as a result of the interaction between the crown of the IMP and the tip of the periplasmic end of the OMP (13), helices H7/H8 of the OMP partially relax and an intra-protomer groove is exposed in the OMP, which forms part of the binding site for the MFP. In the resting state, this groove is less pronounced, and the MFP cannot be accommodated (7). Although our studies indicate that MtrC binds to the intra-protomer groove, formed by helices H7/H8, of MtrE (Fig. 4) even in the absence of MtrD (Fig. 1B), the interaction is weaker than that of MtrD with MtrC (Fig. 2A). The engagement of the MFP with the intra-protomer groove of the OMP, which is in the partially opened state, is thought to be necessary to force further opening of the channel. Consistent with this proposal, when MtrE is expressed with MtrC the channel becomes slightly leaky to vancomycin (Table 3). This behavior is more pronounced for the MtrE E434K mutant, which is insensitive to vancomycin but becomes very leaky to vancomycin when expressed with MtrC (Table 3 and Fig. 7). It is thus possible to suggest a role for the Glu-434 in not-so-much docking of the MFP, but in transmitting the opening signal after the initial dock takes place, possibly by interacting with the second selectivity gate in the OMP (14) and facilitating its unlocking. On the other hand, in the fully assembled tripartite complex, composed of the wild-type proteins, drug binding to MtrD is apparently required before MtrC can unlock the outer gate of MtrE (Table 3). The MtrE E434K derivative likely corresponds to a conformation that is primed for opening upon interaction with MtrC; and as such, may resemble the conformation of wild-type MtrE bound to the MtrD/drug complex. In this conformation of MtrE, presumably the outer gate is locked and the intra-protomer groove is exposed ready for the high affinity binding of the MFP, which would be necessary to unlock the outer gate and stabilize the open state. Considering that the MtrE E434K derivative does not allow the passage of vancomycin (Table 3) is consistent with the outer gate being locked in this conformational state and that binding of MtrC is required to induce unlocking of this gate. Furthermore, the E434K derivative binds the hairpin domain of MtrC with high affinity (Fig. 5), similar to that between MtrC and MtrD (Fig. 2B), consistent with a state in which the MFP becomes buried, and bound tightly, within the intra-protomer groove.
In summary, the MtrCDE proteins form a functional tripartite efflux-pump, similar to the AcrABTolC pump, in which the IMP drives a conformational switch that affects both the destabilization of the OMP gates and the presentation of an intra-protomer groove, to which the MFP binds and becomes buried, transferring the energy provided by the IMP, via several key residues that communicate with residues lining the OMP channel, to drive its full opening.
Supplementary Material
Acknowledgment
We thank Rajeev Misra (Arizona State University) for comments on the manuscript.
This work was supported by grants from the BSAC (British Society for Antimicrobial Chemotherapy) and the Wellcome Trust (to M. I. B.-W. and A. R. W.). This work was also supported in part by a Marie Curie Fellowship from the EU (to V. N. B.), the Wellcome Trust (to C. V. R.), and the BBSRC (to N. P. B., D. M.-V., and C. V. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S3.
- IMP
- inner-membrane protein
- OMP
- outer-membrane protein
- MFP
- membrane fusion protein
- MTR
- multiple transferable resistance
- βDDM
- β-dodecyl-maltoside
- NT-MtrC
- N-terminal-truncated (Δ1–34) MtrC
- hairpin
- 81 residue MtrC α-helical coiled-coil hairpin domain, consisting of residues 103–183 of MtrC
- ITC
- isothermal calorimetry
- MIC
- minimal inhibitory concentration.
REFERENCES
- 1. Pietras Z., Bavro V. N., Furnham N., Pellegrini-Calace M., Milner-White E. J., Luisi B. F. (2008) Curr. Drug Targets 9, 719–728 [DOI] [PubMed] [Google Scholar]
- 2. Misra R., Bavro V. N. (2009) Biochim. Biophys. Acta 1794, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blair J. M., Piddock L. J. (2009) Curr. Opin. Microbiol. 12, 512–519 [DOI] [PubMed] [Google Scholar]
- 4. Kobayashi N., Nishino K., Yamaguchi A. (2001) J. Bacteriol. 183, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borges-Walmsley M. I., McKeegan K. S., Walmsley A. R. (2003) Biochem. J. 376, 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holland I. B., Schmitt L., Young J. (2005) Mol. Membr. Biol. 22, 29–39 [DOI] [PubMed] [Google Scholar]
- 7. Touzé T., Eswaran J., Bokma E., Koronakis E., Hughes C., Koronakis V. (2004) Mol. Microbiol. 53, 697–706 [DOI] [PubMed] [Google Scholar]
- 8. Tikhonova E. B., Zgurskaya H. I. (2004) J. Biol. Chem. 279, 32116–32124 [DOI] [PubMed] [Google Scholar]
- 9. Murakami S., Nakashima R., Yamashita E., Yamaguchi A. (2002) Nature 419, 587–593 [DOI] [PubMed] [Google Scholar]
- 10. Yu E. W., McDermott G., Zgurskaya H. I., Nikaido H., Koshland D. E., Jr. (2003) Science 300, 976–980 [DOI] [PubMed] [Google Scholar]
- 11. Mikolosko J., Bobyk K., Zgurskaya H. I., Ghosh P. (2006) Structure 14, 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. (2000) Nature 405, 914–919 [DOI] [PubMed] [Google Scholar]
- 13. Tamura N., Murakami S., Oyama Y., Ishiguro M., Yamaguchi A. (2005) Biochemistry 44, 11115–11121 [DOI] [PubMed] [Google Scholar]
- 14. Bavro V. N., Pietras Z., Furnham N., Pérez-Cano L., Fernández-Recio, Pei X. Y., Misra R., Luisi B. (2008) Mol. Cell 30, 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lobedanz S., Bokma E., Symmons M. F., Koronakis E., Hughes C., Koronakis V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4612–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stegmeier J. F., Polleichtner G., Brandes N., Hotz C., Andersen C. (2006) Biochemistry 45, 10303–10312 [DOI] [PubMed] [Google Scholar]
- 17. Elkins C. A., Nikaido H. (2003) J. Bacteriol. 185, 5349–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Symmons M. F., Bokma E., Koronakis E., Hughes C., Koronakis V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7173–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Federici L., Du D., Walas F., Matsumura H., Fernandez-Recio J., McKeegan K. S., Borges-Walmsley M. I., Luisi B. F., Walmsley A. R. (2005) J. Biol. Chem. 280, 15307–15314 [DOI] [PubMed] [Google Scholar]
- 20. Andersen C., Koronakis E., Bokma E., Eswaran J., Humphreys D., Hughes C., Koronakis V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11103–11108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bokma E., Koronakis E., Lobedanz S., Hughes C., Koronakis V. (2006) FEBS Lett. 580, 5339–5343 [DOI] [PubMed] [Google Scholar]
- 22. Vediyappan G., Borisova T., Fralick J. A. (2006) J. Bacteriol. 188, 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. (2006) Nature 443, 173–179 [DOI] [PubMed] [Google Scholar]
- 24. Seeger M. A., Schiefner A., Eicher T., Verrey F., Diederichs K., Pos K. M. (2006) Science 313, 1295–1298 [DOI] [PubMed] [Google Scholar]
- 25. Sennhauser G., Bukowska M. A., Briand C., Grütter M. G. (2009) J. Mol. Biol. 389, 134–145 [DOI] [PubMed] [Google Scholar]
- 26. Seeger M. A., von Ballmoos C., Eicher T., Brandstätter L., Verrey F., Diederichs K., Pos K. M. (2008) Nat. Struct. Mol Biol. 15, 199–205 [DOI] [PubMed] [Google Scholar]
- 27. Hagman K. E., Pan W., Spratt B. G., Balthazar J. T., Judd R. C., Shafer W. M. (1995) Microbiology 141, 611–622 [DOI] [PubMed] [Google Scholar]
- 28. Shafer W. M., Qu X., Waring A. J., Lehrer R. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Veal W. L., Nicholas R. A., Shafer W. M. (2002) J Bacteriol. 184, 5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lucas C. E., Hagman K. E., Levin J. C., Stein D. C., Shafer W. M. (1995) Mol. Microbiol. 16, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 31. Rouquette C., Harmon J. B., Shafer W. M. (1999) Mol. Microbiol. 33, 651–658 [DOI] [PubMed] [Google Scholar]
- 32. Jerse A. E., Sharma N. D., Simms A. N., Crow E. T., Snyder L. A., Shafer W. M. (2003) Infect. Immun. 71, 5576–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tzeng Y. L., Ambrose K. D., Zughaier S., Zhou X., Miller Y. K., Shafer W. M., Stephens D. S. (2005) J. Bacteriol. 187, 5387–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Warner D. M., Folster J. P., Shafer W. M., Jerse A. E. (2007) J. Infect. Dis. 196, 1804–1812 [DOI] [PubMed] [Google Scholar]
- 35. Warner D. M., Shafer W. M., Jerse A. E. (2008) Mol. Microbiol. 70, 462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scudamore. R. A., Beveridge T. J., Goldner M. (1979) Antimicrob. Agents Chemother. 15, 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morita Y., Kodama K., Shiota S., Mine T., Kataoka A., Mizushima T., Tsuchiya T. (1998) Antimicrob. Agents Chemother. 42, 1778–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nagakubo S., Nishino K., Hirata T., Yamaguchi A. (2002) J. Bacteriol. 184, 4161–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 40. Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. (2006) Nucleic Acids Res. 34, W604-W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 42. Zemla A., Zhou C. E., Slezak T., Kuczmarski T., Rama D., Torres C., Sawicka D., Barsky D. (2005) Nucleic Acids Res. 33, W111-W115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44. Lovell S. C., Davis I. W., Arendall W. B., de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2002) Proteins: Struct., Funct. Genet. 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 45. Wiederstein M., Sippl M. J. (2007)Nucleic Acids Res. 35, W407-W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barrera N. P., Isaacson S. C., Zhou M., Bavro V. N., Welch A., Schaedler T. A., Seeger M. A., Miguel R. N., Korkhov V. M., van Veen H. W., Venter H., Walmsley A. R., Tate C. G., Robinson C. V. (2009) Nat. Methods. 6, 585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weeks J. W., Celaya-Kolb T., Pecora S., Misra R. (2010) Mol. Microbiol. 75, 1468–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Augustus A. M., Celaya T., Husain F., Humbard M., Misra R. (2004) J. Bacteriol. 186, 1851–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.