Abstract
The hexameric AAA-ATPase, Cdc48p, catalyzes an array of cellular activities, including endoplasmic reticulum (ER)-associated degradation (ERAD), ER/Golgi membrane dynamics, and DNA replication. Accumulating data suggest that unique Cdc48p partners, such as Npl4p-Ufd1p and Ubx1p/Shp1p (p47 in vertebrates), target Cdc48p for these diverse functions. Other Cdc48p-associated proteins have been identified, but the interplay among these factors and their activities is largely cryptic. We now report on a previously uncharacterized Cdc48p-associated protein, Ydr049p, also known as Vms1p, which binds Cdc48p at both the ER membrane and in the cytosol under non-stressed conditions. Loss of YDR049 modestly slows the degradation of the cystic fibrosis transmembrane conductance regulator but does not impede substrate ubiquitination, suggesting that Ydr049p acts at a postubiquitination step in the ERAD pathway. Consistent with Ydr049p playing a role in Cdc48p substrate release, ydr049 mutant cells accumulate Cdc48p-bound ubiquitinated proteins at the ER membrane. Moreover, YDR049 interacts with genes encoding select UBX (ubiquitin regulatory X) and UFD (ubiquitin fusion degradation) proteins, which are Cdc48p partners. Exacerbated growth defects are apparent in some of the mutant combinations, and synergistic effects on the degradation of cystic fibrosis transmembrane conductance regulator and CPY*, which is a soluble ERAD substrate, are evident in specific ydr049-ufd and -ubx mutants. These data suggest that Ydr049p acts in parallel with Cdc48p partners to modulate ERAD and other cellular activities.
Keywords: Chaperone Chaperonin, Endoplasmic Reticulum (ER), Proteasome, Protein Degradation, Ubiquitin, Yeast, Cdc48, ERAD, UBX, p97
Introduction
Endoplasmic reticulum-associated degradation (ERAD)4 clears the early secretory pathway of misfolded proteins and can be subdivided generally into distinct stages of substrate recognition, retrotranslocation from the ER into the cytoplasm, ubiquitin conjugation, and degradation by the 26 S proteasome (1–4). Given the volume of protein traffic through the secretory pathway (5), it is not surprising that ERAD is equipped to handle a wide variety of substrates based on the location of the offending lesion (5, 6). For instance, substrates within the ER lumen are recognized by a distinct subset of molecular chaperones (e.g. BiP, also known as Kar2p in yeast) and chaperone-like lectins and are ubiquitinated by an E3 ubiquitin ligase, Hrd1p, concomitant with retrotranslocation in a process collectively known as ERAD-lumenal, or ERAD-L (7–10). Proteins with misfolded lesions within the ER membrane (ERAD-M substrates) also employ Hrd1p, and it has been proposed that this ligase directly recognizes this type of folding lesion (11). The ERAD of proteins with cytosolic lesions utilizes the ERAD-C (cytosolic) pathway and employs molecular chaperones that are localized in the cytosol as well as a different ubiquitin ligase, Doa10p (6, 12, 13). Interestingly, some terminally misfolded substrates rely on both E3 ligases when expressed in yeast, suggesting that multiple pathways may be required to recognize and dispose of proteins with complex folding lesions (12, 14–16).
Irrespective of the ERAD pathway employed, the retrotranslocation of most substrates relies upon the Cdc48p complex, which consists of the homohexameric Cdc48p and two adaptor proteins, Npl4p and Ufd1p (17–23). Cdc48p, the yeast homolog of p97/valosin-containing protein (p97/VCP), is a multifunctional member of the AAA (ATPases associated with various activities) family (24). In addition to a role for Cdc48 in protein catabolism, Cdc48p is linked to cell division, Golgi and ER membrane dynamics, autophagy, DNA replication, and cell survival (25–30). Regardless of its specific role, Cdc48p (like other AAA proteins) is thought to couple ATP hydrolysis with significant changes in the conformation of its targets (31), and the specificity of Cdc48p function relies on the cofactors to which it binds (32).
To date, many Cdc48p cofactors have been identified, but only a few of these cofactors are functionally defined. For example, Ubx1p, also known as Shp1p, was originally isolated as a suppressor of phosphoprotein phosphatase 1 overexpression but was later found to participate in vesicle fusion, the spindle checkpoint, and ubiquitin-mediated degradation of select substrates (33–36). Ubx1p is one member of a larger group of generally uncharacterized UBX domain-containing proteins (37). The UBX domain mediates binding to Cdc48p, and its tertiary structure resembles ubiquitin. Other members of the UBX family, such as Ubx2p, Ubx4p, Ubx6p, and Ubx7p, have also been linked to ubiquitin-mediated protein degradation (34, 38–40). Some data suggest that Ubx2p may anchor the Cdc48p-Npl4p-Ufd1p complex to the ER membrane, and ubx2Δ yeast exhibit ERAD defects (34, 40–43).
Another group of Cdc48p-associated proteins were isolated as UFD (ubiquitin fusion degradation protein) mutants (44). As noted above, Ufd1p is a Cdc48p partner and is required for ERAD. Ufd2p is a Cdc48p-associated ubiquitin chain assembly factor, catalyzing the extension of ubiquitin chains and increasing the rate of ERAD substrate degradation (45, 46). Ufd3p, also known as Doa1p, is a WD40 repeat protein that binds ubiquitin and can reside in a complex with Cdc48p (47–49). Additionally, a deubiquitinating enzyme, known as Otu1p, binds the Cdc48p complex, and antagonistic interactions among Ufd2p and Ufd3p-Otu1p have been observed (50). Nevertheless, it is unknown how the function of Cdc48p during ERAD is altered by most of these partners and whether additional, ill characterized partners of Cdc48p also impact ERAD.
In this paper, we report on the characterization of a new Cdc48p cofactor that is encoded by the YDR049 open reading frame in the yeast, Saccharomyces cerevisiae. Large scale yeast proteomic studies uncovered Ydr049p in a multiprotein complex that contains Cdc48p (51). Deletion of YDR049 was also observed to exhibit mild synthetic growth defects in yeast compromised for the DOA (degradation of α) pathway (52), and of note, Doa10p and Doa1p (Ufd3p) were first isolated through the DOA screen. Moreover, yeast lacking YDR049 were reported to exhibit synthetic interactions in karmellae-forming yeast, suggesting a role in ER membrane homeostasis (53). Here, we show that YDR049 deletion affects the ERAD of a model transmembrane substrate, the cystic fibrosis transmembrane conductance regulator (CFTR), and that YDR049 exhibits genetic interactions with select members of the UBX and UFD protein families. In some cases, the genetic interaction is evidenced by increased sensitivity to ER and oxidative stressors, and in other cases, it is shown by synergistic ERAD defects. Finally, we show that increased levels of ubiquitinated species are associated with Cdc48p in ydr049Δ yeast.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Growth Assays
Most yeast strains employed in this study (supplemental Table 1) were in the BY4742 background and were obtained from Open Biosystems (Thermo Scientific) or were made by disrupting the gene by homologous recombination using resistance marker cassettes amplified from the plasmid pRS400 (KanMX4) or pFA6-His3MX (His3MX) (54, 55). Synthetic primers used to construct linear DNA strands for gene disruption contained 50–100 bp of sequence homology as well as sequences to amplify the cassette. A complete list of primers used to make the strains in this study is provided in supplemental Table 2. Yeast harboring more than one gene deletion were made by standard genetic techniques (56). In brief, strains were mated on rich medium (YPD: 1% yeast extract, 2% peptone, 2% dextrose), and after 4–6 h at 30 °C, diploids were selected on synthetic complete medium lacking both lysine and histidine. Diploids were nitrogen-starved for 3–7 days in sporulation medium (1% KOAc, 0.1% Bacto yeast extract, 0.05% dextrose) prior to tetrad dissection. The resulting germinated spores were screened on the appropriate medium, and deletions were confirmed by PCR. To introduce the cdc48-3 allele into the BY background, a cdc48-3 strain (MATa, his4-619, leu2,3-112, ura3, pep4Δ::URA3, cdc48-3) in a S288C background (57) was crossed against a BY strain of the opposite mating type. The resulting cdc48-3 strain from this genetic cross was backcrossed three additional times against the BY background. The cdc48-3 strain derived from these backcrosses (supplemental Table 1) was then used.
The plasmids used in the study (supplemental Table 3) were described previously or created by PCR amplification and cloned as follows. For the construction of C-terminal HA- or Myc-tagged Cdc48p constructs, two PCR fragments were generated and ligated together into the desired plasmid. The first fragment was amplified with the forward primer 5′-TTGCGGCCGCGGTGGCCAGCCCAAGAAACGGA and the reverse primer 5′-ACGGATCCACTATACAAATCATCATCTTCC. These primers were synthesized with a NotI restriction site in the forward primer and a BamHI restriction site in the reverse primer (underlined). The resulting PCR product started 437 bp upstream of the ORF and contained the entire CDC48 ORF minus the termination codon. The second fragment was PCR-amplified in a reaction with either the forward primer 5′-ACGGATCCTACCCATACGACGTCCCAGACTACGCTTAGTAGTTATATGCCAGGTATATTTTTATTTTAAATCG for the HA-tagged constructs (the HA tag sequence is in boldface type) or the forward primer 5′-ACGGATCCGAACAAAAACTCATCTCAGAAGAGGATCTGTAGTAGTTATATGCCAGGTATATTTTTATTTTAAATCG for Myc-tagged constructs (the Myc tag sequence is in boldface type), paired with the reverse primer 5′-AGCTCGAGACGACCGAGGTCCTACAGCCT. Both forward primers contained a BamHI site, whereas the reverse primer contained an XhoI site (underlined). The resulting PCR fragments encoded either a single in-frame copy of the HA or Myc epitope tag followed by the termination codon in duplicate and 221 bp of the 3′ intergenic sequence. The PCR fragments were restriction-digested and gel-extracted prior to ligation into a pRS315 plasmid that was similarly digested with NotI and XhoI (58). Sequence-verified constructs were used to create a library of expression vectors in the pRS316, pRS425, and pRS426 backbones (58, 59).
To generate plasmids for the expression of an untagged form of Cdc48p, the forward primer 5′-TTGCGGCCGCGGTGGCCAGCCCAAGAAACGGA and the reverse primer 5′-AGCTCGAGACGACCGAGGTCCTACAGCCT were used in a PCR. The PCR fragment, which contained the full-length, untagged CDC48 gene, was digested with NotI and XhoI (underlined) and cloned into pRS315 (58). Sequence-verified constructs were used to create a library of expression vectors in the pRS316, pRS425, and pRS426 backbones (58, 59). A similar ligation strategy was used to clone the tagged and untagged versions of YDR049. The first fragment was amplified using the forward primer 5′-CTGCGGCCGCTTCTTGGAGGAGTGCCACAG and the reverse primer 5′-ACGGATCCGTATTTCTTTTTCATCCTTTCTTCTGCG. The forward primer contains a NotI site, whereas the reverse primer contains a BamHI site (underlined). This PCR fragment contains 341 bp upstream of the ORF and the entire YDR049 ORF except the termination codon. For the second fragment, either the forward primer 5′-ACGGATCCTACCCATACGACGTCCCAGACTACGCTTGATGAGGAATATCTCATATTCAAATTTTTAGG for the HA-tagged construct (in boldface type) or the forward primer 5′-ACGGATCCGAACAAAAACTCATCTCAGAAGAGGATCTGTGATGAGGAATATCTCATATTCAAATTTTTAGG for the Myc-tagged construct (in boldface type) was used in a PCR with the reverse primer 5′-CGGTCGACGGCGTCATTTTCGCGTTGAG. Both forward primers contained BamHI sites (underlined), whereas the reverse primer contained a SalI restriction site (underlined). The resulting PCR fragment contained the HA or Myc tag followed by a termination codon in duplicate and 227 bp of the 3′ intergenic sequence. For untagged YDR049 constructs, the forward primer 5′-CTGCGGCCGCTTCTTGGAGGAGTGCCACAG and the reverse primer 5′-CGGTCGACGGCGTCATTTTCGCGTTGAG were paired in the PCR. The primers contained a NotI and SalI restriction site, respectively. Cloning was done as described for the tagged and untagged forms of CDC48.
To examine the growth of yeast under various conditions, log phase cells grown at 26 °C were harvested and resuspended in sterile water to a final A600 of 1.0. 10-fold serial dilutions were spot-plated on the indicated medium and grown for 2–6 days at 30 or 38 °C.
Assays for ERAD and the Degradation of Other Substrates
To assess the rate of CFTR and Ste6p* degradation, the appropriately transformed strains (supplemental Table 1) were grown to log phase (A600 = 0.5–1.5) in selective medium at 26 °C. The cells were then harvested and resuspended to a final A600 of 1.0. 1 ml of cells was taken as the zero time point. Next, cycloheximide was added to a final concentration of 100–200 μg/ml, and cells were incubated at 30 °C for Ste6p* or 40 °C for CFTR (45, 60). Aliquots were removed at the indicated time points. Samples were mixed with NaN3 (final concentration, 10 μm) and processed as described previously (61). Data were quantified using ImageJ version 1.42q software (62). The degradation of an HA epitope-tagged form of CPY* (63) was also assessed by cycloheximide chase analysis, as described above, except that the cells were incubated at 30 °C during the chase.
The pulse-chase analysis of CFTR-HA was performed as previously described (14). Briefly, 30 ml of cells were grown overnight to log phase, harvested, and resuspended in medium lacking methionine and cysteine to a final A600 of 6/ml. Cells were labeled for 1 h with 35S-labeled methionine (PerkinElmer Life Sciences) at a final concentration of 100 μCi/ml. A zero time point was then taken, and cells were chased with cold methionine and cysteine. Cells were disrupted by glass bead lysis, and the crude lysate was solubilized in 12.5 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% SDS. The lysate was then diluted with immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% deoxycholic acid, 1% Triton X-100) so that the final concentration of SDS was 0.1%. CFTR-HA was immunoprecipitated from the solubilized lysate with anti-HA-agarose (Roche Applied Science), and immunoprecipitated material was separated by SDS-PAGE and subject to autoradiography.
The degradation of a galactose-inducible version of a ubiquitin-proline β-galactosidase chimera was measured by pulse-chase analysis as described previously (64). In brief, cells were grown to saturation, harvested, and resuspended to an A600 of ∼3/ml in medium lacking methionine. Next, 35S-labeled methionine (PerkinElmer Life Sciences) was added to a final concentration of 11 μCi/μl, and total protein was labeled for 15 min at 30 °C. Cycloheximide was added to a final concentration of 500 μg/ml, aliquots were removed at the indicated time points, and samples were processed. The precipitated chimeric protein was visualized by phosphorimaging analysis on a Fujifilm BAS-2500, and the data were quantified using ImageJ version 1.42q software (62). The degradation of a Deg1-β-galactosidase fusion was assessed by cycloheximide chase analysis as described (48).
Preparation of Yeast Subcellular Fractions and Indirect Immunofluorescence Microscopy
Yeast cytosol was prepared by liquid nitrogen lysis as described previously (65). In brief, 4–6 liters of cells were grown to log phase, harvested, and resuspended in Buffer 88 (20 mm HEPES, pH 6.8, 150 mm KOAc, 250 mm sorbitol, 5 mm MgOAc). The suspension was then poured into liquid nitrogen and pulverized in a Waring blender for 10 min. The resulting mixture was thawed and clarified by centrifugation at 300,000 × g. The final concentration of cytosol was ∼20–30 mg/ml and was stored at −80 °C.
Yeast microsomes were prepared as described previously (45, 66, 67) from 1–2 liters of log phase cells that were spheroplasted, lysed in a Potter-Elvehjem homogenizer, and isolated by differential centrifugation. The enriched ER-derived microsomes were resuspended in Buffer 88 to a final A280 of 40 (as measured in 2% SDS) and stored at −80 °C.
The subcellular localization of Ydr049p was determined by a previously reported method (68) in which log phase cells were harvested, washed, and lysed with glass beads. After unbroken cells were removed by centrifugation at 3,000 × g for 5 min, a crude membrane fraction was obtained as the pellet after a 16,000 × g centrifugation for 15 min, and the supernatant constituted a crude cytosolic fraction. The crude cytosolic fraction was clarified by centrifugation at 150,000 × g for 15 min. 10 μl of total lysate and each membrane and cytosol fraction were subject to SDS-PAGE and Western blotting with the indicated antibodies.
The subcellular residence of Ydr049p was also determined by indirect immunofluorescence microscopy, as described previously (69). Diploid strains lacking YDR049 and expressing an HA-tagged form of Ydr049p from an episomal CEN plasmid were grown to log phase. Cells were fixed by adding a one-tenth volume of 37% formaldehyde and incubating at room temperature for 60 min. Cells were harvested and washed twice with Solution A (1.2 m sorbitol, 50 mm KPO4, pH 7.0) and were then treated with 2 μg/ml zymolyase 20T (MP Biomedicals) resuspended in Solution A for 30–40 min. Following treatment, cells were pelleted and washed twice with Solution A and resuspended in 200 μl of Solution A. A total of 25–30 μl of cells were pipetted onto polylysine-treated slides and allowed to adhere for 30 min at room temperature. Loose cells were aspirated, and the slides were washed once with PBS, 0.1% BSA, followed by two washes with PBS, 0.1% BSA, 0.1% Nonidet P-40. The samples were incubated overnight at 4 °C with primary antibodies against yeast BiP/Kar2p (1:2000) and the HA epitope tag (1:500) that were resuspended in PBS, 0.1% BSA. Next, the samples were washed one time each with PBS, 0.1% BSA and PBS, 0.1% BSA, 0.1% Nonidet P-40. Samples were then incubated with a 1:2000 dilution of AlexaFluor-conjugated secondary antibodies (Invitrogen; goat anti-rabbit 594 and goat anti-mouse 488) resuspended in PBS, 0.1% BSA for 1–2 h at room temperature. The slides were then washed once with PBS, 0.1% BSA; twice with PBS, 0.1% BSA, 0.1% Nonidet P-40; and once with PBS, 0.1% BSA. Anti-fade mounting medium plus DAPI (Invitrogen) was laid on top of each sample before applying a coverslip that was sealed with nail polish. Images were captured by QED software using a Hamamamatsu camera attached to an Olympus BX-60 microscope. Images were analyzed using ImageJ version 1.42q (62).
Measurements of Substrate Ubiquitination
In vitro ubiquitination reactions were performed as described previously (45). In brief, microsomes, cytosol, an ATP-regenerating system, and 125I-labeled ubiquitin were mixed and incubated at room temperature for 1 h. Yeast membranes were then solubilized with detergent, and the ERAD substrate was immunoprecipitated with a mixture of anti-HA antibody and protein A-Sepharose (GE Healthcare). The precipitated material was washed with 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.2% SDS and then released from the beads with SDS-PAGE sample buffer (80 mm Tris-HCl, pH 8.0, 8 mm EDTA, 3.5% SDS, 15% glycerol, 0.08% Tris base, 0.01% bromphenol blue) containing freshly added DTT (100 mm) and heating at 37 °C for 30 min. The resulting 125I-ubiquitinated material was resolved on a 6% SDS-polyacrylamide gel and was detected by phosphorimaging analysis, as described above. Unmodified CFTR was visualized by Western blotting using anti-HA antibody. Data were quantified using Image Gauge version 4.0 or ImageJ version 1.42q (62).
The immunoprecipitation and detection of in vivo ubiquitinated CFTR was carried out as described previously (70). A total of 100 ml of cells were grown overnight to log phase and were harvested and disrupted by glass bead lysis. Unbroken cells were sedimented by low speed centrifugation, and membranes were sedimented at 18,000 × g for 20 min in a refrigerated table top centrifuge and resuspended in Buffer 88 to an A280 of 40 as measured in 2% SDS. Membranes were solubilized in 1% SDS at 37 °C for 30 min, and the solubilized ERAD substrate was immunoprecipitated with anti-HA-conjugated agarose (Roche Applied Science). The immunoprecipitated substrate was released with SDS-PAGE sample buffer containing freshly added DTT and heating at 37 °C for 30 min. The protein was then resolved by SDS-PAGE and transferred to nitrocellulose as in a standard Western blot except with the following modification. After the transfer to nitrocellulose was complete, the nitrocellulose membrane was sandwiched between two sheets of Whatman filter paper and placed in a boiling water bath for 45 min. The sandwiched blot was allowed to cool to room temperature, and after blocking, the ubiquitinated species were detected using anti-ubiquitin antibody.
Immunoprecipitation of Cdc48p and Detection of Cdc48p-associated Ubiquitinated Proteins
To monitor the association between Cdc48p and Ydr049p, cells lacking YDR049 were transformed with CEN plasmids containing a C-terminal Myc-tagged version of Cdc48p and/or a C-terminal HA-tagged version of Ydr049p (supplemental Table 3) or vectors expressing untagged versions. The transformed cells were grown in selective medium to log phase, harvested by low speed centrifugation, resuspended in Buffer 88 (which was supplemented with 1 mm PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A, and 10 mm N-ethylmaleimide), and then disrupted with glass beads by vigorous agitation on a Vortex mixer 10 times for 30 s, followed by a 30-s incubation on ice. The extract was removed and reserved, and the beads were washed with an equal volume of Buffer 88, which was then pooled with the reserved extract. Unbroken cells were removed by centrifugation at 1,500 × g for 5 min at 4 °C. A portion of the resulting supernatant represented the total lysate, and the protein concentration was estimated by adjusting the A280 in 2% SDS such that the absorbance was 40 (i.e. a protein concentration of ∼10 mg/ml). To separate membrane and cytosolic fractions, the total lysate was centrifuged at 150,000 × g for 20 min at 4 °C. The resulting membrane fraction was washed one time with Buffer 88 and then resuspended in Buffer 88 such that the final A280 in 2% SDS was 40 (i.e. a protein concentration of ∼10 mg/ml). The protein concentration of the cytosol was estimated using the Bio-Rad protein assay kit. To immunoprecipitate the tagged proteins, 200 μg of lysate, cytosol, and membrane proteins were treated as follows. The total and membrane fractions were solubilized on ice for 30 min in Buffer A (20 mm HEPES, pH 7.4, 150 mm NaCl, 1% Triton X-100). Then an equal amount of Buffer B (20 mm HEPES, pH 7.4, 150 mm NaCl) was added so that the final concentration of Triton X-100 was 0.5%. For consistency, the cytosolic extract was treated on ice for 30 min by adding Triton X-100 to a final concentration of 0.5%. For each fraction, insoluble material was removed by centrifugation at 16,000 × g for 10 min at 4 °C, and the volume was brought up to 500 μl with Buffer C (20 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% Triton X-100) prior to immunoprecipitation with anti-HA- or anti-Myc-conjugated agarose The immunoprecipitate was washed once with Buffer C and three times with Buffer D (20 mm HEPES, pH 7.4, 300 mm NaCl, 0.5% Triton X-100), and the bound proteins were released with SDS-PAGE sample buffer, as described above.
To examine Cdc48p association with ubiquitinated proteins, yeast cultures containing 100 ml of log phase cells harboring the Cdc48p-HA expression vector or a vector control were either left untreated or treated with 50 μm MG132 for 1 h. Cell disruption was performed using the medium scale preparation, as described previously (45), and all buffers, including SDS-PAGE sample buffer, were supplemented with 1 mm PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A, and 10 mm N-ethylmaleimide. The cells were harvested and disrupted with glass beads, and following the removal of unbroken cells, the crude extract was loaded onto a 1.0 m sucrose cushion, and the mixture was centrifuged at 6,900 × g in a Sorvall HB-6 swinging bucket rotor for 10 min at 4 °C. The material residing above the cushion is enriched for ER-derived microsomes, and this was collected by centrifugation at 20,200 × g for 10 min at 4 °C in a Sorvall SS-34 rotor. The microsomes were washed and resuspended in Buffer 88 to a final A280 (measured in 2% SDS) of 40. To immunoprecipitate epitope-tagged Cdc48p, 20 μl of membranes were solubilized with 1% Triton X-100 in Buffer A (see above) for 30 min on ice. The solubilized material was then diluted with Buffer B (see above) so that the final concentration of Triton X-100 was 0.5%. Insoluble material was pelleted by centrifugation at 16,000 × g for 10 min at 4 °C, and the clarified, soluble material was brought to a final volume of 500 μl with Buffer C (see above) prior to immunoprecipitation with anti-HA-agarose (Roche Applied Science). The immunoprecipitate was washed three times with Buffer C, and the bound proteins were eluted with SDS-PAGE sample buffer, as described above. After SDS-PAGE, the resolved proteins were transferred to a nitrocellulose membrane, and the membrane was treated as described above.
Antibodies and Immunoblot Analysis
Antibodies used in this study included anti-HA (Roche Applied Science); anti-Myc (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); anti-ubiquitin (Santa Cruz Biotechnology, Inc.); anti-Cdc48p (Schizosaccharomyces pombe, a kind gift from Dr. Rasmus Hartmann-Petersen); anti-Ufd2p, -Ufd3p, -Ubx1p, and -Ubx2p (a kind gift from Dr. Alexander Buchberger); anti-Otu1p and -Ufd1p (a kind gift from Dr. Stefan Jentsch); and monoclonal anti-yeast BiP/Kar2p (a generous gift from Dr. Mark Rose). Immunoblots were decorated with the indicated primary antibodies and appropriate HRP-conjugated anti-mouse or anti-rabbit IgG secondary antibodies. The HRP-chemiluminescent signal was visualized by enhanced chemiluminescence (Pierce). Images were captured on a Kodak Image Station 440CF (Eastman Kodak Co.) and were analyzed using ImageJ version 1.42q (62).
RESULTS
Ydr049p/Vms1p Resides Primarily in the Cytoplasm
Concomitant with the work presented here, the S. cerevisiae YDR049 gene was implicated in maintaining the viability of cells exposed to mitochondrial and oxidative stressors. Although the protein was primarily cytoplasmic, during mitochondrial stress, Vms1p was found associated with the mitochondrial membrane (71). Additionally, it was shown that deletion and knockdown, respectively, of the yeast and Caenorhabditis elegans homolog decreased life span (71). Based on the fact that the gene product functions in the context of Cdc48p/p97/VCP, the gene was named VMS1 (VCP-associated mitochondrial stress-responsive-1). However, because of the attributes of Ydr049p/Vms1p, as outlined in the Introduction, we chose to explore its function as a component of the Cdc48p complex and during ERAD. We also wished to explore whether VMS1 interacted with genes encoding other Cdc48p-associated factors. For simplicity, we will refer hereafter to the protein/gene as Vms1p/VMS1.
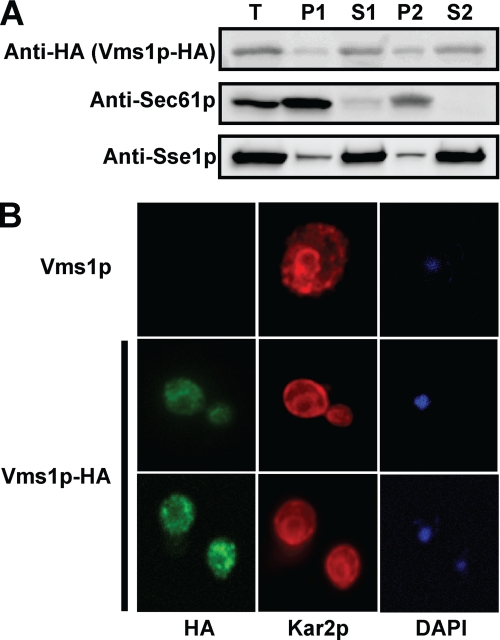
To probe the localization of Vms1p in yeast during normal cell growth, we constructed a C-terminal, HA epitope-tagged version of Vms1p. The protein's expression was controlled by the endogenous promoter, and the gene was maintained on a single copy CEN plasmid. By taking advantage of the fact that yeasts lacking VMS1 are cycloheximide-sensitive (72), we established that Vms1p-HA was functional because it rescued the growth of vms1Δ cells in the presence of cycloheximide (supplemental Fig. S1A). Next, we used differential centrifugation to assess the localization of Vms1p. As shown in Fig. 1A, Vms1p appears to reside in both membrane-associated and soluble fractions but is predominantly a cytosolic protein. In this experiment, we also monitored the localization of Sec61p, which is an ER membrane protein (73), and Sse1p, which like Vms1p is primarily a cytosolic protein but exhibits partial membrane residence (74). To further analyze the residence of Vms1p, we performed indirect immunofluorescence microscopy. As seen in Fig. 1B, Vms1p-associated fluorescence was primarily cytoplasmic and in some cases enriched in large intracellular punctae. In some cells, these bodies appeared to reside just at the periphery of the ER, as evidenced by the signal corresponding to yeast BiP, Kar2p. These bodies did not colocalize with mitochondria-derived DAPI staining (Fig. 1B, bottom). As assessed by indirect immunofluorescence, the localization of Cdc48p is also primarily cytosolic, except in late G1, and can also be visualized in large bodies that reside peripherally to the ER in some cells (75).
FIGURE 1.
VMS1 encodes a cytosolic protein with limited membrane association. Yeasts deleted for VMS1 were transformed with a low copy VMS1-HA expression vector whose expression is under the control of the endogenous promoter (see “Results” for details). A, yeast cells were disrupted, and lysates were processed for SDS-PAGE and immunoblot analysis as described under “Experimental Procedures.” The ER membrane protein, Sec61p, and the cytosolic chaperone, Sse1p, served as controls. T, total lysate; P1, membrane pellet after a 16,000 × g centrifugation; S1, supernatant after 16,000 × g centrifugation; P2, membrane pellet derived from S1 after 150,000 × g centrifugation; S2, supernatant derived from S1 after the 150,000 × g spin. A total of 10 μl of each fraction was analyzed. B, yeast cells were fixed and decorated with anti-HA antibody to detect Vms1p and anti-BiP (Kar2p), an ER-resident protein, and the appropriate secondary fluorescence-conjugated antibodies. DAPI staining was used to detect nuclear and mitochondrial DNA.
Vms1p Is Associated with Cdc48p in both Cytosol and Membrane Fractions
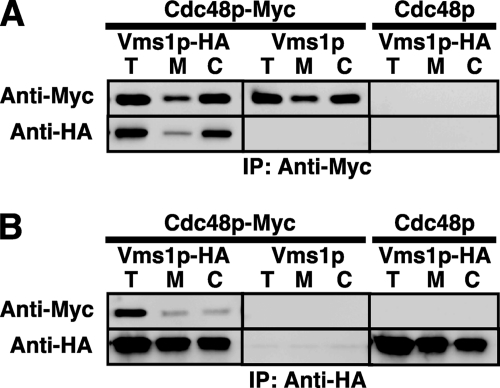
Large scale proteomic studies identified Vms1p as a component of a multiprotein complex that included Cdc48p (51). Because a large number of false-positives can arise from proteomic analyses, we next wished to confirm that Vms1p and Cdc48p can be co-precipitated. We therefore performed reciprocal co-immunoprecipitations with a C-terminal Myc-tagged version of Cdc48p and the HA-tagged form of Vms1p. Expression of the tagged version of Cdc48p rescued the growth of cdc48-3 yeast at a non-permissive temperature (supplemental Fig. S1B), indicating that Cdc48p-Myc is active. We then isolated cytosolic and membrane fractions from cells grown in the absence of stress to determine whether Cdc48p and Vms1p co-precipitate. As shown in Fig. 2, Vms1p and Cdc48p can be co-precipitated when a total yeast lysate is examined (T) and when both membrane (M) and cytosolic (C) fractions are examined. As controls for this experiment, co-precipitation was absent when cells expressed untagged versions of either protein. We also noted that significantly more Cdc48p resided in the cytosol than at the membrane and that the amount of Vms1p associated with Cdc48p reflected this distribution (Fig. 2A). In contrast, when Vms1p was precipitated (Fig. 2B), approximately equal amounts of the protein resided at the membrane and in the cytosol, but in both cases again Cdc48p co-precipitated with Vms1p. Further evidence for the functional significance of Vms1p-Cdc48p interaction is provided by a synthetic growth phenotype in vms1Δ yeast that simultaneously harbors a temperature-sensitive allele of CDC48 (supplemental Fig. S1C). Given that Vms1p can localize and associate with the Cdc48p complex at the mitochondrial membrane in the presence of mitochondrial and oxidative stressors (71), we also tested directly whether Vms1p associates with the Cdc48p complex at the ER membrane. To this end, Vms1p-HA was immunoprecipitated from highly enriched ER fractions, and the resulting precipitate was blotted for Cdc48p. As shown in supplemental Fig. S1D, Cdc48p was coimmunoprecipitated with Vms1p-HA from ER-enriched fractions but not from strains that contained an untagged version of Vms1p. Because Vms1p contains a VCP/p97-interacting motif domain (71), which is known to mediate direct interactions with VCP/p97, it is likely that Vms1p and Cdc48p directly associate. We also observed reproducible interactions between Vms1p-HA and Ubx2p and Ufd3p (supplemental Fig. S2). Together, these data indicate that Vms1p is a component of a Cdc48p complex that exists both in the cytosol and at the ER membrane.
FIGURE 2.
Vms1p and Cdc48p co-precipitate. VMS1-deleted cells were transformed with a plasmid engineered for the expression of a functional, Myc-tagged version of Cdc48p and/or with the HA-tagged version of Vms1p. As controls, untagged versions of each protein were introduced into each strain. The resulting transformants were disrupted, and portions of the total lysate (T), the membrane fraction (M), and the cytosolic fraction (C) were subject to immunoprecipitation (IP) with anti-Myc (A) or anti-HA (B) antibodies and were processed as described under “Experimental Procedures.” The immunoprecipitated proteins were resolved by SDS-PAGE and subjected to immunoblot analysis with the indicated antibodies.
Yeast Lacking VMS1 Exhibit Slowed Degradation of CFTR, an Integral Membrane ERAD Substrate
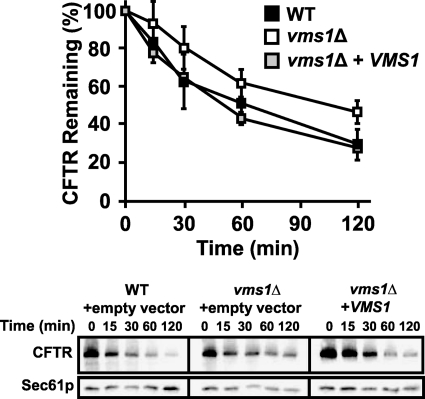
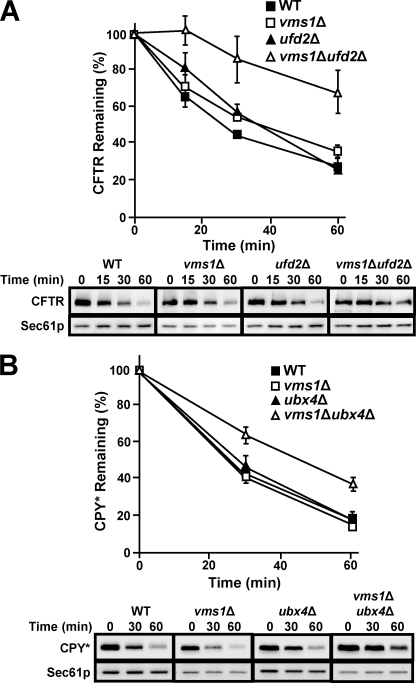
As described in the Introduction, Cdc48p plays a well defined role during ERAD. To test if Vms1p function is also important for ERAD, we performed degradation assays with a panel of well established ERAD substrates. We found first that CFTR degradation was modestly slowed in vms1Δ yeast (Fig. 3, compare filled versus unfilled squares) and that the CFTR degradation defect was fully recoverable when an extrachromosomal copy of VMS1 was introduced (Fig. 3, gray squares). The CFTR degradation defect caused by VMS1 loss was reproducible by pulse-chase analysis (supplemental Fig. S3). Interestingly, vms1Δ yeast proficiently degraded several other ERAD substrates, including Ste6p* and CPY* (supplemental Fig. S4, A and B). Moreover, the degradation of an N-end rule substrate, the ubiquitin-proline β-galactosidase chimera, and the DOA pathway substrate, Deg1-β-galactosidase, were unaffected (data not shown). Based on these results, we conclude that loss of Vms1p modestly affects the ERAD of select substrates.
FIGURE 3.
Loss of VMS1 results in compromised ERAD efficiency. The degradation of CFTR was assessed by cycloheximide chase assay, as described under “Experimental Procedures.” Each cell type was transformed with a CFTR-HA expression plasmid and was co-transformed with either an empty vector or a plasmid containing VMS1 driven by its endogenous promoter. Wild-type cells containing the empty vector control are represented by black squares, vms1Δ cells containing an empty vector are represented by white squares, and vms1Δ yeasts containing the extrachromosomal copy of VMS1 are represented by gray squares. Data represent the means of six independent determinations ± S.D. (error bars); p < 0.01 for the 120 min time point, and p < 0.05 for the 60 min time point, when comparing the extent of degradation in the wild type and the vms1Δ strains. Representative images corresponding to this experiment are shown in the bottom panels.
Cdc48p was recently linked to autophagy (27, 76, 77). To rule out the possibility that the degradation defects were a result of other catabolic mechanisms, such as autophagy, we looked at the processing of Ape1p/Lap4p as a measure of cytoplasmic to vacuole transport/autophagic induction under optimal and nitrogen-starved conditions (78). As seen in supplemental Fig. S5, Ape1p/Lap4p processing was apparent in both wild-type and vms1Δ cells and under both optimal growth and nitrogen-starved conditions. As anticipated, processing was absent in a strain lacking ATG8. Thus, it appears that the induction of autophagy is not affected by the loss of VMS1.
VMS1 Genetically Interacts with Members of the UBX and UFD Gene Families during ERAD
Cdc48p is a multifunctional protein that physically interacts with a large number of cofactors. It is unlikely that all of these cofactors are bound simultaneously to Cdc48p (37). Indeed, evidence indicates competition between distinct cofactors (50, 79). However, the many complex genetic interactions between Cdc48p partners have not been fully explored.
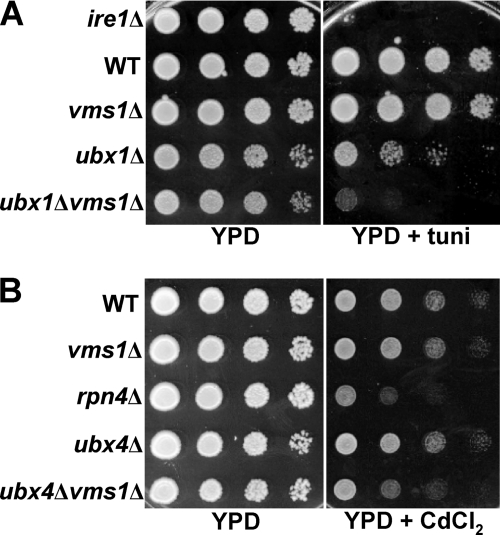
Based on the data presented above, we hypothesized that Vms1p might act in parallel and/or complementary with established Cdc48p partners during ERAD and in the presence of specific stress-inducing agents. Therefore, we performed genetic crosses between vms1Δ yeast and strains lacking the genes encoding select Cdc48p cofactors (Table 1). We focused on UBX domain-containing proteins because yeasts lacking the genes encoding some members of the UBX family (UBX1 and UBX4) share phenotypes with vms1Δ (e.g. cycloheximide and rapamycin sensitivity) and because most of the UBX and UFD proteins have not been examined for their roles during ERAD. Therefore, the resulting tetratype progeny were first screened for growth on tunicamycin- and cadmium-containing media, which induce ER and heavy metal stress, respectively. Under each condition, an exaggerated requirement for the protein quality control machinery is evident. In these assays, yeasts deleted for IRE1, which is required to induce the unfolded protein response (80), were used as a positive control for sensitivity on tunicamycin, and yeasts deleted for RPN4, which is required for the induction of genes encoding proteasome subunits (81), were used as a positive control for sensitivity on cadmium. Consistent with previous data, we observed that yeast deleted for UBX1 were sensitive to tunicamycin (72), but we also noted that deletion of VMS1 exacerbated the growth defect (see Table 1 (growth defects are denoted by a plus sign, and a heightened defect is denoted by a double plus sign); also see Fig. 4 for select examples of synergistic growth defects). This result suggests that Ubx1p, which is the yeast homolog of the p97 partner, p47, may contribute to ER homeostasis (see below). In addition, we discovered that VMS1 interacted genetically with other genes encoding Cdc48p cofactors, including UBX2, UBX3, UBX4, and UFD3, when growth was assessed in the presence of cadmium. These data suggest that Vms1p functions in parallel with several Cdc48p-associated proteins and helps mitigate the toxic effects of oxidative stress.
TABLE 1.
Genetic crosses between vms1Δ yeast and strains lacking genes encoding select Cdc48p cofactors
| Strain | Tunicamycin |
Cadmium |
||
|---|---|---|---|---|
| Single | +vms1Δ | Single | +vms1Δ | |
| ubx1Δ | + | ++ | + | + |
| ubx2Δ | − | − | − | + |
| ubx3Δ | − | − | − | + |
| ubx4Δ | − | − | − | + |
| ubx5Δ | − | − | − | − |
| ubx6Δ | − | − | − | − |
| ubx7Δ | − | − | − | − |
| ufd2Δ | − | − | − | − |
| ufd3Δ | − | − | − | + |
FIGURE 4.
VMS1 interacts with genes encoding several Cdc48p partners. Yeast cells with the indicated genotypes were grown to log phase and harvested. The cells were then serially diluted and spot-plated on medium containing either tunicamycin (tuni; A) or cadmium (CdCl2; B). Plates lacking stressors and on rich medium (YPD) were grown for 2–3 days, plates containing tunicamycin were grown for 2–3 days, and plates containing cadmium were grown for 4–6 days. All plates were incubated at 30 °C. The images are representative of several independent experiments.
Because loss of VMS1 led to a substrate-specific ERAD defect (see above), we reasoned that the simultaneous deletion of VMS1 and other Cdc48p partners would result in synthetic effects on ERAD. To date, the coordinated action of Cdc48p partners during ERAD has not been examined. Therefore, the degradation of CFTR, as a representative misfolded membrane protein was examined, and the turnover of CPY*, a model misfolded soluble ERAD substrate, was assessed in each of the tetratype progeny. The most striking example of a synergistic ERAD defect was observed when VMS1 and UFD2 were simultaneously deleted. Specifically, we found that CFTR was significantly stabilized when UFD2 was disrupted in vms1Δ cells (Fig. 5A, open triangles). This strong effect is comparable with the CFTR degradation defect observed when Cdc48p function is disabled or when the Hrd1p and Doa10p ubiquitin ligases are both absent (14). Furthermore, we found that ufd2Δ yeasts exhibit a modest delay during the CFTR degradation time course (Fig. 5A, compare closed triangles and closed squares), which is reminiscent of the ufd2Δ defect when Ste6p* degradation was assessed (45). Finally, we discovered that vms1Δufd2Δ yeast proficiently degraded CPY*, and robust growth was noted on stress-inducing media (data not shown and Table 1). One explanation for these data is that the ERAD defect in this mutant strain is confined to ERAD-C substrates. Consequently, the UPR is not induced under normal growth conditions, and tunicamycin has no impact on cell growth.
FIGURE 5.
ERAD defects are exacerbated in yeast lacking VMS1 and UFD2 or UBX4. The stability of the indicated ERAD substrate was examined by cycloheximide chase analysis as described under “Experimental Procedures” at either 40 (A) or 30 °C (B). Anti-HA antibody was used to detect each epitope-tagged ERAD substrate, and Sec61p served as a loading control. Data represent the means of three independent experiments ± S.D. (error bars). For the graphs in A and B, wild type cells are represented by black squares, and vms1Δ yeast is represented by white squares. Black triangles represent ufd2Δ in A and ubx4Δ in B, whereas white triangles represent vms1Δufd2Δ in A and vms1Δubx4Δ in B. Shown at the bottom of each panel are representative immunoblots that were used to obtain the data in each graph.
That Vms1p contributes to the ERAD pathway in a substrate-specific manner was also evident in vms1Δubx4Δ yeast. Although CPY* degradation proceeded maximally when VMS1 and UBX4 were individually deleted, the proteolysis of CPY* was slowed in the double mutant (Fig. 5B). In other cases, we observed that ERAD defects were notable in mutants lacking a single Cdc48p cofactor, but the absence of VMS1 had no further effect on ERAD efficiency. For example, the degradation of CPY* is significantly attenuated in ubx2Δ yeast (43), but the magnitude of the defect was not exacerbated in vms1Δubx2Δ yeast (supplemental Fig. S6). This phenomenon was also apparent when the degradation of CFTR and CPY* were examined in ubx1Δ and ubx1Δvms1Δ yeast (supplemental Fig. S7, A and B). Both ERAD substrates were stabilized in yeast lacking UBX1, regardless of whether VMS1 was present. An ERAD defect in ubx1Δ yeast has not, to our knowledge, previously been reported, and the effect on the degradation of an ERAD-C and ERAD-L substrate in the ubx1Δ strain is consistent with the tunicamycin-sensitive growth phenotype observed in this strain (Table 1 and Fig. 4A). The data in supplemental Figs. S6 and S7 represent the strongest effects observed, but a variety of other vms1Δ combined mutants were also examined. A summary of the degradation assays for the ERAD substrates Ste6p* and CPY* (supplemental Fig. S8, A and B) and the N-end rule substrate, Ub-Pro β-galactosidase (supplemental Fig. S9), in these strains is provided in the supplemental materials. In brief, we confirmed first that the deletion of UFD2 slows the degradation of Ste6p* (45), and we found that the loss of VMS1 in the strain had no further effect on degradation. Second, we discovered that the loss of both UBX1 and VMS1 slowed the degradation of Ste6p* (supplemental Fig. S8A). Finally, as anticipated, loss of UBX1, UBX2, or UFD2 slowed degradation of Ub-Pro β-galactosidase and the results indicated that the deletion of VMS1 in these backgrounds was without an added effect on degradation (supplemental Fig. S9).
Substrate Ubiquitination Is Proficient in Yeast Deleted for VMS1
The results presented above implicate Vms1p as a contributing factor during the ERAD of specific substrates and highlight the complex functional interplay among Cdc48p-associated factors. One mechanism by which Vms1p might inhibit ERAD is by impeding substrate ubiquitination. However, because Cdc48p acts after ERAD substrates have been ubiquitinated, we instead predicted that the loss of Vms1p would have no effect on substrate ubiquitination; nor would the loss of VMS1 lead to synergistic effects on ubiquitination in mutant strains with known defects in the ubiquitin pathway.
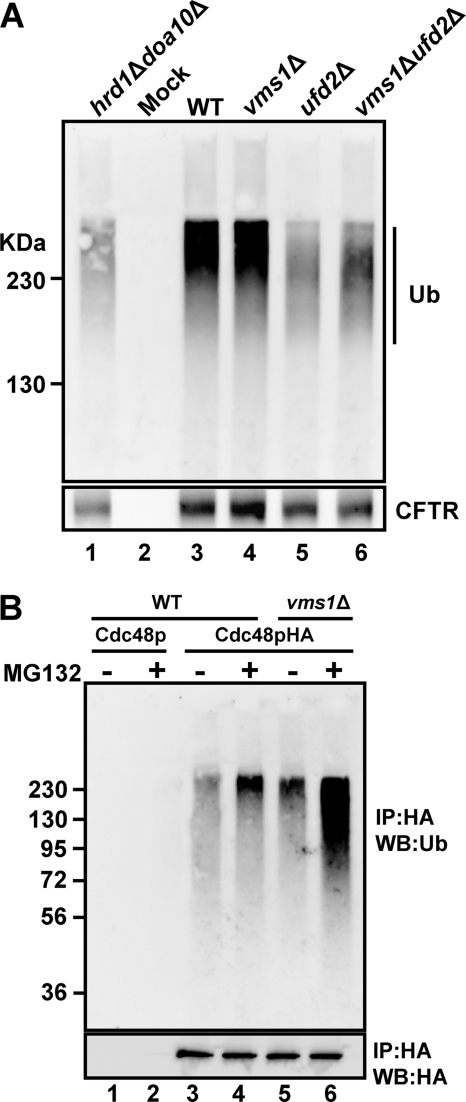
To test this hypothesis, we assessed the extent of CFTR ubiquitination in the ufd2Δvms1Δ strain due to the significant stabilization of this substrate in the double mutant (Fig. 5A). We also wished to investigate whether the extent of CFTR ubiquitination decreased in ufd2Δ yeast because Ufd2p is required for ubiquitin chain extension, and the loss of UFD2 results in reduced Ste6p* ubiquitination (45). To this end, CFTR was immunoprecipitated from wild type cells and from vms1Δ, ufd2Δ, and ufd2Δvms1Δ yeast (Fig. 6A). As a positive control for this experiment, the amount of ubiquitinated CFTR in doa10Δhrd1Δ yeast was also measured. As anticipated, we first noted that CFTR ubiquitination decreased in the doa10Δhrd1Δ strain compared with wild type cells (compare lanes 1 and 3) and that a signal corresponding to ubiquitinated CFTR was absent when immunoprecipitations in the absence of anti-HA antibody were performed (Mock, lane 2). In addition, we noted that the amount of polyubiquitinated CFTR decreased by ∼30%, and there appeared to be a shift to lower molecular weight ubiquitinated species when CFTR was examined in ufd2Δ-derived lysates. As predicted, the loss of VMS1 did not decrease the extent of CFTR ubiquitination, consistent with Vms1p acting in the ERAD pathway after substrate ubiquitination. In fact, the loss of VMS1 led to a small but reproducible increase in the amount of ubiquitinated CFTR, in either the presence (compare lanes 3 and 4) or absence (compare lanes 5 and 6) of Ufd2p. Similar results were obtained when the ubiquitination of Ste6p* was monitored in these strains and in an in vitro ubiquitination assay (data not shown) (45). Together, these data are consistent with the notion that Vms1p acts in the ERAD pathway after substrate ubiquitination.
FIGURE 6.
Increased levels of ubiquitinated proteins are associated with the Cdc48p complex in yeast lacking Vms1p. A, the indicated yeast strains expressing CFTR were grown to log phase, collected, and processed for an anti-ubiquitin (Ub; top) and anti-HA (CFTR; bottom) immunoprecipitation as described under “Experimental Procedures.” Note that polyubiquitinated CFTR resolves by SDS-PAGE as a smear ≥140 kDa, which is the approximate molecular mass of CFTR. B, WT and vms1Δ cells in which the PDR5 locus was also disrupted were transformed with either an untagged or HA-tagged version of Cdc48p. After treatment of each indicated cell type with MG132 or an equal volume of DMSO for 1 h, ER fractions were isolated, and the Cdc48p complex was immunoprecipitated (IP) with anti-HA-agarose from Triton X-100-solubilized fractions. The isolated proteins were resolved by SDS-PAGE and processed for immunoblot analysis (WB) as described under “Experimental Procedures.” The top panel depicts the results of the immunoblot with anti-ubiquitin antibody, and the bottom panel depicts the results of the immunoblot with anti-HA antibodies.
Polyubiquitinated Species Associated with Cdc48p Increase in vms1Δ Yeast
Based on the data presented in Fig. 6A and because Vms1p associates with the Cdc48p complex (Fig. 2), we next asked whether Vms1p regulates the ability of the Cdc48p complex to bind ubiquitinated substrates or the ability of the complex to release substrates. If Vms1p is required for the association of ubiquitinated substrates with the Cdc48p complex, then decreased amounts of these substrates should be associated with the Cdc48p complex in the vms1Δ mutant. In contrast, if Vms1p aids in the transfer of substrates to the proteasome, then increased levels of ubiquitinated proteins should associate with the Cdc48p complex in vms1Δ yeast.
To differentiate between these models, we immunoprecipitated Cdc48p under native conditions from ER-enriched membranes. We then performed an anti-ubiquitin immunoblot to detect Cdc48p-associated ubiquitinated species. This protocol was conducted in a wild type and vms1Δ strain, and to minimize the potential for artifacts, expression vectors that overexpress ubiquitin were not employed. In addition, to bias the analysis toward the spectrum of proteasome-targeted, Cdc48p substrates, the wild type and vms1Δ strains also lacked the gene encoding the multidrug resistance pump, PDR5, which sensitizes yeast to proteasome inhibitors (82). In a control experiment, a signal corresponding to ubiquitinated proteins was absent when an anti-HA immunoprecipitate from lysates containing an untagged version of Cdc48p was examined (Fig. 6B, lanes 1 and 2). Also, as expected, the addition of MG132 resulted in increased association of ubiquitinated species with the Cdc48p complex (compare lanes 3 and 4). Most striking, we observed an increase in the amount of ubiquitinated species in association with Cdc48p when solubilized ER membranes from vms1Δ yeast were examined (compare lanes 3 and 5), an effect that was significantly enhanced when proteasome-mediated degradation was blocked with MG132 (compare lanes 4 and 6). When the immunoprecipitation was performed in the presence of SDS, no ubiquitin signal was detected (data not shown), indicating that the ubiquitin profile was not attributable to ubiquitin-modified Cdc48p. We also found that the levels of free, unconjugated ubiquitin were constant, regardless of the strain that was used in this experiment (data not shown). We therefore conclude that Vms1p plays a role in regulating the population of proteasome-targeted, ubiquitinated species in association with Cdc48p.
DISCUSSION
A large and increasing number of Cdc48p-associated partners in yeast have been identified, and with few exceptions, the activities of most of these partners are not clear. Notable partners with established activities include Otu1p, a deubiquitinating enzyme; Ufd2p, a ubiquitin extension enzyme (also referred to as an E4); and Ufd3p, which competes with Ufd2p and can consequently rescue ubiquitinated proteins from being destroyed (50, 83). Ubx2p has been reported to help anchor the Cdc48p complex to the ER membrane, although the magnitude of Cdc48p release from the membrane in ubx2Δ yeast is variable (40, 43). The Cdc48p complex has also been found in association with multiprotein membrane complexes that include Hrd1p and Doa10p (9, 10), and Ufd1p-Npl4p, which compete with Ubx1p/p47, aid in the binding of ubiquitinated substrates and designate Cdc48p function for ERAD. In contrast, the functions of most other Cdc48p partners are ill defined, and the list of Cdc48p/p97 partners is more complex in higher eukaryotes.
In this study, we report on the characterization of a new Cdc48p partner, the product of the YDR049 gene, which was recently termed Vms1p (71). Vms1p is a conserved, soluble, cytosolic protein that exhibits limited residence on membranes in non-stressed cells and is found in the Cdc48p-containing complex. Although the loss of VMS1 leads to a modest ERAD defect, this phenotype is significantly enhanced when another Cdc48p partner, UFD2, is deleted. Based on the growth phenotypes of the vms1Δ allele in the context of mutations in the genes encoding diverse Cdc48p partners (i.e. UBX1, UBX2, UBX3, UBX4, and UFD3), we suggest that Vms1p participates in other quality control and stress relief pathways in yeast besides ERAD. Indeed, the recent discovery of Vms1p as a contributor to Cdc48p-associated mitochondrial protection under conditions of mitochondrial stress (71) is consistent with this hypothesis. Of note, an increasing body of data indicates that cellular quality control processes that operate under both stressed and unstressed conditions affect life span (84).
By developing new genetic and biochemical tools, we have also obtained data consistent with a role for Vms1p as a Cdc48p substrate release factor in unstressed cells. Namely, we find that there is an increase in the amount and possibly the population of ubiquitinated Cdc48p-associated proteins in vms1Δ cells. Other factors that may contribute to this critical step are Rad23p and Dsk2p, which contain both ubiquitin-like domains and ubiquitin-binding domains (79), and help link the Cdc48p complex and the proteasome. Future efforts may indicate whether Vms1p functions in tandem with Rad23p-Dsk2p to affect the release of ubiquitinated substrates from the Cdc48p complex. In addition, future efforts may identify the spectrum of substrates that are enriched in complex with Cdc48p when the proteasome is disabled and Vms1p function is ablated (Fig. 6B). We suspect that ERAD substrates as well as cytoplasmic substrates en route to the proteasome will be present in association with the Cdc48p complex under these conditions. An identification of these substrates could lead to the designation of unique classes of Cdc48p-associated partners and/or substrates: those that require Vms1p function for release and those that rely on other factors. Alternatively, the loss of VMS1 might lead to the absence of the bona fide release factor(s). Given the synthetic interactions between VMS1 and some of the UBX proteins, it is even possible that Vms1p and the UBX proteins function in parallel at this step in the ubiquitin-proteasome pathway.
In accordance with this proposal, one UBX protein that was recently shown to contribute to Cdc48p function and to alter ERAD efficiency is Ubx4p. Wolf and co-workers (39) recently reported that ubx4Δ yeast exhibit a defect in the degradation of CPY* and Ste6p*, and (as observed in our experiments with the vms1Δ allele) loss of UBX4 results in a build-up of ubiquitinated proteins on Cdc48p. Thus, Ubx4p may cooperate with Vms1p as a release factor. Notably, we observed a genetic interaction between VMS1 and UBX4, and a CPY* degradation defect was enhanced in the double mutant. However, we failed to observe an ERAD defect for CPY* or CFTR in the ubx4Δ mutant (Table 1), possibly because of the unique strain backgrounds used in our study and in the previously published report. The importance of strain background in analyzing the phenotypes in strains deleted for Cdc48p partners is highlighted by the fact UBX1 is an essential gene in the W303 strain (85), but the knock-out is viable in the BY background, as employed in this study. Additionally, the single deletion of UBX2 is not sensitive to cadmium in the BY background but is sensitive in another genetic background (34).
In our hands, the strongest effect on ERAD was apparent in the vms1Δufd2Δ strain. The loss of VMS1 did not exacerbate the decrease in ubiquitinated proteins or alter the profile of ubiquitinated proteins observed in the ufd2Δ strain. However, the synergistic effect may best be explained by positing that Ufd2p and Vms1p regulate unique steps in the ERAD pathway. For example, Ufd2p is required for ubiquitin chain elongation, and consistent with its role as a processivity factor, the absence of this enzyme does not alter the extent of ubiquitination but only the rate at which polyubiquitination is achieved; therefore, ERAD is initially delayed in the ufd2Δ strain, but over time, the amount of degradation catches up with that observed in wild type yeast (Fig. 5) (45). If Vms1p also catalyzes a relatively slow but non-essential step in the ERAD pathway, then the combined effect of deleting VMS1 and UFD2 may result in synergism. It is also formally possible that the loss of Vms1p reduces the cellular levels of Ufd2p or that its absence prevents Ufd2p association with the Cdc48p complex. We tested these hypotheses but observed wild type levels of Ufd2p in vms1Δ yeast and in the Cdc48p complex (data not shown).
In sum, we provide genetic evidence that a previously uncharacterized Cdc48p partner performs an important housekeeping function during ER and cellular homeostasis. Consistent with our genetic data that suggest a role for VMS1 in the ubiquitin-proteasome system, Boone and colleagues (86) showed that VMS1, UBX1, UBX4, and UFD2 negatively interact with genes that encode non-essential components of the 19 S proteasome particle and PRE9, the only nonessential component of the 20 S core. These data suggested that Vms1p, Ubx1p, Ubx4p, and Ufd2p may play a role in substrate recognition, transfer of substrate to the proteasome, and/or deubiquitination. Considering this proposal in combination with our genetic and biochemical data, we favor the view that Vms1p is a regulator of an important but poorly defined step in the ubiquitin-proteasome pathway, substrate release from Cdc48p.
Supplementary Material
Acknowledgments
We thank Karen Arndt and Margaret Shirra for help with genetic analysis; Bob Duda and Roger Hendrix for reagents; Alexander Buchberger, Stefan Jentsch, Mark Rose, and Rasmus Hartmann-Petersen for generous gifts of antibodies; the Pittsburgh Center for Kidney Research for use of core microscopy facilities (supported by National Institutes of Health Grant DK 79307); and Ora Weisz and Christopher J. Guerriero for help with microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant GM75061. This work was also supported by Cystic Fibrosis Foundation Therapeutics Grant BRODSK08XXO (to J. L. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. S1–S3.
- ERAD
- endoplasmic reticulum-associated degradation
- ER
- endoplasmic reticulum
- CFTR
- cystic fibrosis transmembrane conductance regulator.
REFERENCES
- 1. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xie W., Ng D. T. (2010) Semin. Cell Dev. Biol. 21, 533–539 [DOI] [PubMed] [Google Scholar]
- 3. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 4. Hoseki J., Ushioda R., Nagata K. (2010) J. Biochem. 147, 19–25 [DOI] [PubMed] [Google Scholar]
- 5. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 6. Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanehara K., Xie W., Ng D. T. (2010) J. Cell Biol. 188, 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gauss R., Jarosch E., Sommer T., Hirsch C. (2006) Nat. Cell Biol. 8, 849–854 [DOI] [PubMed] [Google Scholar]
- 9. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 10. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 11. Sato B. K., Schulz D., Do P. H., Hampton R. Y. (2009) Mol Cell 34, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 13. Arteaga M. F., Wang L., Ravid T., Hochstrasser M., Canessa C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11178–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gnann A., Riordan J. R., Wolf D. H. (2004) Mol. Biol. Cell 15, 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kota J., Gilstring C. F., Ljungdahl P. O. (2007) J. Cell Biol. 176, 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buck T. M., Kolb A. R., Boyd C. R., Kleyman T. R., Brodsky J. L. (2010) Mol. Biol. Cell 21, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeHoratius C., Silver P. A. (1996) Mol. Biol. Cell 7, 1835–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 19. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) Mol. Cell. Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. (2001) Mol. Biol. Cell 12, 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. (2002) EMBO J. 21, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 23. Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. (2002) Nat. Cell Biol. 4, 134–139 [DOI] [PubMed] [Google Scholar]
- 24. Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. (2004) J. Struct. Biol. 146, 11–31 [DOI] [PubMed] [Google Scholar]
- 25. Kondo H., Rabouille C., Newman R., Levine T. P., Pappin D., Freemont P., Warren G. (1997) Nature 388, 75–78 [DOI] [PubMed] [Google Scholar]
- 26. Latterich M., Fröhlich K. U., Schekman R. (1995) Cell 82, 885–893 [DOI] [PubMed] [Google Scholar]
- 27. Ju J. S., Fuentealba R. A., Miller S. E., Jackson E., Piwnica-Worms D., Baloh R. H., Weihl C. C. (2009) J. Cell Biol. 187, 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao K., Nakajima R., Meyer H. H., Zheng Y. (2003) Cell 115, 355–367 [DOI] [PubMed] [Google Scholar]
- 29. Braun R. J., Zischka H., Madeo F., Eisenberg T., Wissing S., Büttner S., Engelhardt S. M., Büringer D., Ueffing M. (2006) J. Biol. Chem. 281, 25757–25767 [DOI] [PubMed] [Google Scholar]
- 30. Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeLaBarre B., Brunger A. T. (2005) J. Mol. Biol. 347, 437–452 [DOI] [PubMed] [Google Scholar]
- 32. Jentsch S., Rumpf S. (2007) Trends Biochem. Sci. 32, 6–11 [DOI] [PubMed] [Google Scholar]
- 33. Cheng Y. L., Chen R. H. (2010) J. Cell Sci. 123, 2025–2034 [DOI] [PubMed] [Google Scholar]
- 34. Schuberth C., Richly H., Rumpf S., Buchberger A. (2004) EMBO Rep. 5, 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seeley E. S., Kato M., Margolis N., Wickner W., Eitzen G. (2002) Mol. Biol. Cell 13, 782–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang S., Guha S., Volkert F. C. (1995) Mol. Cell. Biol. 15, 2037–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schuberth C., Buchberger A. (2008) Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Decottignies A., Evain A., Ghislain M. (2004) Yeast 21, 127–139 [DOI] [PubMed] [Google Scholar]
- 39. Alberts S. M., Sonntag C., Schäfer A., Wolf D. H. (2009) J. Biol. Chem. 284, 16082–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilson J. D., Liu Y., Bentivoglio C. M., Barlowe C. (2006) Traffic 7, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 41. Schuberth C., Buchberger A. (2005) Nat. Cell Biol. 7, 999–1006 [DOI] [PubMed] [Google Scholar]
- 42. Hartmann-Petersen R., Wallace M., Hofmann K., Koch G., Johnsen A. H., Hendil K. B., Gordon C. (2004) Curr. Biol. 14, 824–828 [DOI] [PubMed] [Google Scholar]
- 43. Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005) Nat. Cell Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 44. Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. (1995) J. Biol. Chem. 270, 17442–17456 [DOI] [PubMed] [Google Scholar]
- 45. Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., Jentsch S. (1999) Cell 96, 635–644 [DOI] [PubMed] [Google Scholar]
- 47. Ghislain M., Dohmen R. J., Levy F., Varshavsky A. (1996) EMBO J. 15, 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- 48. Hochstrasser M., Varshavsky A. (1990) Cell 61, 697–708 [DOI] [PubMed] [Google Scholar]
- 49. Mullally J. E., Chernova T., Wilkinson K. D. (2006) Mol. Cell. Biol. 26, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rumpf S., Jentsch S. (2006) Mol. Cell 21, 261–269 [DOI] [PubMed] [Google Scholar]
- 51. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 52. Ravid T., Kreft S. G., Hochstrasser M. (2006) EMBO J. 25, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wright R., Parrish M. L., Cadera E., Larson L., Matson C. K., Garrett-Engele P., Armour C., Lum P. Y., Shoemaker D. D. (2003) Yeast 20, 881–892 [DOI] [PubMed] [Google Scholar]
- 54. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 55. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 56. Adams A., Kaiser C. (1998) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, pp. 19–29, Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 57. Moir D., Stewart S. E., Osmond B. C., Botstein D. (1982) Genetics 100, 547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y., Nijbroek G., Sullivan M. L., McCracken A. A., Watkins S. C., Michaelis S., Brodsky J. L. (2001) Mol. Biol. Cell 12, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y., Michaelis S., Brodsky J. L. (2002) Methods Mol. Med. 70, 257–265 [DOI] [PubMed] [Google Scholar]
- 62. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 63. Bhamidipati A., Denic V., Quan E. M., Weissman J. S. (2005) Mol. Cell 19, 741–751 [DOI] [PubMed] [Google Scholar]
- 64. Bachmair A., Finley D., Varshavsky A. (1986) Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 65. McCracken A. A., Brodsky J. L. (1996) J. Cell Biol. 132, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deshaies R. J., Schekman R. (1989) J. Cell Biol. 109, 2653–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brodsky J. L., Hamamoto S., Feldheim D., Schekman R. (1993) J. Cell Biol. 120, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kabani M., Beckerich J. M., Brodsky J. L. (2002) Mol. Cell. Biol. 22, 4677–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Coughlan C. M., Walker J. L., Cochran J. C., Wittrup K. D., Brodsky J. L. (2004) J. Biol. Chem. 279, 15289–15297 [DOI] [PubMed] [Google Scholar]
- 70. Ahner A., Nakatsukasa K., Zhang H., Frizzell R. A., Brodsky J. L. (2007) Mol. Biol. Cell 18, 806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heo J. M., Livnat-Levanon N., Taylor E. B., Jones K. T., Dephoure N., Ring J., Xie J., Brodsky J. L., Madeo F., Gygi S. P., Ashrafi K., Glickman M. H., Rutter J. (2010) Mol. Cell 40, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Parsons A. B., Brost R. L., Ding H., Li Z., Zhang C., Sheikh B., Brown G. W., Kane P. M., Hughes T. R., Boone C. (2004) Nat. Biotechnol. 22, 62–69 [DOI] [PubMed] [Google Scholar]
- 73. Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. (1992) Mol. Biol. Cell 3, 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. (2002) Mol. Biol. Cell 13, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Madeo F., Schlauer J., Zischka H., Mecke D., Fröhlich K. U. (1998) Mol. Biol. Cell 9, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tresse E., Salomons F. A., Vesa J., Bott L. C., Kimonis V., Yao T. P., Dantuma N. P., Taylor J. P. (2010) Autophagy 6, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Krick R., Bremer S., Welter E., Schlotterhose P., Muehe Y., Eskelinen E. L., Thumm M. (2010) J. Cell Biol. 190, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cheong H., Klionsky D. J. (2008) Methods Enzymol. 451, 1–26 [DOI] [PubMed] [Google Scholar]
- 79. Raasi S., Wolf D. H. (2007) Semin. Cell Dev. Biol. 18, 780–791 [DOI] [PubMed] [Google Scholar]
- 80. Cox J. S., Shamu C. E., Walter P. (1993) Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 81. Xie Y., Varshavsky A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fleming J. A., Lightcap E. S., Sadis S., Thoroddsen V., Bulawa C. E., Blackman R. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005) Cell 120, 73–84 [DOI] [PubMed] [Google Scholar]
- 84. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 85. Cheng Y. L., Chen R. H. (2010) J. Cell Sci. 123, 2025–2034 [DOI] [PubMed] [Google Scholar]
- 86. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.