Abstract
Most bacteria use free enzymes to degrade plant cell walls in nature. However, some bacteria have adopted a different strategy wherein enzymes can either be free or tethered on a protein scaffold forming a complex called a cellulosome. The study of the structure and mechanism of these large macromolecular complexes is an active and ongoing research topic, with the goal of finding ways to improve biomass conversion using cellulosomes. Several mechanisms involved in cellulosome formation remain unknown, including how cellulosomal enzymes assemble on the scaffoldin and what governs the population of cellulosomes created during self-assembly. Here, we present a coarse-grained model to study the self-assembly of cellulosomes. The model captures most of the physical characteristics of three cellulosomal enzymes (Cel5B, CelS, and CbhA) and the scaffoldin (CipA) from Clostridium thermocellum. The protein structures are represented by beads connected by restraints to mimic the flexibility and shapes of these proteins. From a large simulation set, the assembly of cellulosomal enzyme complexes is shown to be dominated by their shape and modularity. The multimodular enzyme, CbhA, binds statistically more frequently to the scaffoldin than CelS or Cel5B. The enhanced binding is attributed to the flexible nature and multimodularity of this enzyme, providing a longer residence time around the scaffoldin. The characterization of the factors influencing the cellulosome assembly process may enable new strategies to create designers cellulosomes.
Keywords: Biophysics, Carbohydrate-binding Protein, Computer Modeling, Enzymes, Molecular Dynamics, Bacteria, Cellulases, Cellulosome, Coarse-grained Model, Self-assembly
Introduction
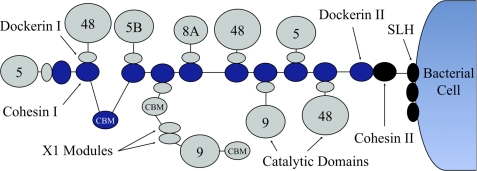
Many bacteria degrade cellulose in the biosphere via enzyme complexes called cellulosomes, a concept originally proposed by Bayer and Lamed from studies on the thermophilic cellulolytic anaerobe, Clostridium thermocellum (1–3). Cellulosomes are composed of two major units: long, putatively flexible scaffoldin proteins that contain specific binding sites, called cohesins, and enzymes that contain dockerin modules, which bind to the cohesins (Fig. 1).
FIGURE 1.
Concept of a cellulosome from C. thermocellum. The scaffoldin subunit (dark blue) contains nine cohesins and a carbohydrate binding module. The cellulolytic enzymes (gray) bind to cohesin partners with their dockerins. Another set of dockerin/cohesin interaction connects the scaffoldin to cell wall via a S-layer homologous (SLH) protein.
The self-assembly of the cellulosome complex is facilitated by the high-affinity recognition between cohesins and the enzyme-borne dockerin modules. The scaffoldin usually contains multiple cohesins, thereby enabling multiple enzymes containing glycosyl hydrolases (GH)3 to assemble into the cellulosome complex. In C. thermocellum, the cellulosome is composed of a primary scaffoldin subunit that can integrate up to nine enzymes. The cellulosome-integrating protein (CipA) of this cellulosome contains a single cellulose-specific family 3 carbohydrate binding module (CBM) that binds to the cellulose component of plant cell wall polysaccharides. In general, however, CBMs often reside in both the scaffoldin and enzymes for binding for selective binding to the variety of polysaccharide components of the plant. The cellulosome is a complex macromolecular system whose components are hypothesized to work synergistically to degrade plant cell walls efficiently.
The “plasticity theory” of the quaternary structure of the cellulosome is the main rationale for synergism (4–7). The plasticity theory is the hypothesized ability of the cellulosome to adjust to the substrate due to linker flexibility and to provide a more refined tuning to the substrate using the enzymatic linkers. However, the exact mechanism of attack of the cellulosome for plant cell wall is not well characterized. Another feature of cellulosomes is that different types of cohesins and dockerins exist in different microbial species and that recognition between cohesin and dockerin is type- and species-specific (8). This discovery permitted the development of “designer cellulosomes” for which controlled inclusion of selected enzymes into desired positions of artificial complexes is possible.
With the ability to make designer cellulosomes, Bayer et al. (9–11) were able to probe the following hypotheses; 1) the proximity of different enzymes may provide synergistic action on the crystalline substrate, which would perform better than their free forms when positioned in a designed pattern, and 2) enzymes from different species that have superior activities on given substrates can be assembled into one complex successfully and synergistically enhance conversion rates.
In support of the first hypothesis mentioned above are a series of studies aimed at building and testing engineered cellulosomes (1, 9–11). The first engineered cellulosome constructed by Fierobe et al. (12, 13) was composed of two cohesins and was, thus, “bi-functional” due to two cellulases being incorporated into the same cellulosome. Compared with the mixture of free cellulases, the resultant cellulosome chimeras exhibited enhanced synergistic action on crystalline cellulose. However, making comparisons to free cellulases, several of which lack CBMs, is difficult because the catalyst concentration at the biomass surface may be significantly different in the cellulosomes and the free cellulases. In 2005, Fierobe and coworkers created a new trifunctional engineered cellulosome by developing a third divergent cohesin-dockerin pair (4). The tri-functional engineered cellulosome was found to be superior in function compared with the bi-functional one. When the tri-functional engineered cellulosome was decorated with one hemicellulase (GH10) and two cellulases, it performed with superior activity on hatched straw. Cha et al. (15) also studied the effect of cohesin number (scaffold size) on synergistic performance of Clostridium cellulovorans minicellulosomes acting on cellulosic and hemicellulosic substrates. The results showed that as the number of cohesins engineered increased (up to four cohesins), a greater synergism was observed. However, the concentration of enzymes on the substrate surface, which is typically mediated by the CBM, is a key quantity for making direct comparisons of intrinsic conversion rates. Hence, comparing the activities of two enzymes where only one enzyme possesses a CBM and the other does not is not an equivalent comparison for intrinsic conversion rate. Mingardon et al. (16) later studied the effect of the symmetry of the minicellulosome using designer cellulosomes with various geometries for examining the targeting, proximity, and flexibility effects. This work suggested that increasing restriction of the relative mobility of the enzymes on the scaffold within the complexes negatively affects the cellulase activity; therefore, enzyme mobility is likely a critical parameter for cellulosome efficiency in degrading cellulose. Other studies have shown that recombining a single cellulosomal enzyme with a full-length scaffoldin was enough to increase hydrolytic activity over their free forms. This effect was partly attributed to enzyme proximity and better targeting of the substrate (17–19). Additionally, Bhat et al. (20) created a simplified cellulosome out of selected enzymes and a full-length scaffoldin and observed better synergy than in the free enzyme mixture. Unfortunately, a feature these studies have in common is the use of protein loadings too low to hydrolyze the crystalline content in the substrates chosen.
In support of the second hypothesis wherein enzymes from different species can be assembled in a complex enhancing activity, Caspi et al. (21) reported that the CBM modules of two free Thermobifida fusca family-6 cellulases, an endoglucanase Cel6A and an exoglucanase Cel6B, could be replaced by divergent dockerin modules. These modules could then be used to assemble engineered cellulosomes. The resultant chimeric proteins appeared to retain cellulase activity on cellulose. This result suggests that a free bacterial cellulase enzyme system could be transferred to a cellulosome-type scaffold, thus, providing more opportunities for engineered cellulosomes to use a diversity of free enzymes for assembly.
In addition, Raman et al. (22) observed that the contents of cellulosomes are adjustable, depending on the substrate. However, the feedback mechanisms used by the bacterium or the way the bacterium controls the population of enzymes on cellulosomes is not clear currently. The study of cellulosome assembly and enzyme binding competition are essential to understand how the bacterium adjusts its digestion mechanism.
Despite this considerable body of experimental work, a mechanistic understanding of cellulosome assembly and action remains elusive. Many questions remain including the following: 1) Do bacteria control the stoichiometry and relative positioning of the enzymes populating the scaffoldin? 2) Is such control useful for optimal conversion of plant cell wall polymers? 3) Does the microbial cell react to biomass chemistry and structure by optimizing the specificity of dockerin borne enzymes? 4) Can conversion rates be increased relative to the native configuration by engineering the enzymatic decoration of bacterial cellulosomes?
Here we use simulation to address the first question above regarding the self-assembly of the enzyme-scaffoldin complex. We examine the relative population of enzymes on the scaffoldin as a function of relative enzyme concentration in solution and the physical characteristics of the enzymes. Understanding the self-assembly process is important for behavior of natural cellulosomes because the cohesins on natural scaffolds can bind any dockerin within the same species. Thus, unlike designer cellulosomes that have specific, engineered binding sites, the location and proximity of different enzymes from natural (or unmodified) cellulosomes is unknown a priori.
Computer simulation has long been used to gain direct insight into biological self-assembly processes across multiple length and time scales. The model resolution is typically tuned to the length and time scale of interest. Models over a huge range of resolutions have been constructed to describe self-assembly of protein complexes. Atomistic protein models treat each atom in the system explicitly and have been typically limited to the aggregation of small peptides (23–26). Many groups have applied this approach to examine the aggregation of amyloid fibrils. Coarse-grained models of proteins start from these atomistic representations and include a huge body of literature too large to cover here. Several reviews (27–33) include discussions of such models, such as united atom models, implicit solvent representations, lattice and off-lattice descriptions where residues are modeled as beads, and native topology-based (e.g. Go) models up to nearest neighbor-type models where nodes represent one or more proteins. The lattermost types of models are often used to describe virus capsid assembly (34–38) for which the full complex consists of hundreds to thousands of individual protein molecules. Clearly, the level of coarse-graining for protein self-assembly will depend strongly on the system and hypothesis to address.
In this study we aim to understand the assembly of a system of cellulosomal enzymes on a protein scaffold with simulation. We describe an approach combining elements of the native-based topology methods with off-lattice protein simulations. Because cellulosomal proteins are known experimentally to populate the scaffold via the cohesin-dockerin interaction (39, 40) and these interactions have been quantified with simulation (41), we tune the cohesin-dockerin interactions to known values. All other interactions between the proteins and the scaffold are approximated as weak Lennard-Jones interactions (i.e. nearly hard sphere interactions). To our knowledge this is the first description of cellulosomal self-assembly and is a general means to model self-assembly in protein complexes up to hundreds or thousands of protein molecules with known specific interactions.
Our model is comprised of the C. thermocellum scaffoldin and three enzymes known to associate with this cellulosome. Below we list the names of the enzymes as referred to on the CAZy database (42), which in these cases are different from the names used in the original publications describing the enzymes. Here, we refer to the enzymes as listed in parentheses: exocellulase Cel48A (CelS) (43), endoglucanase Cel5A (Cel5B) (44), and endoglucanase Cbh9A (CbhA) (45, 46). The relative abundance of these enzymes in cellulosomes on different substrates can be found in Raman et al. (22).
The main questions addressed in this study are the following. 1) Is shape, mass, and architecture of the enzymes and the scaffold relevant in the self-assembly process? 2) Can we predict cellulosome composition from known, initial enzyme concentrations? Based on the results of this study, the shape and modularity of the enzymes are the most important factors in the their ability to bind to the scaffoldin. Volume, mass, and enzyme concentration are irrelevant, within the ranges and the simulation box size that was selected in this study as a compromise between a realistic volume surrounding the scaffoldin and a volume in which the binding events would occur in a tractable simulation timescale.
MATERIALS AND METHODS
Architecture
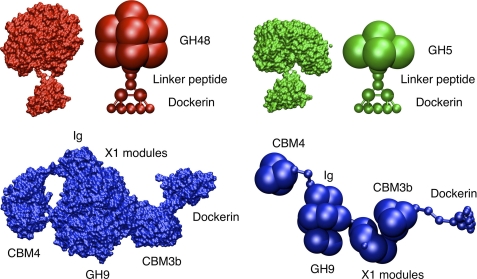
Cellulosomes from C. thermocellum can adopt different structures from the simplest nine-cohesin scaffoldin protein to more complex assemblies of seven scaffoldin proteins organized on an additional scaffold. Our discussion will be solely based on the simpler nine-cohesin structure of CipA (Fig. 2). A list of the components of CipA and the cellulosomal enzymes considered in this study can be found in Table 1.
FIGURE 2.
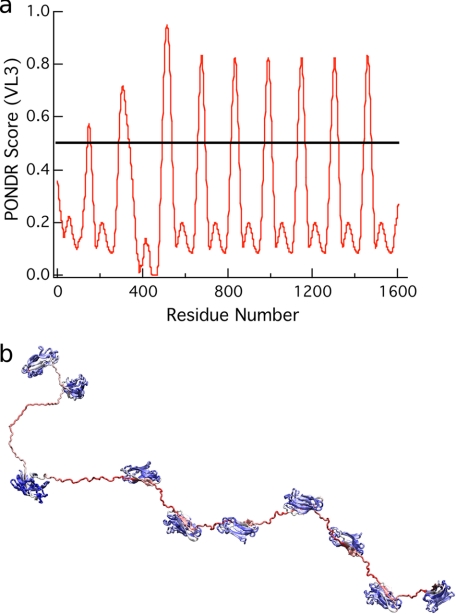
Sequence-based PONDR screen for protein disorder applied to CipA. a, the VL3 algorithm (58) predicts the CipA linkers to be disordered regions; these are the regions with scores greater than 0.5. b, the CipA scaffoldin colored by VL3 scores where the minimum score is 0 (blue) and the maximum VL3 score is 1.0 (red) show the predicted ordered and disordered regions of the scaffoldin. The X domain and dockerin module at the C terminus of the scaffoldin were omitted.
TABLE 1.
Architecture of the cellulosome protein complex
The following modules are included in the model: CBM, cohesin (COH), dockerin (DOC), X1 module (X1), and glycosyl hydrolase (GH). The signal peptide was not included in the calculation of the mass that was based on amino acid content.
| Protein | Modules | Molecular mass |
|---|---|---|
| kDa | ||
| CipA | 2COH-CBM3–7COH | 197 |
| Cel5B | GH5-DOC | 34 |
| CelS | GH48-DOC | 83 |
| CbhA | CBM4-Ig-GH9-2X1-CBM3-DOC | 138 |
The linkers between CipA modules vary in length and may be important contributors to the cellulosome plasticity. The linker glycosylation is not explicitly considered in our model but could be added by varying force-constant parameters and adding extra beads on the linker. Cellulosomal enzymes can have general structures of two modules, a dockerin and a catalytic module connected by a linker, or the enzymes may contain several modules. The cellulosome binds to cellulose with the CipA-borne CBM (47, 48) and other multi-component enzymes that include CBMs (45). Moreover, many cellulosomal enzymes contain different types of CBMs, which may provide the ability to bind to different substrates. Some CBMs seem to have an anchor function, whereas others have been hypothesized to be “helper” CBMs capable of holding a single cellulose chain and feeding it to its catalytic module partner (49). The immunoglobulin-like modules on Family 9 enzymes are believed to stabilize the catalytic modules (50). The function of X1 modules, is not understood but has been purported to disrupt cellulose from microscopy observations (51).
Despite the number of different modules present in the cellulosome, the overall assembly remains relatively intact (quasi-irreversible binding at physiological conditions) due to the high affinity between cohesins and dockerins. As mentioned earlier, in C. thermocellum, this affinity is nonspecific, and each dockerin can equally bind to any cohesin (8). This interaction requires Ca2+, which is essential for the complex to maintain structure (52, 53).
Functional Form and Parameters
The model for protein structures consists of spheres (beads) that represent regions of protein volume. The beads are connected by a network of restraints to mimic the shape and flexibility of globular proteins, dockerins, cohesins, and linkers. The beads interact via a weak Lennard-Jones potential. Each bead represents from three amino acids for linkers to hundreds of amino acids in large globular proteins. The restraints between beads are defined as bonds between beads in a linker to networks of bonds between beads in globular proteins. Additional interactions are included to mimic the cohesin-dockerin interaction. The model was developed within CHARMM (54). CHARMM offers flexibility in creating new pseudo atoms, has functionality for specific nonbond interactions between particular atom types, and allows the specification of additional parameters in the topology and parameters.
Within our template, the interactions between coarse-grained beads can be expressed as a sum of traditional classical bonded and non-bonded terms as follows. The non-bonded interactions are represented by a 6–12 Lennard-Jones potential,
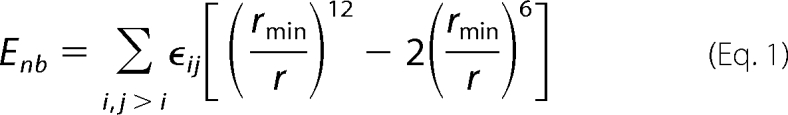 |
where rmin is the equilibrium distance between two particles, ϵij is the strength of their interaction, and r is the distance between two pseudo atom centers. The rmin for two pseudo-atoms was defined to accurately reproduce the sum of the radii of the beads and was chosen to mimic the size of the protein atoms the beads were approximating. Electrostatic effects were neglected due to the limited number of beads per protein.
A specific interaction was added between the beads in the binding site of the cohesins and dockerins with an additional set of non-bonded parameters. The binding energy was 13 kcal mol−1, which is between the experimental (12 kcal mol−1) (40) and theoretical value of 14.5 kcal mol−1 (41).
The bonded interactions are defined by the internal energy terms,
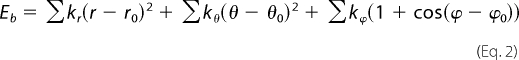 |
where r, θ, and φ are the distance, angle, and torsion angles between connected beads, r0, θ0, and φ0 are the coarse-grained bond, angle, and torsion angle equilibrium values, and kr, kθ, and kφ are the force constants (56). The force constants between beads of the same module are high to ensure rigidity, whereas intermodule linker regions have a wide range of flexibility. The distance, angles, and torsion angles were chosen to fit the original (all-atom) structures.
Description of the Cellulosomal Enzymes and Scaffold subunit
Scaffold Subunit
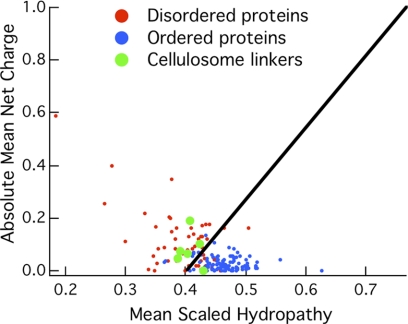
The polymeric scaffoldin of C. thermocellum, CipA, consists of nine cohesin proteins connected by linkers of 10–30 amino acid residues and an additional Family 3 CBM (CBM3). The X1 domain and dockerin module at the C terminus of the scaffoldin was omitted because the focus of this study is solely based on scaffoldin-enzyme binding. The linkers were analyzed with the VL3 algorithm, which is a feed-forward neural network trained on 152 disordered proteins characterized experimentally (57–61). Also, the charge-hydropathy relationship described by Uversky et al. (62) was used to predict the relative disorder of regions in the scaffoldin protein. Figs. 2 and 3 show the predictions from the VL3 algorithm and the charge-hydrophobicity relationship, respectively. The linker regions are disordered in both measures.
FIGURE 3.
Mean net charge as a function of hydropathy for the CipA linkers with an average length of 25 residues. The training sets for disordered and ordered proteins are shown in red and blue, respectively.
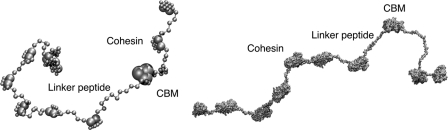
Each linker bead in the coarse-grained representation represents three amino acids to provide the flexibility of the all-atom structure. The all-atom and the coarse-grained representations of the full-length CipA are shown in Fig. 4. The linker regions are hypothesized to offer the plasticity required by the cellulosome to assume the most appropriate configuration given a particular substrate. Our study assumes that the linkers are flexible, which is substantiated by several experimental studies, including small angle x-ray scattering experiments conducted by Hammel et al. (7) and also crystallographic studies from Noach et al. (63). Also, recent computational work using replica exchange molecular dynamics simulations has shown that similar linkers are inherently flexible (64).
FIGURE 4.
Coarse-grained representation and all atom representation of CipA from C. thermocellum. The structures of the CBM and one of the cohesins are known and reported in the literature (48, 55). The other cohesins were obtained from homology modeling.
Cohesin and Dockerin
The coarse-grained cohesin was constructed to describe the binding interaction and create a flat binding surface while conserving the volume of the protein (Fig. 5). The dockerin is constructed with a binding surface to match the cohesin. There are three special attractor beads in a row across the center of the binding surface of the cohesin and dockerin, which are given special attracting properties for each other. The attracting beads are surrounded on the backside of the binding surface by beads that prevent multiple bindings to the same cohesin or dockerin simply by steric hindrance.
FIGURE 5.
All atom and coarse-grained representations of the cohesin from CipA with the attractive beads shown in red.
Cellulosomal Enzymes
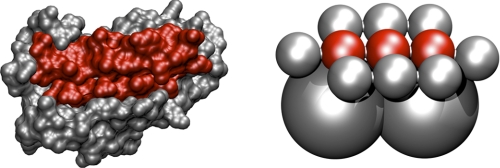
As mentioned earlier, C. thermocellum produces a variety of enzymes with different architectures. Three of these enzymes were selected in our study: the exocellulase, CelS, the endoglucanase, Cel5B, and the endoglucanase, CbhA. These enzymes have different masses, volumes, architectures, linker lengths, and radii of gyration and, thus encompass the complexity of the cellulosomal enzymes found in C. thermocellum. The modular architectures of these enzymes and the scaffoldin protein are shown in Table 1. The linkers between modules vary in length between 3 to 12 amino acids. Cel5B and CelS include a catalytic module, a linker, and a dockerin, whereas CbhA contains multiple modules with mostly unknown functions, such as X1 modules (45, 51) an immunoglobulin-like module (Ig) (45, 50), and two types of CBMs, CBM3b and CBM4 (45). One should note that the Cel5B all-atom structure was obtained from a homology model based on the endoglucanase C (65, 66). All of the enzymes studied here have a dockerin module able to bind to any cohesin on the scaffoldin, and in the coarse-grained model, dockerins, similar to cohesins, have an engineered binding face. The coarse-grained representations of these enzymes are shown in Fig. 6 along with their all atom counterparts. Note that the shape of the enzymes is reproduced and captures the effects of size and shapes of the enzymes in dynamics simulations.
FIGURE 6.
All atom and coarse-grained representations of CelS (GH48), Cel5B (GH5), and CbhA (GH9).
Simulation Setup
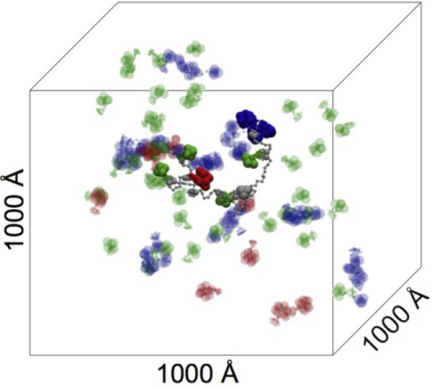
The periodic simulation box has a volume of 1 × 109 Å3 (1000 × 1000 × 1000Å) (Fig. 7). The enzyme concentration varies from 30 to 60 total enzyme molecules per scaffoldin molecule, equivalent to 50 and 100 μm. The concentration used in our simulations is higher than the one usually found in bacterial cells (1–10 μm). However, researchers have suggested that in the vicinity of the cell wall there could exist regions where the local enzyme concentration is increased to facilitate certain cell functions (67). The initial configurations were randomly generated, and velocities were drawn from the Boltzmann distribution. Initial simulations were performed with the full-length scaffoldin (nine cohesin). The second part of this study used a four-cohesin scaffoldin. The non-bonded cutoff distance was 99 Å, and frames were saved every 500 steps. Each trajectory was equilibrated for 100,000 steps with a time step of 2 fs, and trajectories were run for 30–100 ns. We performed 30 simulations of 30 ns duration for each of the different configurations to achieve meaningful statistical analysis in which total concentration, ratio of enzymes, masses, and shape were varied. These scans are summarized below and in Fig. 8.
FIGURE 7.
Simulation box with a scaffoldin molecule and some cellulosomal enzymes. The enzymes bound on the scaffoldin have solid colors. The color coding is the following: CelS (red), Cel5B (green), CbhA (blue).
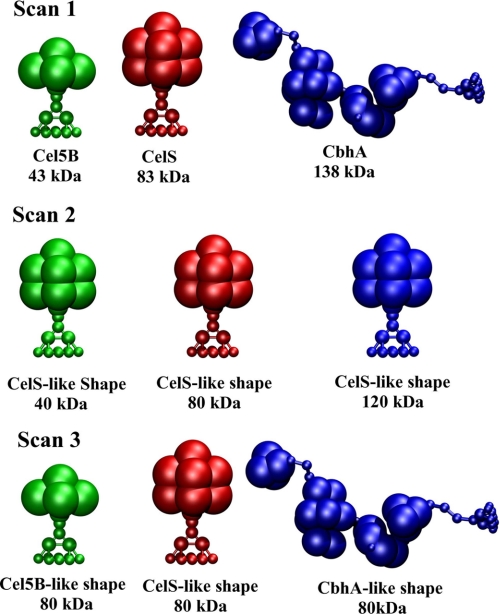
FIGURE 8.
Summary of the binding studies conducted. Scan 1, original masses and volumes/shapes are shown. Scan 2, same volumes/shapes and different masses are shown; Scan 3, different volumes/shapes and same masses are shown.
Scan 1
Two total enzyme concentrations were considered, 30 and 60 enzymes per 109 Å3, and the individual enzyme counts were varied with an increment of 10 to scan the possible concentration ratios at the same initial total enzyme concentration.
Scan 2
One total enzyme concentration (60 enzymes per 109 Å3) was used with three CelS-like enzymes of different masses. Individual enzyme counts were varied with an increment of 20 to scan concentration ratios with the same initial total enzyme concentration.
Scan 3
One enzyme concentration (60 enzymes per 109 Å3) was used with Cel5B, CelS, CbhA-like shapes and the same mass. Individual enzyme counts were varied with an increment of 20.
RESULTS
Behavior of the Component Enzymes
The 9-cohesin scaffoldin model was initially constructed to study the stability of the coarse-grained model developed in this work and to ensure that this scaffold possessed the geometrical and dynamical characteristics of the all-atom scaffold. In our simulations, the scaffold adopts compact configurations reminiscent of the TEM images by Mayer et al. (68). Starting from an extended configuration, the scaffold will tend to adopt a more compact form, as measured by the distribution of the radius of gyration, shown in the supplemental information along with a representative trajectory (supplemental Fig S2 and Movie 1). These data are directly comparable with small-angle x-ray scattering or light scattering experiments. It should be noted that in a more compact configuration the scaffold may be more shielded from the outside, and this might explain the results found by Morag et al. (69), where it was shown that removing enzymes docked on the scaffold was easier when the cellulosome was bound on cellulose where it may adopt a more extended configuration but was much harder when free in solution.
Additionally, we calculated the radius of gyration for the three component enzymes for comparison to experimental data. These distributions are shown in supplemental Fig. S1). Up to 100 ns were collected for each Rg distribution for a box of component enzymes. As mentioned above, the remaining simulations were conducted with a 4-cohesin scaffold for comparison to experimental binding studies on a 4-cohesin scaffold being conducted in our laboratory. The results from these binding studies can be found in the supplemental Tables S1–S4.
Scan with Original Enzymes (Scan 1)
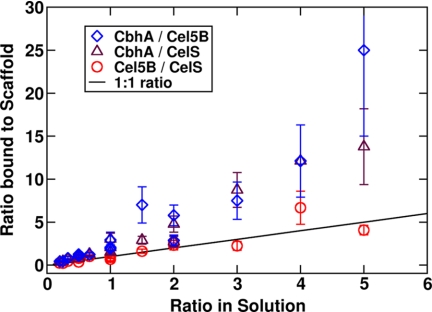
In the first binding study we used an enzyme concentration of 60 enzymes/box (100 μm) and the three enzymes with native properties as in Fig. 8. The results are shown in Figs. 9 and 10. This scan was designed to probe binding of these enzymes on the scaffoldin based solely on their physical characteristics: volume, shape, flexibility, and mass. The binding affinities of each of the dockerins for the CipA-borne cohesins were identical.
FIGURE 9.
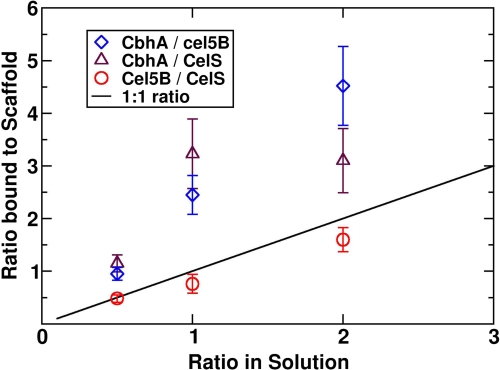
The ratio of the solution fraction to the 4-cohesin-scaffold-bound fraction for total enzyme concentration of 60 enzymes per box with original masses and shapes. The solid line represents an equal enzyme ratio in both solution and on the scaffoldin.
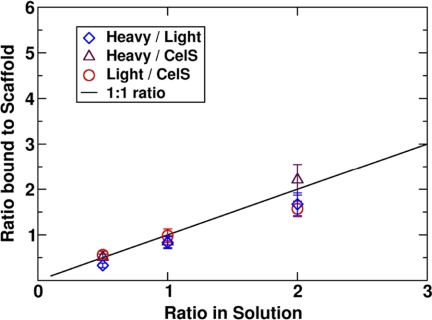
FIGURE 10.
The ratio of solution fraction to the 4-cohesin-scaffold-bound fraction for a total enzyme concentration of 60 enzymes per box with original masses and shapes for a ratio between 0 and 3. The solid line represents an equal enzyme ratio in both solution and on the scaffoldin.
CelS and Cel5B have similar shapes and flexibility characteristics. Even though CelS has twice the mass of Cel5B, the relative binding to the scaffoldin is similar; the ratio of Cel5B/CelS bound to the scaffoldin is approximately equal to the ratio in bulk solution for all concentrations tested. However, CbhA tends to bind more frequently to the scaffoldin in these simulations and even more so when they are predominant in solution. For a ratio of CbhA to CelS or to Cel5B of around 5 in solution, the ratio on the cellulosome is 15 or 25, respectively.
The importance of the modular nature of CbhA becomes clear upon closer inspection of the trajectories. The binding mechanism is different from the smaller, lighter, and more rigid CelS and Cel5B. Several dynamical properties of CbhA work together to increase its binding propensity. The larger mass of the whole CbhA makes its diffusion rate much slower than the lighter enzymes. When in the vicinity of the scaffoldin, CbhA remains within binding distance for a longer period of time than the lighter, less flexible enzymes. Because of its extended and flexible configuration, it has a larger cross-section that is on the order of the scaffoldin cross-section, thus, inhibiting its free diffusion past the scaffoldin. These factors extend the residence time near the scaffoldin and increase its binding probability. Finally, the dockerin has the ability to diffuse within the volume of CbhA, giving it a higher likelihood of encountering a cohesin. That is, the entire CbhA molecule does not have to rotate to put the dockerin near a cohesin. The dockerin visits many more positions within the volume of the CbhA molecule than it would if the molecule was rigid, and its position was determined by the rotation and translation of the entire CbhA enzyme. We suggest based on these results that all proteins with less flexible shapes will be less likely to bind than those with more flexible shapes in the limit of microscopic diffusion.
The same scan was conducted with a concentration of 30 enzymes per box and is shown in Fig. 11. Cel5b and CelS bind equally to the scaffold, whereas CbhA dominates the bound fraction. The only noticeable differences are in the ratio of CbhA/Cel5B and CbhA/CelS, especially for high CbhA concentrations.
FIGURE 11.
The ratio of solution fraction to the 4-cohesin-scaffold-bound fraction for a total enzyme concentration of 30 enzymes per box with original masses and shapes. The solid line represents an equal enzyme ratio in both solution and on the scaffoldin.
Crucial to the design of optimized cellulosomes is understanding the physical characteristics of an enzyme that enhances its binding affinity for the scaffoldin. From both scans that used the native physical characteristics of each enzyme, it is clear that CbhA is a preferred component of the cellulosome when its concentration in solution is similar to that of other enzymes. The next two scans were designed to isolate two important features of the enzymes presented in this study; that is, mass and shape/architecture.
Scan with Different Masses and Similar Volumes/Architectures (Scan 2)
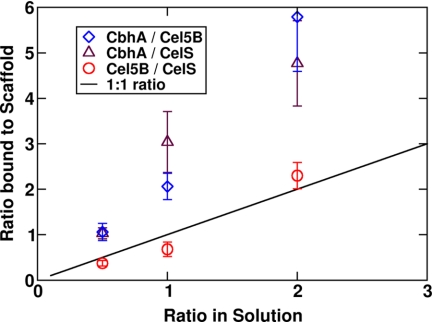
For this scan, the architecture and volume of CelS were selected with three different masses. Although the enzymes constructed in our model are not realistic for enzymes in nature, this approach allows the decoupling of the mass from the volume and shape of a given enzyme (i.e. this scan changes the diffusion coefficient).
The results (Fig. 12) reveal that the masses, at least in the range of masses selected here, do not appear to affect the way these “artificial enzymes” bind on the scaffold. The large mass of CbhA is not the reason for its predominance in the composition of the enzyme-bound cellulosomes in Scan 1.
FIGURE 12.
The ratio of solution fraction to the 4-cohesin-scaffold-bound fraction for a total enzyme concentration of 60 enzymes per box with same volumes/shapes and different masses. The system is described in Fig. 8.
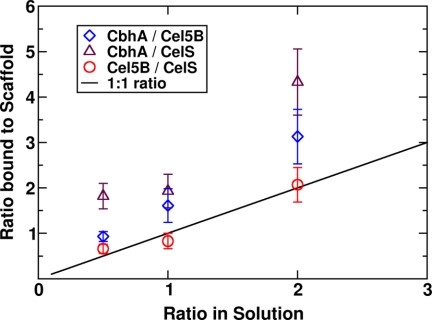
Scan with Different Volumes/Architectures and Similar Masses (Scan 3)
Scan 3 was designed to decouple the effects of architecture and volume from the effect of enzyme mass on the binding probability. Enzymes were all given a mass of 80 kDa, similar to CelS, therefore, assigning both CbhA and Cel5B masses different from their native masses. The results shown in Fig. 13 are strikingly similar to the Scan 1 with 60 enzymes (Fig. 10). Cel5B and CelS bind equally to the scaffold as observed in the Scans 1 and 2. CbhA again dominates the population of enzymes in cellulosomes even with a much-smaller-than-native mass. The importance of the architecture of CbhA is supported by this result. The diffusion rate in the vicinity of a CipA is reduced by the extended configuration of CbhA, and the local mobility of the dockerin is enhanced by a flexible multimodular architecture, enabling a higher probability of dockerin-cohesin encounter.
FIGURE 13.
The ratio of solution fraction to the 4-cohesin-scaffold-bound fraction for a total enzyme concentration of 60 enzymes per box with different volumes/shape and same masses. The system is described in Fig. 8.
Residence Time of the Enzymes around the Scaffoldin
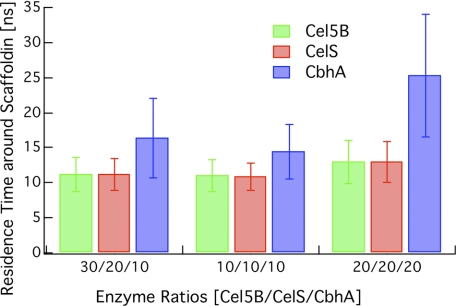
From the results in Figs. 9–13, we hypothesized that one of the factors contributing to disparate binding to the scaffoldin is the relative residence times of these enzymes around the scaffoldin. We calculated the residence times as the time for which the dockerin of an enzyme was within a certain distance, set here to 200 Å, of any of the cohesins on the scaffoldin. The residence time was averaged over all replicas and enzymes. This residence time was calculated for three different cases considered in Scan 1 (Fig. 14). The residence time for CbhA is in all cases longer than it is for the other enzymes and is similar for Cel5B and CelS independent of the total enzyme concentration. Also, the residence time for CbhA is longer for a higher total concentration of CbhA. The results reported here highlight again the importance of the architecture of CbhA and are consistent with the binding scans conducted in the previous sections.
FIGURE 14.
Residence time around the scaffoldin for Cel5B, CelS, and CbhA using three cases from Scan 1 with S.D. (1 S.D.).
DISCUSSION
Here we present a new coarse-grained model to study the assembly of bacterial cellulosomes at long length scales and time scales. The model was developed in CHARMM for ease of defining new potentials and pseudo-atoms. This model enables direct comparisons to experimental studies on cellulosomes including properties of individual enzymes obtained from small angle x-ray scattering and light scattering experiments. For cellulosome assemblies, this model can potentially be used to predict the composition of cellulosome assemblies based on the relative concentrations of enzymes produced by a given expression host or growth on different substrates, which has been shown to vary experimentally (22).
Our model shows that the large, modular enzyme CbhA from C. thermocellum binds preferentially to a small scaffoldin over the smaller, more rigid Cel5B and CelS enzymes due to its flexibility. It is likely that these results will extend to other cellulosomal systems because of the high sequence and structural similarity between the enzymes and enzyme complexes observed here for cellulosomal bacteria.
One important consideration not taken into account in this study is the difference between microscopic and macroscopic diffusion. Here we have simulated a box size of 109 Å3 with enzyme concentrations potentially higher than those found globally in the secretome. However, a significant open experimental question remains in cellulosomal self-assembly regarding the enzyme concentrations and overall volume of space between the plant cell wall and the bacterium. This will have a significant impact on the self-assembly process in that larger enzymes will diffuse more slowly to a scaffold, which in the limit of large volumes and low enzyme concentrations will impact the bound composition. However, with this model, or potentially with a lattice or a transport model, we can probe this question of scaffold composition at much lower enzyme concentrations and much larger volumes. Thus, this present model will lead to another layer of coarse graining for even longer length and time scales.
Additionally, the linkers examined here are assumed to be flexible. Evidence supporting the observation of peptide linker flexibility in cellulases comes from small angle x-ray scattering experiments (7), the typical inability to crystallize intact enzymes containing linkers, the sequence-based protein disorder screens (Figs. 2 and 3), and from recent simulations of other cellulase linkers from our group (64). For future iterations of this model, we will apply atomistic simulations of the linkers to measure the intrinsic flexibility of each linker of interest to more accurately treat the linker flexibility.
Another interesting question that relates to the construction of this model is the behavior and role of the X1 module in CbhA. The X1 modules have been suggested based on microscopy studies to disrupt the cell wall, although the mechanism has yet to be elucidated. Additionally, studies of homologous domains have shown that they are able to unfold under the application of biologically relevant forces (70–72). This observation suggests the application of additional experimental and computational work on Fn-III domains from CbhA, such as steered molecular dynamics, light scattering, atomic force microscopy pulling, and FRET, to refine the behavior of the Fn-III domains in CbhA in solution is advisable.
The study presented here suggests several direct experimental comparisons. For example, from mixing studies of purified enzyme components and scaffoldins, one can essentially conduct the same experiments as simulated here. Mass spectrometry, analytical ultracentrifugation, gel filtration, and other experimental techniques can be used to quantify the enzyme compositions on the scaffold over a range of initial concentrations (relative and absolute). An interesting extension to both a computational and experimental study with free scaffoldins would be to tether the scaffoldins to a surface, similar to the bacterial cell membrane.
CONCLUSIONS
We have developed a new coarse-grained model to study cellulosomal behavior at much longer length and time scales than are accessible by conventional atomistic simulation. Here, we used this model to simulate the self-assembly of three component enzymes on a four-cohesin scaffoldin. The results indicate that the large enzyme complex, CbhA, binds to the cellulosome scaffolding preferentially over the smaller enzymes Cel5B and CelS. The enhanced binding of CbhA is shown to be primarily driven by significant flexibility in the multimodular domain and the higher residence time of CbhA around the scaffoldin within the limit of local diffusion. This suggests that bacteria may need to produce less CbhA than the smaller enzyme components. Generally, this model provides insight into the biological self-assembly process of cellulosomes and will be a useful tool for the design of engineered cellulosomes for biomass conversion processes.
Supplementary Material
Acknowledgments
We thank the Texas Advanced Computing Center Ranger cluster under National Science Foundation Teragrid Grant MCB090159. We thank Qi Xu for helpful discussions. Access to PONDR® was provided by Molecular Kinetics. Computational time for this research was supported in part by the Golden Energy Computing Organization at the Colorado School of Mines using resources acquired with financial assistance from the National Science Foundation and the National Renewable Energy Laboratory.
This work was supported by the Department of Energy (DOE) Office of Science, Office of the Biological and Environmental Research through the Bioenergy Science Center, a DOE Bioenergy Research Center, and by the DOE Office of Science ASCR SciDAC program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4, Figs. S1 and S2, and Movie 1.
- GH
- glycosyl hydrolase
- CBM
- carbohydrate binding module.
REFERENCES
- 1. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) Annu. Rev. Microbiol. 58, 521–554 [DOI] [PubMed] [Google Scholar]
- 2. Demain A. L., Newcomb M., Wu J. H. D. (2005) Microbiol. Mol. Biol. Rev. 69, 124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doi R. H., Kosugi A. (2004) Nat. Rev. Microbiol. 2, 541–551 [DOI] [PubMed] [Google Scholar]
- 4. Fierobe H. P., Mingardon F., Mechaly A., Bélaïch A., Rincon M. T., Pagès S., Lamed R., Tardif C., Bélaïch J. P., Bayer E. A. (2005) J. Biol. Chem. 280, 16325–16334 [DOI] [PubMed] [Google Scholar]
- 5. Lamed R., Bayer E. A. (1988) Adv. Appl. Microbiol. 33, 1–46 [Google Scholar]
- 6. Gilbert H. J. (2007) Mol. Microbiol. 63, 1568–1576 [DOI] [PubMed] [Google Scholar]
- 7. Hammel M., Fierobe H. P., Czjzek M., Kurkal V., Smith J. C., Bayer E. A., Finet S., Receveur-Bréchot V. (2005) J. Biol. Chem. 280, 38562–38568 [DOI] [PubMed] [Google Scholar]
- 8. Pagès S., Bélaïch A., Bélaïch J. P., Morag E., Lamed R., Shoham Y., Bayer E. A. (1997) Proteins. 29, 517–527 [PubMed] [Google Scholar]
- 9. Bayer E. A., Chanzy H., Lamed R., Shoham Y. (1998) Curr. Opin. Struct. Biol. 8, 548–557 [DOI] [PubMed] [Google Scholar]
- 10. Bayer E. A., Lamed R., Himmel M. E. (2007) Curr. Opin. Biotechnol. 18, 237–245 [DOI] [PubMed] [Google Scholar]
- 11. Bayer E. A., Shimon L. J., Shoham Y., Lamed R. (1998) J. Struct. Biol. 124, 221–234 [DOI] [PubMed] [Google Scholar]
- 12. Fierobe H. P., Bayer E. A., Tardif C., Czjzek M., Mechaly A., Bélaïch A., Lamed R., Shoham Y., Bélaïch J. P. (2002) J. Biol. Chem. 277, 49621–49630 [DOI] [PubMed] [Google Scholar]
- 13. Fierobe H. P., Mechaly A., Tardif C., Belaich A., Lamed R., Shoham Y., Belaich J. P., Bayer E. A. (2001) J. Biol. Chem. 276, 21257–21261 [DOI] [PubMed] [Google Scholar]
- 14. Deleted in proof.
- 15. Cha J., Matsuoka S., Chan H., Yukawa H., Inui M., Doi R. H. (2007) J. Microbiol. Biotechnol. 17, 1782–1788 [PubMed] [Google Scholar]
- 16. Mingardon F., Chanal A., Tardif C., Bayer E. A., Fierobe H. P. (2007) Appl. Environ. Microbiol. 73, 7138–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ciruela A., Gilbert H. J., Ali B. R., Hazlewood G. P. (1998) FEBS Lett. 422, 221–224 [DOI] [PubMed] [Google Scholar]
- 18. Wu J. H. D., Ormejohnson W. H., Demain A. L. (1988) Biochemistry 27, 1703–1709 [Google Scholar]
- 19. Kataeva I., Guglielmi G., Béguin P. (1997) Biochem. J 326, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bhat S., Goodenough P. W., Bhat M. K., Owen E. (1994) Int. J. Biol. Macromol. 16, 335–342 [DOI] [PubMed] [Google Scholar]
- 21. Caspi J., Irwin D., Lamed R., Shoham Y., Fierobe H. P., Wilson D. B., Bayer E. A. (2006) Biocatal. Biotransform. 24, 3–12 [Google Scholar]
- 22. Raman B., Pan C., Hurst G. B., Rodriguez M., McKeown C. K., Lankford P. K., Samatova N. F., Mielenz J. R. (2009) PLoS One 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bellesia G., Shea J. E. (2009) J. Chem. Phys. 130, 145103. [DOI] [PubMed] [Google Scholar]
- 24. Bellesia G., Shea J. E. (2009) Biophys. J. 96, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strodel B., Wales D. J. (2008) J. Chem. Theory Comput. 4, 657–672 [DOI] [PubMed] [Google Scholar]
- 26. Strodel B., Whittleston C. S., Wales D. J. (2007) J. Am. Chem. Soc. 129, 16005–16014 [DOI] [PubMed] [Google Scholar]
- 27. Onuchic J. N., Luthey-Schulten Z., Wolynes P. G. (1997) Annu. Rev. Phys. Chem. 48, 545–600 [DOI] [PubMed] [Google Scholar]
- 28. Shea J. E., Brooks C. L., 3rd (2001) Annu. Rev. Phys. Chem. 52, 499–535 [DOI] [PubMed] [Google Scholar]
- 29. Hills R. D., Jr., Brooks C. L., 3rd (2009) Int. J. Mol. Sci. 10, 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dill K. A., Ozkan S. B., Shell M. S., Weikl T. R. (2008) Annu. Rev. Biophys. 37, 289–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J., Brooks C. L., 3rd, Khandogin J. (2008) Curr. Opin. Struct. Biol. 18, 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Snow C. D., Sorin E. J., Rhee Y. M., Pande V. S. (2005) Annu. Rev. Biophys. Biomol. Struct. 34, 43–69 [DOI] [PubMed] [Google Scholar]
- 33. Levy Y., Cho S. S., Onuchic J. N., Wolynes P. G. (2005) J. Mol. Biol. 346, 1121–1145 [DOI] [PubMed] [Google Scholar]
- 34. Hagan M. F., Chandler D. (2006) Biophys. J. 91, 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwartz R., Shor P. W., Prevelige P. E., Jr., Berger B. (1998) Biophys. J. 75, 2626–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen H. D., Reddy V. S., Brooks C. L., 3rd (2007) Nano Lett. 7, 338–344 [DOI] [PubMed] [Google Scholar]
- 37. Nguyen H. D., Reddy V. S., Brooks C. L., 3rd (2009) J. Am. Chem. Soc. 131, 2606–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kivenson A., Hagan M. F. (2010) Biophys. J. 99, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carvalho A. L., Dias F. M., Prates J. A., Nagy T., Gilbert H. J., Davies G. J., Ferreira L. M., Romão M. J., Fontes C. M. G. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13809–13814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carvalho A. L., Dias F. M., Nagy T., Prates J. A., Proctor M. R., Smith N., Bayer E. A., Davies G. J., Ferreira L. M.., Romão M. J., Fontes C. M., Gilbert H. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3089–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu J., Crowley M. F., Smith J. C. (2009) Protein Sci. 18, 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guimarães B. G., Souchon H., Lytle B. L., David, Wu J. H., Alzari P. M. (2002) J. Mol. Biol. 320, 587–596 [DOI] [PubMed] [Google Scholar]
- 44. Grépinet O., Béguin P. (1986) Nucleic Acids Res. 14, 1791–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zverlov V. V., Velikodvorskaya G. V., Schwarz W. H., Bronnenmeier K., Kellermann J., Staudenbauer W. L. (1998) J. Bacteriol. 180, 3091–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schubot F. D., Kataeva I. A., Chang J., Shah A. K., Ljungdahl L. G., Rose J. P., Wang B. C. (2004) Biochemistry 43, 1163–1170 [DOI] [PubMed] [Google Scholar]
- 47. Kruus K., Lua A. C., Demain A. L., Wu J. H. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9254–9258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., Steitz T. A. (1996) EMBO J. 15, 5739–5751 [PMC free article] [PubMed] [Google Scholar]
- 49. Jindou S., Xu Q., Kenig R., Shulman M., Shoham Y., Bayer E. A., Lamed R. (2006) FEMS Microbiol. Lett. 254, 308–316 [DOI] [PubMed] [Google Scholar]
- 50. Kataeva I. A., Uversky V. N., Brewer J. M., Schubot F., Rose J. P., Wang B. C., Ljungdahl L. G. (2004) Protein. Eng. Des. Sel. 17, 759–769 [DOI] [PubMed] [Google Scholar]
- 51. Kataeva I. A., Seidel R. D., 3rd, Shah A., West L. T., Li X. L., Ljungdahl L. G. (2002) Appl. Environ. Microbiol. 68, 4292–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lytle B. L., Volkman B. F., Westler W. M., Wu J. H. D. (2000) Arch. Biochem. Biophys. 379, 237–244 [DOI] [PubMed] [Google Scholar]
- 53. Chauvaux S., Beguin P., Aubert J. P., Bhat K. M., Gow L. A., Wood T. M., Bairoch A. (1990) Biochem. J. 265, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comput. Chem. 4, 187–217 [Google Scholar]
- 55. Tavares G. A., Béguin P., Alzari P. M. (1997) J. Mol. Biol. 273, 701–713 [DOI] [PubMed] [Google Scholar]
- 56. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchninr L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 57. Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradović Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 58. Radivojac P., Iakoucheva L. M., Oldfield C. J., Obradovic Z., Uversky V. N., Dunker A. K. (2007) Biophys. J. 92, 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., Dunker A. K. (2001) Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 60. Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. (2001) J. Mol. Graph. Model. 19, 26–59 [DOI] [PubMed] [Google Scholar]
- 61. Dunker A. K., Obradovic Z. (2001) Nat. Biotechnol. 19, 805–806 [DOI] [PubMed] [Google Scholar]
- 62. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 63. Noach I., Frolow F., Alber O., Lamed R., Shimon L. J., Bayer E. A. (2009) J. Mol. Biol. 391, 86–97 [DOI] [PubMed] [Google Scholar]
- 64. Beckham G. T., Bomble Y. J., Matthews J. F., Taylor C. B., Resch M. G., Yarbrough J. M., Decker S. R., Bu L., Zhao X., McCabe C., Wohlert J., Bergenstråhle M., Brady J. W., Adney W. S., Himmel M. E., Crowley M. F. (2010) Biophys. J. 99, 3773–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Domínguez R., Souchon H., Lascombe M., Alzari P. M. (1996) J. Mol. Biol. 257, 1042–1051 [DOI] [PubMed] [Google Scholar]
- 66. Dominguez R., Souchon H., Spinelli S., Dauter Z., Wilson K. S., Chauvaux S., Béguin P., Alzari P. M. (1995) Nat. Struct. Biol. 2, 569–576 [DOI] [PubMed] [Google Scholar]
- 67. Oehler S., Müller-Hill B. (2010) J. Mol. Biol. 395, 242–253 [DOI] [PubMed] [Google Scholar]
- 68. Mayer F., Coughlan M. P., Mori Y., Ljungdahl L. G. (1987) Appl. Environ. Microbiol. 53, 2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Morag E., Yaron S., Lamed R., Kenig R., Shoham Y., Bayer E. A. (1996) J. Biotechnol. 51, 235–242 [DOI] [PubMed] [Google Scholar]
- 70. Baugh L., Vogel V. (2004) J. Biomed. Mater. Res. A 69, 525–534 [DOI] [PubMed] [Google Scholar]
- 71. Gao M., Craig D., Lequin O., Campbell I. D., Vogel V., Schulten K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14784–14789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klotzsch E., Smith M. L., Kubow K. E., Muntwyler S., Little W. C., Beyeler F., Gourdon D., Nelson B. J., Vogel V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18267–18272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.