Abstract
Choline acetyltransferase (ChAT) synthesizes the neurotransmitter, acetylcholine, at cholinergic nerve terminals. ChAT contains nuclear localization signals and is also localized in the nuclei of neural and non-neuronal cells. Nuclear ChAT might have an as yet unidentified function, such as transcriptional regulation. In this study, we investigated the alteration of candidate gene transcription by ChAT. We chose high affinity choline transporter (CHT1) and vesicular acetylcholine transporter (VACHT) as candidate genes, which function together with ChAT in acetylcholine production. Using SH-SY5Y human neuroblastoma cells stably expressing wild-type human ChAT, we found that overexpressed ChAT enhanced transcription of the CHT1 gene but not the VACHT gene. In contrast, nuclear localization signal disrupted, and catalytically inactive mutant ChATs could not induce, CHT1 expression. Additionally, ChAT did not alter CHT1 expression in non-neuronal HEK293 cells. Our results suggest that ChAT activates the transcription of selected target genes in neuronal cells. Both enzymatic activity and nuclear translocation of ChAT are required for its transcriptional enhancement.
Keywords: DNA Transcription, Neurodifferentiation, Neurotransmitters, Nuclear Translocation, Phosphatidylinositol 3-Kinase, SH-SY5Y Neuroblastoma Cell, Choline Acetyltransferase, High-affinity Choline Transporter, Nuclear Localizing Signal, Transcriptional regulation
Introduction
Choline acetyltransferase (ChAT)2 (E.C. 2.3.1.6) catalyzes acetylation of choline to synthesize a neurotransmitter acetylcholine (ACh), which mediates neuronal transmission in both the peripheral and central nervous systems. Cholinergic nervous systems are involved in neuromuscular transmission and in the central cholinergic projection implicated in cognitive function. Several mutations of the CHAT gene have been reported to cosegregate with congenital myasthenic syndrome with episodic apnea (1). Moreover, cholinergic neurons are preferentially affected in several neurodegenerative diseases such as Alzheimer disease and amyotrophic lateral sclerosis (2).
The major isoform of human ChAT is a 69-kDa soluble protein, referred to as the common type of ChAT (cChAT), although alternative translation initiation generates two minor isoforms of 74- and 82-kDa with distinct N-terminal extension sequences (3–5). In addition, our group has previously reported an alternatively spliced variant of a rat ChAT homologue, which is predominantly localized in the peripheral nervous system and is therefore named peripheral ChAT (pChAT) (6).
ACh is synthesized in the neuronal cytoplasm including the nerve terminal, and then transported into synaptic vesicles by the vesicular acetylcholine transporter (VAChT). Upon neuronal depolarization, ACh is released into the synaptic cleft and binds to its specific receptors. Subsequently, intrasynaptic ACh is hydrolyzed by acetylcholinesterase to liberate choline and acetate. The liberated choline is immediately recycled into the presynaptic terminals by reuptake mainly mediated through the high-affinity choline transporter (CHT1). The rate-limiting step for ACh synthesis is the supply of choline (7, 8). Thus, the regulation of ACh synthesis in presynaptic neurons depends on VAChT and CHT1 as well as on ChAT (9). Despite the many studies carried out following the molecular cloning of these proteins (10–13), regulation of the expression of these coordinately functioning proteins remains to be clarified (14).
ChAT is also expressed in various types of non-neuronal cells including epithelial, mesothelial, endothelial, muscle, and immune cells (15). This observation suggests that ChAT is implicated in as yet unidentified function(s) besides neurotransmitter synthesis. Furthermore, Resendes et al. (17) have described nuclear localization of ChAT isoforms. This group has also defined one nuclear localization signal (NLS) in the 69-kDa isoform, and two NLSs in the 82-kDa isoform of human ChAT (hChAT) (16). An NLS acts like a “tag” on the exposed surface of a protein and targets it to the nucleus through the nuclear pore complex. The 69-kDa hChAT isoform is shuttled between the cytoplasmic and nuclear compartments, whereas the 82-kDa isoform is predominantly localized in the nucleus. Furthermore, we previously reported that a rat pChAT isoform, in contrast to the 69-kDa cChAT isoform, is preferentially localized in the cytoplasm and that inhibition of cellular protein kinase C activity leads to increased accumulation of nuclear pChAT (18). These observations led us to examine the functional relevance of the distinct subcellular localizations of ChAT.
To date the functional significance of nuclear ChAT remains unresolved. However, the 82-kDa nuclear isoform is expressed in human brains of normal aged subjects and in patients with Alzheimer disease (19). Additionally, because the molecular weight of ChAT is larger than that of proteins that can passively diffuse across the nuclear envelope, the nuclear translocation of ChAT should be an energy requiring process. This fact may suggest this process has some physiological significance. To elucidate possible functions of nuclear ChAT, we investigated transcriptional changes of genes related to ACh production, which we considered likely to respond to ChAT overexpression in neuronal cell lines. We report here that ChAT overexpression induces transcription of the CHT1 gene in SH-SY5Y cells and that this induction requires the nuclear localization and catalytic activity of ChAT.
EXPERIMENTAL PROCEDURES
Construction of Expression Plasmids for Human ChAT
To obtain the full-length cDNA encoding the human 69-kDa isoform of ChAT (hChAT), we performed the polymerase chain reaction (PCR) using a human small intestine Marathon-Ready cDNA library (Clontech Laboratories, Otsu, Japan) as a template. For construction of hChAT fused with green fluorescent protein (hChAT-GFP), an EcoRI restriction site and the Kozak sequence were introduced into the 5′-end of the cDNA, and a BamHI restriction site was introduced into the 3′-end. The stop codon TGA was substituted for TGC. The cDNA fragment was ligated into the pEGFP-N1 vector (Clontech).
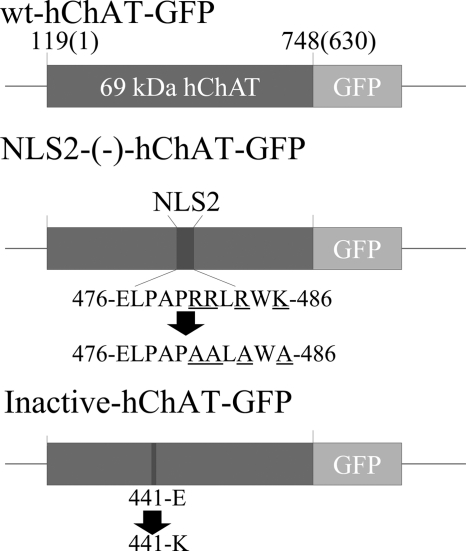
The catalytically inactive mutant, inactive-hChAT-GFP, and the NLS-disrupted mutant, NLS2-(−)-hChAT-GFP (Fig. 1, inactive and NLS2-(−)), were generated by PCR-based site-directed mutagenesis using the QuikChange XL-II Mutagenesis kit (Stratagene, Cedar Creek, TX) according to the manufacturer's instructions. The numbering of amino acid residues described below is according to the 82-kDa hChAT isoform 2 (GenBankTM accession number NP_065574). For the inactive-hChAT-GFP, Glu at amino acid residue 441 (Glu441) was substituted by Lys using the sense-strand oligonucleotide: GCGGTGTGGTGTGCAAACACTCCCCATTC. For NLS2-(−)-hChAT-GFP, Arg481, Arg482, Arg484, and Lys486 were all substituted by Ala using the sense-strand oligonucleotide: CGAGCTCCCCGCCCCCGCGGCGCTGGCGTGGGCATGCTCCCCGGAAATT. Additionally, we also made another candidate for NLS-disrupted mutant, NLS1-(−)-hChAT, Arg322, Arg323, Arg334, and Lys335 were all substituted by Ala with two processes of PCR-based site-directed mutagenesis using the sense strand oligonucleotides: GTCTTGGATGTTGTCATTAATTTCGCCGCTCTCAGTGAGGGGGATC and GGGGGATCTGTTCACTCAGTTGGCAGCGATAGTCAAAATGGCTTCCAAC. DNA sequences of all plasmids were confirmed by sequencing using the ABI 3100 DNA Sequencer (Applied Biosystems, Foster City, CA).
FIGURE 1.
Schematic illustration of the hChAT (CHAT) constructs used in this study. wt-hChAT-GFP is the 69-kDa isoform of human ChAT fused at the C terminus with a GFP tag. NLS2-(−)-hChAT-GFP has an Ala substituted for each of Arg481, Arg482, Arg484, and Lys486 in the nuclear localization NLS2 sequence. In inactive hChAT-GFP, a Lys residue has been substituted for Glu441. The amino acid residue positions are numbered to correspond to the 82-kDa human ChAT sequence, which differs from the 69-kDa isoform by the presence of a 118-amino acid extension on the amino terminus. The numbers in parentheses indicate the amino acid number in 69-kDa hChAT.
Establishment of Polyclonal Stable Cell Lines
For selection of stable transformants of SH-SY5Y cells, the transfected cells were maintained in culture medium containing 300 μg/ml of G418 (Nacalai Tesque, Kyoto, Japan). After 3 weeks of culture, the GFP-positive cells were collected using a fluorescence-activated cell sorter (FACS; BD FACSAria II Cell Sorter, BD Biosciences). SH-SY5Y cells expressing wild-type hChAT-GFP (wt-hChAT-GFP), mutant hChAT-GFP, or GFP alone were selected based on the fluorescence intensity of the GFP they expressed. According to the fluorescence intensity, we divided wt-hChAT-GFP into two types: highly expressing cells (wt-hChAT-GFP) and moderately expressing cells (wt-hChAT-GFP-M), for further comparisons by using the FACS. Stable expression of each construct was verified using RT-PCR and Western blot analysis. We also prepared human embryonic kidney (HEK293) cells stably expressing wt-hChAT-GFP, mutant hChAT-GFP, or GFP alone for preliminary experiments and further comparisons with the SH-SY5Y cell lines.
ChAT Activity Assay
Cultured cells were washed twice with cold phosphate-buffered saline (PBS) (pH 7.4), and treated with 0.02% EDTA and 0.25% trypsin. The cells were pelleted by centrifugation at 80 × g for 5 min and then homogenized in 5–10 volumes (v/w) of 10 mm phosphate buffer (PB) (pH 7.4) containing 0.5% Triton X-100. After centrifugation at 12,000 × g for 20 min at 4 °C, 10 μl of the supernatant was used for ChAT activity measurement using the radiometric method with a slight modification (20). The reaction mixture consisted of 300 mm NaCl, 8 mm choline, 0.12 mm eserine, and 0.3 mm cold acetyl-CoA, and an appropriate quantity of [3H]acetyl-CoA in 50 mm PB (pH 7.4). Each reaction was performed in a 0.6-ml plastic tube at 37 °C for 30 min. After stopping the reaction by ice cooling, the reaction mixture (20 μl total volume) was transferred to another tube containing 4.5 ml of 0.1 m PB (pH 7.4), followed by the addition of 2 ml of acetonitrile containing 5 mg/ml of sodium tetraphenylboron (Dojindo, Kumamoto, Japan) and 8 ml of a toluene-based scintillation mixture (Scintiblender II, Nacalai Tesque). The radioactivity of [3H]ACh, collected by means of this liquid cation-exchange method into the organic phase, was counted using a liquid scintillation counter (Packard Tri-CARB 3100, Meridian, CT). The ChAT activity was expressed as nanomoles of [3H]ACh formed per minute per milligram of protein (21). Protein concentrations were determined using the Bradford assay (Bio-Rad).
Confocal Laser Scanning Microscopy
Images were collected as previously reported (18). For comparison of the fluorescence intensity of individual images, identical parameters were set for imaging. Image processing and data analysis were performed using Photoshop or ImageJ software.
To evaluate transient nuclear translocation, we used the Crm-1-dependent nuclear export inhibitor, leptomycin B (Sigma) (22). Cultured cells were treated with 10 ng/ml of leptomycin B for up to 24 h. Analysis by confocal laser scanning microscopy was carried out as described above.
Immunocytochemistry for Cell Cycle Markers
We observed the relationship between the subcellular localization of ChAT-GFP and the cell cycle by immunocytochemistry for cell cycle markers. Immunocytochemistry was performed as previously reported (18). In brief, the cells were cultured for 2 days in a glass-bottom dish. After rinsing twice with cold PBS, the cells were fixed for 5 min with 4% paraformaldehyde in 100 mm phosphate buffer at 4 °C. The fixed cells were rinsed gently three times with 100 mm PBS containing 0.3% Triton X-100 (PBST). They were incubated overnight with anti-cyclin E antibody conjugated with rhodamine (rabbit polyclonal, Sc-481, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, 1:50) or anti-cyclin B1 antibody conjugated with Alexa 647 (mouse monoclonal, Santa Cruz Biotechnology, Inc., 1:50) in PBST at 4 °C. After washing 3 times with PBST for 10 min each, the cells were mounted with 4′,6-diamidino-2-phenylindole (DAPI)-Fluoromount-G (Southern Biotech, Birmingham, AL). Analysis by confocal laser scanning microscopy was carried out as described above.
Acetylcholine Antagonist and Agonists Treatment
To evaluate involvement of acetylcholine muscarinic receptors, cultured cells were treated with 10 nm to 10 μm atropine (Wako, Tokyo, Japan), a nonspecific muscarinic antagonist, for 72 h. We also tested the effects of nonspecific muscarinic agonists on CHT1 induction in the cultured cells, by using pilocarpine (10 μm to 1 mm, Wako) and carbachol (10 μm to 1 mm, Wako) for 72 h. Fresh aliquots of the agonist or antagonists were added to the culture at 24-h intervals with medium change. The treated cells were collected and analyzed using real time PCR as described below.
PI3K Inhibitor Treatment
Cultured cells were treated with 1 to 50 μm LY294002 (Cayman Chemical, Ann Arbor, MI), a phosphatidylinositol 3-kinase (PI3K) inhibitor, for 24 or 72 h (23). The treated cells were collected and analyzed using real time PCR as described below.
Real Time PCR
Real time reverse transcription PCR assays were performed to quantify the mRNA levels of CHT1, CHAT, and VACHT. Total RNA was extracted from cultured cells using the FastPure RNA kit (Takara-Bio, Otsu, Japan). Typically, 500 ng of total RNA was reverse-transcribed using the PrimeScript RT reagent kit (Takara-Bio) and random hexamers and oligo(dT) primers (Takara-Bio). To make external cDNA standards for real time PCR, the PCR products were subcloned into the pCR2.1 vector using a TA cloning kit (Invitrogen Japan Ltd., Tokyo, Japan). The plasmid cDNA was amplified in a One Shot TOP10 chemically competent Escherichia coli (Invitrogen Japan Ltd.), and isolated using a Quantum Prep Plasmid Miniprep kit (Bio-Rad Laboratories). The primers used for PCR of hChAT (CHAT), CHT1, VACHT, β-actin (ACTB), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown under supplemental Table S1.
Real time PCR was carried out using a Light Cycler system or the Light Cycler 480 instrument (Roche Diagnostics K.K., Tokyo, Japan) and the SYBR Premix Ex Taq (Perfect Real Time) II kit (Takara-Bio). To confirm amplification specificity, the PCR products from each primer pair were subjected to melting curve analysis and subsequent agarose gel electrophoresis. The results were analyzed using the second derivative maximum method of the LightCycler Software (Roche Diagnostics). Relative quantification of cDNA was performed based on these fluorescence measurements by comparison with a standard curve that was generated during the course of each PCR run. We confirmed a good correlation between the expression level of GAPDH mRNA and that of ACTB mRNA. These genes were therefore used as reference genes for normalization. The values were normalized to the levels of GAPDH cDNA determined from the same cDNA sample in the same PCR run. All experiments were performed independently at least three times. The levels of target mRNA were expressed relative to levels of GAPDH mRNA (arbitrary units).
Preparation of Cell Homogenates and Subcellular Fractions
For the preparation of total cell homogenates cultured cells were washed twice with cold PBS, and harvested using trypsin/EDTA. The cells were pelleted by centrifugation at 80 × g for 5 min and then lysed in 5–10 volumes (v/w) of 10 mm PB (pH 7.4) containing 0.5% Triton X-100 and a protease inhibitor mixture (Complete-mini, Roche Applied Science). The lysates were centrifuged at 12,000 × g for 20 min at 4 °C, and the supernatants were collected as total soluble fractions.
Nuclear fractions of cultured cells were obtained using a previously described method (24). Briefly, the SH-SY5Y cells were harvested in a buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.5 mm DTT) containing a protease inhibitor mixture. After rinsing the cells, 1/10 volume of 0.5% Nonidet-P 40 was added to the cell suspension. After the lysate was centrifuged at 1,000 × g for 2 min at 4 °C, the supernatant was collected as the cytoplasmic fraction. The resultant pellets were resuspended in a high-salt buffer (20 mm HEPES, 0.4 m NaCl, 1 mm EDTA, 10% glycerol, 0.5 mm DTT) supplemented with protease inhibitors, and then placed on ice for 15 min. The supernatant was cleared by centrifugation at 12,000 × g for 15 min at 4 °C and collected as the nuclear fraction. CHT1 is functionally a plasma protein, which is also known to be constantly cycled through the plasma membrane and intracellular compartments. Because previous studies showed that CHT1 is predominantly intracellular, we used the cytoplasmic fractions for Western blotting of CHT1, as described below (25–29).
Western Blotting
Aliquots of cell lysates containing equal amounts of protein were subjected to Western blotting (18). The primary antibodies used were anti-GFP (mouse monoclonal, Roche, diluted 1:2500), anti-β-actin (mouse monoclonal, MAB1501, Chemicon International Inc., Billerica, MA, diluted 1:2000), anti-GAPDH (mouse monoclonal, MAB374, Chemicon, diluted 1:2000), anti-ChAT (rabbit polyclonal generated in our laboratory (30), diluted 1:5000), and anti-CHT1 (rabbit polyclonal, AB5966, Chemicon, diluted 1:1000). Bands were visualized using the Super-Signal West Pico chemiluminescent substrate (Thermo Fisher Scientific K.K, Yokohama, Japan), and analyzed using a LAS-4000 IR multicolor image analyzer (Fujifilm, Tokyo, Japan). The intensity of each band was semi-quantified using ImageJ software.
Statistical Analysis
The results are expressed as mean ± S.E. For statistical analysis, an unpaired two group t test, a non-parametric Kruskal-Willis test, a Mann-Whitney test, or a post-hoc Tukey-Kramer test was used to determine the significance of differences between means.
RESULTS
Expression, Catalytic Activity, and Subcellular Localization of wt-hChAT-GFP
We first determined if the hChAT-GFP construct was useful for examination of ChAT activity and subcellular localization. The expected molecular weight of wt-hChAT fused with the 27-kDa GFP is 96 kDa. On immunoblotting of wt-hChAT-GFP-transfected HEK293 cell lysates, polyclonal anti-ChAT and monoclonal anti-GFP antibodies both detected wt-hChAT-GFP of the expected size (data not shown). A high level of ChAT-specific activity was detected in the homogenate of HEK293 cells transiently transfected with wt-hChAT-GFP. Furthermore, as we previously reported for a rat homolog of ChAT fused with GFP (18), wt-hChAT-GFP was observed in both the cytoplasm and nucleus in HEK293 cells by fluorescence microscopy (data not shown).
Characterization of Polyclonal SH-SY5Y Cell Lines Stably Expressing wt-hChAT-GFP
SH-SY5Y cells stably expressing wt-hChAT-GFP or GFP alone were selected based on the intensity of their GFP fluorescence. To avoid clonal deviation, we collected GFP-positive cells using FACS. The level of CHAT mRNA in each cell line was confirmed using RT-PCR. As expected, wt-hChAT-GFP cells expressed abundant CHAT mRNA, whereas GFP alone cells expressed only a trace level of CHAT mRNA (Fig. 2A).
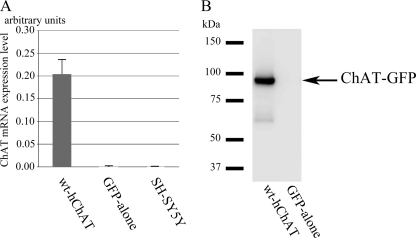
FIGURE 2.
The mRNA and protein expression of hChAT-GFP in a stable polyclonal cell line. A, the hChAT (CHAT) mRNA level in wt-hChAT-GFP cells. Total RNA was extracted from wt-hChAT-GFP (wt-hChAT), GFP alone, and native SH-SY5Y cells, and the relative levels of CHAT mRNA were evaluated using real time PCR assays. Native and GFP alone cells showed a trace level of CHAT mRNA expression that was less than 0.25% that of wt-hChAT-GFP cells. Values are the mean ± S.E. B, immunoblot of hChAT-GFP protein expression. Aliquots of cell homogenates containing the same amount of protein were subjected to immunoblotting. The blots were probed with an anti-ChAT antibody.
Western blot analysis using polyclonal anti-ChAT or monoclonal anti-GFP antibodies confirmed the expression of a protein of the expected size in this cell line (Fig. 2B and data not shown). To ensure that the expressed ChAT-GFP was catalytically active, specific ChAT activities were measured. The wt-hChAT-GFP cells exhibited a high level of ChAT activity (wt-hChAT-GFP, 97.9 ± 7.16 nmol/mg of protein/min, mean ± S.E., and wt-hChAT-GFP-M, 12.6 ± 3.94), whereas no activity was detected in homogenates of GFP alone cells. ChAT activity of HEK293 cells stably expressing wt-hChAT-GFP was 158.68 ± 11.45 nmol/mg/min.
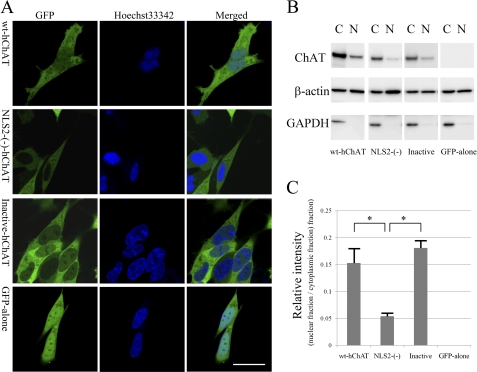
Subcellular distribution of wt-hChAT-GFP and GFP alone in these living cells was investigated using a confocal laser scanning microscope. As we previously reported for the rat ChAT-GFP homolog in HEK293 cells (18), wt-hChAT-GFP was located both in the cytoplasm and nucleus. Although it was more strongly expressed in the cytoplasm, wt-hChAT-GFP nuclear localization was assessed by its co-localization with the nuclear fluorescent dye Hoechst 33342 (Fig. 3A) and was confirmed using a biochemical cell fractionation assay (Fig. 3, B and C). The fluorescence intensity of GFP alone was the same in both the cytoplasm and nucleus, which may be a result of nonspecific passive diffusion.
FIGURE 3.
Subcellular localization of hChAT-GFP. A, representative images of wt-hChAT-GFP (wt-hChAT), NLS2-(−)-hChAT-GFP (NLS2-(−)-hChAT), inactive hChAT-GFP (Inactive-hChAT), and GFP alone cells. GFP fluorescence was detected in living SH-SY5Y cells using a confocal laser scanning microscope. The cells were co-stained with the nuclear dye Hoechst 33342. Merged images are at the right. Bar = 20 μm. B, subcellular fractionation assays. Nuclear (N) and cytoplasmic (C) fractions were obtained from wt-hChAT-GFP (wt-hChAT), NLS2-(−)-hChAT-GFP (NLS2-(−)), inactive hChAT-GFP (Inactive), and GFP alone cells, and subjected to immunoblotting using anti-ChAT, anti-β-actin, and anti-GAPDH antibodies (from top to bottom). GAPDH served as a marker for cytoplasmic proteins and β-actin immunoblotting was used as a loading control. C, relative intensity was measured by ImageJ software, and ratios between the nuclear fraction and the cytoplasmic fraction in each clone were calculated. Values are expressed as the mean ± S.E. A post-hoc Tukey-Kramer test was used for statistical analysis. * indicates p < 0.05 significant difference.
We observed ChAT-GFP subcellular localization in wt-hChAT-GFP cells with cell-cycle markers: cyclin B1 (G2/M phase marker) and cyclin E (G1/S phase marker). In the cell culture condition we used in this study, strong nuclear staining with the anti-cyclin E antibody was detected in most of the tested wt-hChAT-GFP cells (supplemental Fig. S1), however, cyclin B1 staining was not detected.
Up-regulation of CHT1 mRNA Transcription in wt-hChAT-GFP Cells
To determine whether CHAT affects transcriptional activation of the CHT1 or VACHT genes, we analyzed RNA extracted from wt-hChAT-GFP and GFP alone cells for mRNA expression of these genes using real time PCR. The CHT1 mRNA level in wt-hChAT-GFP cells was significantly higher than that of GFP alone cells (Fig. 4B). VACHT mRNA in the wt-hChAT-GFP cells was comparable with that in GFP alone cells (relative ratio of wt-hChAT-GFP to GFP alone cells: 0.96 ± 0.02). These results suggested that transcription of CHT1 was selectively up-regulated by overexpression of wt-hChAT-GFP.
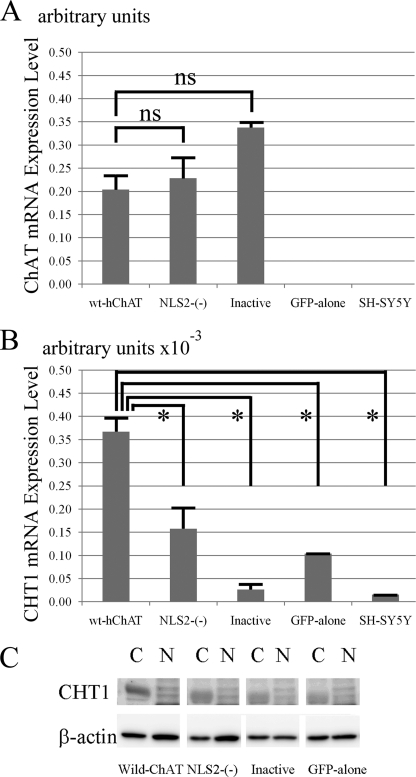
FIGURE 4.
Both of the nuclear translocation and catalytic activity of ChAT are required for CHT1 induction by ChAT. A, hChAT mRNA level in wt-hChAT-GFP (wt-hChAT), NLS2-(−)-hChAT-GFP (NLS2-(−)), inactive hChAT-GFP (Inactive), GFP alone, and nontransfected SH-SY5Y cells. Total RNA was extracted from the cultured cells and used as a template for a real time PCR assay. Values are the mean ± S.E. The non-parametric Mann-Whitney test was used for statistical analysis. ns represents no significant difference. B, real time RT-PCR analysis of CHT1 mRNA expression in wt-hChAT-GFP (wt-hChAT), NLS2-(−)-hChAT-GFP (NLS2-(−)), inactive hChAT-GFP (Inactive), GFP alone, and nontransfected SH-SY5Y cells. *, p < 0.05 by the non-parametric Mann-Whitney test. C, immunoblot of CHT1 protein expression. Nuclear (N) and cytoplasmic (C) fractions were obtained from each clone, and subjected to immunoblotting with anti-CHT1 and anti-β-actin antibodies. Immunoblotted β-actin was used as a loading control.
To examine whether the up-regulated CHT1 mRNA in the wt-hChAT-GFP cells was translated into protein, we performed Western blot analysis for CHT1. The anti-CHT1 antibody strongly detected CHT1-protein expression in the cytoplasmic fraction of wt-hChAT-GFP cells, whereas only a faint band was detected in GFP alone cell homogenates (Fig. 4C), suggesting that exogenous wt-hChAT-GFP induced CHT1 protein expression. In contrast, stable expression of wt-hChAT-GFP (or of GFP alone) in HEK293 cells, which do not express CHT1 mRNA or protein, did not induce CHT1 mRNA or protein expression (data not shown), suggesting that CHT1 induction by wt-hChAT-GFP may be specific to neuronal cell types.
CHT1 mRNA Induction Depends on the Nuclear Localization of hChAT
To examine whether nuclear translocation of hChAT is required for its up-regulation of CHT1 expression, we prepared a mutant hChAT construct in which the NLS was disrupted. It has been reported that hChAT bears two putative NLSs: NLS1 (322RRLSEGDLFTQLRKIVKM339) and NLS2 (476ELPAPRRLRWK486) (16). We constructed two mutants: one of which was NLS1-(−)-hChAT-GFP, in which all four of the Arg322, Arg323, Arg334, and Lys335 residues of the NLS1 sequence were replaced by Ala, and the second was NLS2-(−)-hChAT-GFP, in which all four of the Arg481, Arg482, Arg484, and Lys486 residues of the NLS2 sequence were substituted by Ala (Fig. 1). Using fluorescence microscopy, we confirmed that mutation of NLS2 but not mutation of NLS1, inhibits hChAT-GFP translocation to the nucleus upon transient transfection into HEK293 cells (data not shown). This observation was consistent with a previous report showing that the C-terminal NLS2 is more important for hChAT nuclear translocation than NLS1 (16). We also confirmed that NLS2-(−)-hChAT-GFP exhibited ChAT activity equivalent to that of wt-hChAT-GFP in HEK293 cells (data not shown).
We next established a polyclonal SH-SY5Y cell line stably expressing NLS2-(−)-hChAT-GFP using a procedure similar to that used for establishment of wt-hChAT-GFP cell lines. We then compared the mRNA expression level of CHAT in these cells using real time PCR. The results indicated that equivalent levels of hChAT were expressed in wt-hChAT-GFP and NLS2-(−)-hChAT-GFP cells (Fig. 4A). In addition, NLS2-(−)-hChAT-GFP cells exhibited a significant level of CHAT activity (6.6 ± 1.05 nmol/mg/min). The ChAT activity of these cells was not as high as that of wt-hChAT-GFP cells, but was comparable with that of rat striatum homogenate (3.56 ± 0.12) (21).
Using confocal laser scanning microscopy, we determined that NLS2-(−)-hChAT-GFP fluorescence was almost exclusively localized in the cytoplasm, whereas nuclear fluorescence was similar to the background level (Fig. 3A). Co-staining with the Hoechst 33342 nuclear dye clearly showed that NLS2-(−)-hChAT-GFP did not co-localize with this dye. This cellular localization of NLS2-(−)-hChAT-GFP was in clear contrast to that of wt-hChAT-GFP. We further confirmed almost exclusive cytoplasmic localization of NLS2-(−)-hChAT-GFP using a biochemical cell fractionation assay. The isolated cytoplasmic and nuclear fractions from these cells were subjected to immunoblotting using antibodies against ChAT, GAPDH (as a cytoplasmic marker), or β-actin (as a loading control) (Fig. 3B). The nuclear fraction of NLS2-(−)-hChAT-GFP cells contained a very small amount of this hChAT protein compared with the cytoplasmic fraction. In contrast, obvious bands corresponding to hChAT-GFP were observed in the nuclear fractions of wt-hChAT-GFP cells, although the intensity of these bands was lower than the intensity of those in the cytoplasmic fraction (Fig. 3C).
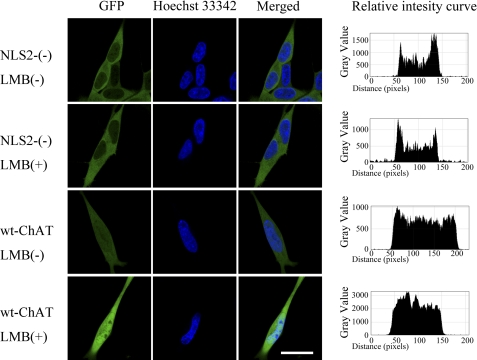
To further examine the nuclear translocation of the NLS2-(−)-hChAT-GFP construct, we treated these cells with 10 ng/ml of leptomycin B, a potent inhibitor of nuclear export. Following treatment of wt-hChAT-GFP-expressing cells with leptomycin B for 8 h, the accumulation of hChAT-GFP in the nucleus was clearly enhanced (Fig. 5). In contrast, treatment with leptomycin B did not result in a significant increase in nuclear NLS2-(−)-hChAT-GFP even after 24 h of treatment (8-h treatment is shown in Fig. 5). This result suggested that NLS2-(−)-hChAT-GFP did not even transiently translocate into the nucleus.
FIGURE 5.
NLS2-(−)-hChAT-GFP did not accumulate in the nucleus after leptomycin B treatment. The wt-hChAT-GFP (wt-hChAT) and NSL2-(−)-hChAT-GFP (NLS2-(−)) cells were treated with (+) or without (−) 10 ng/ml of leptomycin B (LMB), a potent inhibitor of nuclear export, for 8 h. GFP fluorescence and the fluorescence of the nuclear dye Hoechst 33342 were detected using a confocal laser scanning microscope. Bar = 20 μm. The relative intensity curves were made using ImageJ software.
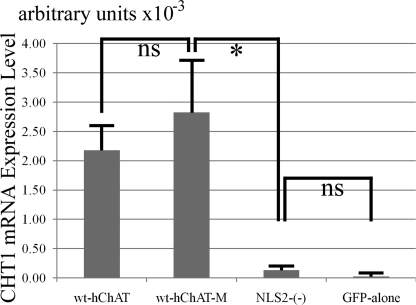
We then compared CHT1 expression in wt-hChAT-GFP and NLS2-(−)-hChAT-GFP cells using quantitative RT-PCR. The CHT1 mRNA level in NLS2-(−)-hChAT-GFP cells was significantly less than that in wt-hChAT-GFP cells and was comparable with that in GFP alone cells (Fig. 4B). Immunoblotting of CHT1 protein expression also revealed that the level of CHT1 protein expressed in NLS2-(−)-hChAT-GFP cells was clearly lower than that of wt-hChAT-GFP cells (Fig. 4C). These findings indicated that nuclear translocation of hChAT is required for transcriptional activation of CHT1.
To exclude the possibility that the catalytic activity of NLS2-(−)-hChAT-GFP was not sufficient to induce CHT1 expression, we prepared another wt-hChAT-GFP cell with a moderate catalytic activity (wt-hChAT-GFP-M, 12.6 ± 2.27 nmol/mg/min), which was not significantly different from that of NLS2-(−)-hChAT-GFP (6.6 ± 1.05), and then compared CHT1 expression using quantitative RT-PCR. The CHT1 mRNA level in NLS2-(−)-hChAT-GFP cells was again significantly less than that in wt-hChAT-GFP-M cells. The CHT1 mRNA level in wt-hChAT-GFP-M cells was on the same level as wt-ChAT-GFP cells (Fig. 6).
FIGURE 6.
Real time RT-PCR analysis of CHT1 mRNA expression in wt-hChAT-GFP-M and NLS2-(−)-hChAT-GFP cells. Total RNA was extracted from wt-hChAT-GFP (wt-hChAT), wt-hChAT-GFP-M (wt-hChAT-M), NLS2-(−)-hChAT-GFP ((NLS2-(−)), and GFP alone cells and was used as a template for a real time PCR assay. Values are the mean ± S.E. * indicates p < 0.05 significant difference by the non-parametric Mann-Whitney test, and ns represents not significant. Note that the catalytic activity of wt-hChAT-GFP-M cells was not significantly different from that of NLS2-(−)-hChAT-GFP cells.
ChAT Activity Is Necessary for Up-regulation of CHT1 mRNA Expression
To address the question as to whether transcriptional activation of CHT1 by hChAT-GFP requires catalytic activity of ChAT or not, we prepared an enzymatically inactive ChAT-GFP construct harboring a E441K substitution, termed inactive-hChAT-GFP (Fig. 1). This Glu441 corresponds to Glu332 of the rat homologue, which is very close to one of the possible active sites (31). A E441K mutation in the CHAT gene is the cause of congenital myasthenic syndrome and results in the complete abolition of ChAT activity (1). We confirmed that no significant ChAT activity could be detected in homogenates of HEK293 cells transiently transfected with inactive hChAT-GFP.
We next established a polyclonal SH-SY5Y cell line stably expressing inactive hChAT-GFP using a method similar to that used for establishment of wt-hChAT-GFP cell lines. We then compared the expression levels of inactive hChAT-GFP in these cell lines using real time PCR (Fig. 4A). An equivalent level of CHAT mRNA was expressed in wt-hChAT-GFP and inactive hChAT-GFP cells. Using confocal microscopy and a biochemical subcellular fractionation assay, we further confirmed that the cytoplasmic and nuclear localization of inactive hChAT-GFP was similar to that of wt-hChAT-GFP (Fig. 3A). No ChAT activity was detected in homogenates of inactive hChAT-GFP cells.
Finally, we evaluated CHT1 expression in inactive hChAT-GFP cells. A real time PCR assay showed that the CHT1 mRNA level in these cells was equivalent to that in GFP alone cells (Fig. 4B). Western blotting of CHT1 protein levels revealed bands of approximately the same intensity in both inactive hChAT-GFP cells and GFP alone cells (Fig. 4C). These findings suggested that ChAT activity is indispensable for CHT1 induction by hChAT-GFP.
Muscarinic Antagonist Did Not Reverse the Up-regulation of CHT1 by ChAT
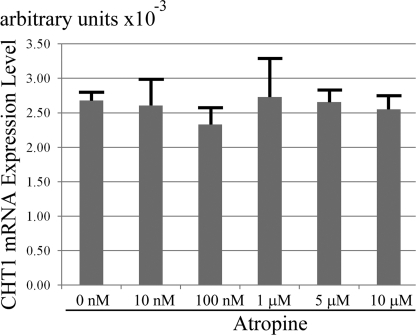
Because SH-SY5Y cells are known to be expressing muscarinic receptors on their cell surface (32, 33), we investigated whether the muscarinic receptors participate in the process of the observed CHT1 induction. Nonspecific muscarinic antagonist treatments did not effect the up-regulation of CHT1 by ChAT overexpression even with the highest concentration at 10 μm for 72 h (Fig. 7). Furthermore, the two tested agonists did not significantly change the level of CHT1 mRNA expression in ether wt-hChAT-GFP cells or GFP alone cells (data not shown).
FIGURE 7.
A muscarinic antagonist, atropine, did not reverse CHT1 induction by ChAT. Real time RT-PCR analysis of CHT1 in wt-hChAT-GFP cells treated with various concentrations of atropine for 72 h. Total RNA was then extracted from the cells and the relative level of CHT1 mRNA was evaluated by a real time PCR assay. Values are the mean ± S.E. No significant difference was detected by using a post hoc Tukey-Kramer test.
Inhibition of PI3K Reduced the Up-regulation of CHT1 by ChAT
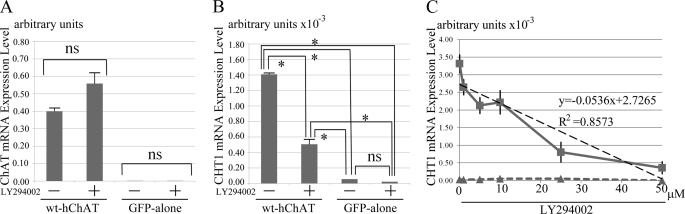
A previous study showed that CHT1 expression is induced in primary septal cells by stimulation with nerve growth factor (NGF) (34). It has also been reported that treatment with an inhibitor of phosphatidylinositol 3-kinase (PI3K) prevents this effect of NGF on CHT1 expression (34). To determine whether PI3K inhibition could reverse CHT1 induction by ChAT, we treated wt-hChAT-GFP cells with 10 μm LY294002 in DMSO for 72 h. Although LY294002 treatment reduced the cell number of both wt-hChAT-GFP and GFP alone cell cultures, the mRNA expression level of CHAT mRNA in the surviving cells was not significantly different to that in cells treated with the same amount of control DMSO (Fig. 8A). The treatment of the wt-hChAT-GFP cells with LY294002 at 10 μm for 72 h induced a significant reduction in CHT1 mRNA expression (Fig. 8B). To determine whether PI3K inhibition could reverse CHT1 induction in a dose-response manner, we treated wt-hChAT-GFP cells with 1–50 μm LY294002 in DMSO for 24 h. We shortened the duration of treatment, because at a concentration higher than 25 μm LY294002, the treated cells did not survive for 2 days. A significant correlation between the LY294002 concentration and reduction of the CHT1 mRNA level was observed (Fig. 8C).
FIGURE 8.
Inhibition of PI3K reduced CHT1 induction by ChAT. A, real time RT-PCR analysis of hChAT in wt-hChAT-GFP (wt-hChAT) and GFP alone cells treated with either the PI3K inhibitor LY294002 in DMSO or with DMSO alone for 48 h. Total RNA was then extracted from the cells and the relative level of hChAT mRNA evaluated by a real time PCR assay. Values are expressed as the mean ± S.E. A post hoc Tukey-Kramer test and the non-parametric Mann-Whitney test were used for statistical analysis. ns represents no significant difference. B, real time RT-PCR analysis of CHT1 in wt-hChAT-GFP and GFP alone cells treated with either LY294002 in DMSO or with DMSO alone for 48 h. Total RNA was then extracted from the cells and the relative level of CHT1 mRNA was evaluated by a real time PCR assay. Values are expressed as the mean ± S.E. A post hoc Tukey-Kramer test and the non-parametric Mann-Whitney test were used for statistical analysis. ns represents no significant difference. *, p < 0.05 by the non-parametric Mann-Whitney test. wt-hChAT-GFP cells treated with LY294002 continued to express CHT1 mRNA at a level that was ∼8-fold higher than that of GFP alone cells. Activation of CHT1 transcription was partially reduced in LY294002-treated wt-hChAT-GFP cells compared with DMSO alone-treated wt-hChAT-GFP cells. C, dose-response curves obtained by using real time RT-PCR analysis of CHT1 in wt-hChAT-GFP and GFP alone cells with 0 nm to 50 μm LY294002 in DMSO for 24 h. Squares indicate values of wt-hChAT cells, and triangles represent values of GFP alone cells. There is an approximate straight line relationship between the reduction of CHT1 induction in wt-hChAT-GFP and the dose of LY294002. y = −0.0536x + 2.7265, R2 = 0.8573.
DISCUSSION
Here we showed that overexpressed ChAT enhanced transcriptional activation of the CHT1 gene in human neuronal cell lines. This induction required nuclear translocation and the catalytic activity of ChAT. Our data are consistent with a previous paper by De Jaco et al. (35) who reported that an FB5 subclone of N18TG2 murine neuroblastoma cells in which a rat ChAT homologue was overexpressed showed a higher level of high affinity choline uptake compared with that in a ChAT-negative clone. Choline uptake, mediated by high affinity choline transporters, was assayed in that study rather than CHT1 expression because CHT1 had not yet been cloned at the time of the study. To our knowledge, the present study is the first report demonstrating that nuclear ChAT acts as a transcriptional activator of selected target genes.
The underlying mechanism by which CHT1 gene expression is regulated remains unresolved. The CHT1 gene is located on chromosome 2; 2q12 (GenBank accession number NC_000002; region 107969427.107996876), whereas CHAT and VACHT are encoded by the cholinergic gene locus on chromosome 10; 10q11.2 (36). Many reports have demonstrated that the two genes encoding CHAT and VACHT are coordinately regulated by extracellular stimuli such as NGF, retinoic acids (RAs), and cytokines of the bone morphogenetic protein family (37–40). Synergic up-regulation of CHT1 and ChAT is also observed when superior cervical ganglia are treated with leukemia inhibitory factor (41). It has been shown that simultaneous up-regulation of CHT1 and VACHT is induced when primary septal cells are stimulated with NGF (34). The effects of NGF on cholinergic gene expression are known to be dependent on PI3K signaling (42), and treatment with an inhibitor of PI3K inhibits the effect of NGF on CHT1 expression (34). Madziar et al. (43) also revealed that NGF regulates the expression of the cholinergic locus and CHT1 via the Akt/protein kinase B signaling pathway in pheochromocytoma 12 cells and mouse primary septal cells. On the other hand, it has been reported that regulation of CHT1 expression is independent of the cholinergic gene locus under some conditions. Treatment of superior cervical ganglia neurons with RA up-regulated ChAT expression but down-regulated CHT1 mRNA expression (41). Berse et al. (34) also reported that all-trans-RA treatment increased VACHT expression in cultured mouse septal neurons, but that CHT1 expression was unaffected. Brock et al. (44) showed a differential regulation of CHT1 and the cholinergic locus by cAMP signaling pathways in NSC-19 cells. Up-regulated CHT1 expression in vivo has been reported to compensate for cholinergic dysfunction due to ChAT haploinsufficiency in mutant mice (45). In the present report, we showed that ChAT itself could induce the expression of CHT1, suggesting that ChAT might act in a positive feedback loop to maintain cholinergic phenotypes during neuronal differentiation. Our results further suggest that activation of CHT1 transcription by ChAT depends on PI3K signaling. The reduction of ChAT-induced CHT1 transcription was significantly correlated with the concentration of LY294002 used for the treatment. A previous report showed that LY294002 treatment reduces endogenous CHT1 expression in mouse primary septal culture as well as NGF-induced CHT1 expression (34). Our observation of reduction in CHT expression by LY294002 appears to be consistent with the LY294002 effect on endogenous CHT1 expression. The expression of CHT1 in non-neuronal tissues has also been reported (46–51). However, because we did not detect this positive feedback loop in non-neuronal HEK293 cells, the regulation of CHT1 expression might differ between neuronal and non-neuronal cells. We speculated that one of the following two potential molecular mechanisms might underlie CHT1 induction by ChAT: 1) ChAT enhances the production of ACh, which acts on ACh receptors on the cell surface via an autocrine loop and results in induction of CHT1 expression, or 2) nuclear ChAT directly activates the transcriptional machinery important for CHT1 gene transcription.
Our result that enzymatically active but exclusively cytoplasmic NLS2-(−)-hChAT could not induce CHT1 expression indicated that the former mechanism may be implausible. The fact that wt-ChAT-GFP-M cells, which have the same level of enzymatic activity as NLS2-(−)-hChAT, induced CHT1 mRNA expression indicates that NLS2-(−)-hChAT may satisfy the conditions for the induction of CHT1 in terms of enzymatic activity. In the opposite situation, inactive nuclear hChAT also could not enhance CHT1 transcription at all. These results suggested that nuclear ChAT activity is required for up-regulation of CHT1 transcription, although we cannot exclude the possibility that Glu441 is critical for both ChAT activity and CHT1 transcriptional induction.
SH-SY5Y cells are reported to be expressing muscarinic receptors (32, 33). To clarify whether ACh receptors on the cell surface did not participate in the CHT1 mRNA induction by ChAT overexpression, we performed the pharmacologic study using a muscarinic antagonist and muscarinic agonists. Atropine up to 10 μm could not reverse the up-regulation of CHT1 expression by ChAT, and either pilocarpine or carbachol up to 1 mm did not enhance the induction of CHT1. These results indicate that cell surface ACh receptors do not modulate this induction.
If this is the case, then, how is it possible that nuclear ChAT activates CHT1 transcription? One possible mechanism is that ACh generated by ChAT acts via acetylcholine receptors existing inside of the cells. G-protein-coupled muscarinic ACh receptors have been detected in the nuclear envelope by use of specific substrate binding assays (52–54). Nicotinic ACh receptors have been also detected in the nuclear envelope using immunoelectron microscopy (55, 56). However, even if ACh receptors reside on the inner membrane of the nuclear envelope, ACh binding sites are predicted to face to the luminal side. Therefore, ACh cannot access these binding sites. ACh receptor-independent functions of ACh have not been described.
A second possibility is that the intranuclear ChAT may acetylate novel substrates other than choline in the nuclei. Possible acetylation of chromosome-associated proteins or transcriptional regulators could influence gene transcription. Another possible mechanism is that ChAT might directly bind to certain domains of genomic DNA and function directly as a transcriptional factor. This possibility is supported by data obtained by three-dimensional structure analysis, which suggested that the periodic basic patches along the ChAT surface ridges might interact with two turns of DNA strands (57). However, direct binding of ChAT to DNA has not been examined to date. We are currently planning to screen for ChAT-binding genomic domains using a chromatin immunoprecipitation assay.
A third possible explanation is that ChAT, overexpressed in the nucleus, may utilize significant amounts of nuclear acetyl-CoA, thus reducing its pool used by nuclear protein acetyltransferases. This could alter the acetylation state of key proteins (e.g. histones) and modulate gene transcription.
In the present study, we demonstrated a possible function of intranuclear ChAT. Our findings support the notion that ACh and/or ChAT may be a modulator of neuronal differentiation (58). We predict that ChAT might regulate the transcription of a selected gene population other than the CHT1 gene in neuronal and non-neuronal cells. Future studies are needed to elucidate the overall functions of ACh and/or ChAT, especially in cell bodies.
Supplementary Material
Acknowledgments
We thank T. Yamamoto, Y. Mori, and M. Suzaki (Central Research Laboratory Shiga University of Medical Science) for technical assistance.
This work was supported, in part, by a presidential Grant-in-aid from the Shiga University of Medical Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- ChAT
- choline acetyltransferase
- ACh
- acetylcholine
- NLS
- nuclear localization signal
- CHT1
- high affinity choline transporter
- VAChT
- vesicular acetylcholine transporter
- cChAT
- common type of ChAT
- pChAT
- peripheral ChAT
- hChAT
- human ChAT
- wt-hChAT
- wild-type hChAT.
REFERENCES
- 1. Ohno K., Tsujino A., Brengman J. M., Harper C. M., Bajzer Z., Udd B., Beyring R., Robb S., Kirkham F. J., Engel A. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oda Y. (1999) Pathol. Int. 49, 921–937 [DOI] [PubMed] [Google Scholar]
- 3. Oda Y., Nakanishi I., Deguchi T. (1992) Brain Res. Mol. Brain Res. 16, 287–294 [DOI] [PubMed] [Google Scholar]
- 4. Chireux M. A., Le Van Thai A., Weber M. J. (1995) J. Neurosci. Res. 40, 427–438 [DOI] [PubMed] [Google Scholar]
- 5. Misawa H., Matsuura J., Oda Y., Takahashi R., Deguchi T. (1997) Brain Res. Mol. Brain Res. 44, 323–333 [DOI] [PubMed] [Google Scholar]
- 6. Tooyama I., Kimura H. (2000) J. Chem. Neuroanat. 17, 217–226 [DOI] [PubMed] [Google Scholar]
- 7. Kuhar M. J., Murrin L. C. (1978) J. Neurochem. 30, 15–21 [DOI] [PubMed] [Google Scholar]
- 8. Simon J. R., Atweh S., Kuhar M. J. (1976) J. Neurochem. 26, 909–922 [DOI] [PubMed] [Google Scholar]
- 9. Ferguson S. M., Blakely R. D. (2004) Mol. Interv. 4, 22–37 [DOI] [PubMed] [Google Scholar]
- 10. Apparsundaram S., Ferguson S. M., George A. L., Jr., Blakely R. D. (2000) Biochem. Biophys. Res. Commun. 276, 862–867 [DOI] [PubMed] [Google Scholar]
- 11. Apparsundaram S., Ferguson S. M., Blakely R. D. (2001) Biochem. Soc. Trans. 29, 711–716 [DOI] [PubMed] [Google Scholar]
- 12. Okuda T., Haga T., Kanai Y., Endou H., Ishihara T., Katsura I. (2000) Nat. Neurosci. 3, 120–125 [DOI] [PubMed] [Google Scholar]
- 13. Okuda T., Haga T. (2000) FEBS Lett. 484, 92–97 [DOI] [PubMed] [Google Scholar]
- 14. Ribeiro F. M., Black S. A., Prado V. F., Rylett R. J., Ferguson S. S., Prado M. A. (2006) J. Neurochem. 97, 1–12 [DOI] [PubMed] [Google Scholar]
- 15. Wessler I., Kilbinger H., Bittinger F., Unger R., Kirkpatrick C. J. (2003) Life Sci. 72, 2055–2061 [DOI] [PubMed] [Google Scholar]
- 16. Gill S. K., Bhattacharya M., Ferguson S. S., Rylett R. J. (2003) J. Biol. Chem. 278, 20217–20224 [DOI] [PubMed] [Google Scholar]
- 17. Resendes M. C., Dobransky T., Ferguson S. S., Rylett R. J. (1999) J. Biol. Chem. 274, 19417–19421 [DOI] [PubMed] [Google Scholar]
- 18. Matsuo A., Bellier J. P., Hisano T., Aimi Y., Yasuhara O., Tooyama I., Saito N., Kimura H. (2005) Neurochem. Int. 46, 423–433 [DOI] [PubMed] [Google Scholar]
- 19. Gill S. K., Ishak M., Dobransky T., Haroutunian V., Davis K. L., Rylett R. J. (2007) Neurobiol. Aging 28, 1028–1040 [DOI] [PubMed] [Google Scholar]
- 20. Fonnum F. (1975) J. Neurochem. 24, 407–409 [DOI] [PubMed] [Google Scholar]
- 21. Bellier J. P., Kimura H. (2007) J. Neurochem. 101, 1607–1618 [DOI] [PubMed] [Google Scholar]
- 22. Kudo N., Wolff B., Sekimoto T., Schreiner E. P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. (1998) Exp. Cell Res. 242, 540–547 [DOI] [PubMed] [Google Scholar]
- 23. Vlahos C. J., Matter W. F., Hui K. Y., Brown R. F. (1994) J. Biol. Chem. 269, 5241–5248 [PubMed] [Google Scholar]
- 24. Nishimura M., Yu G., Levesque G., Zhang D. M., Ruel L., Chen F., Milman P., Holmes E., Liang Y., Kawarai T., Jo E., Supala A., Rogaeva E., Xu D. M., Janus C., Levesque L., Bi Q., Duthie M., Rozmahel R., Mattila K., Lannfelt L., Westaway D., Mount H. T., Woodgett J., St. George-Hyslop P. (1999) Nat. Med. 5, 164–169 [DOI] [PubMed] [Google Scholar]
- 25. Okuda T., Okamura M., Kaitsuka C., Haga T., Gurwitz D. (2002) J. Biol. Chem. 277, 45315–45322 [DOI] [PubMed] [Google Scholar]
- 26. Xie J., Guo Q. (2004) J. Biol. Chem. 279, 28266–28275 [DOI] [PubMed] [Google Scholar]
- 27. Ribeiro F. M., Alves-Silva J., Volknandt W., Martins-Silva C., Mahmud H., Wilhelm A., Gomez M. V., Rylett R. J., Ferguson S. S., Prado V. F., Prado M. A. (2003) J. Neurochem. 87, 136–146 [DOI] [PubMed] [Google Scholar]
- 28. Ribeiro F. M., Black S. A., Cregan S. P., Prado V. F., Prado M. A., Rylett R. J., Ferguson S. S. (2005) J. Neurochem. 94, 86–96 [DOI] [PubMed] [Google Scholar]
- 29. Ribeiro F. M., Pinthong M., Black S. A., Gordon A. C., Prado V. F., Prado M. A., Rylett R. J., Ferguson S. S. (2007) Eur. J. Neurosci. 26, 3437–3448 [DOI] [PubMed] [Google Scholar]
- 30. Kimura S., Bellier J. P., Matsuo A., Tooyama I., Kimura H. (2007) Neurochem. Int. 50, 251–255 [DOI] [PubMed] [Google Scholar]
- 31. Carbini L. A., Hersh L. B. (1993) J. Neurochem. 61, 247–253 [DOI] [PubMed] [Google Scholar]
- 32. Lambert D. G., Ghataorre A. S., Nahorski S. R. (1989) Eur. J. Pharmacol. 165, 71–77 [DOI] [PubMed] [Google Scholar]
- 33. Adem A., Mattsson M. E., Nordberg A., Påhlman S. (1987) Brain Res. 430, 235–242 [DOI] [PubMed] [Google Scholar]
- 34. Berse B., Szczecinska W., Lopez-Coviella I., Madziar B., Zemelko V., Kaminski R., Kozar K., Lips K. S., Pfeil U., Blusztajn J. K. (2005) Brain Res. Dev. Brain Res. 157, 132–140 [DOI] [PubMed] [Google Scholar]
- 35. De Jaco A., Ajmone-Cat M. A., Baldelli P., Carbone E., Augusti-Tocco G., Biagioni S. (2000) J. Neurochem. 75, 1123–1131 [DOI] [PubMed] [Google Scholar]
- 36. Erickson J. D., Varoqui H., Schäfer M. K., Modi W., Diebler M. F., Weihe E., Rand J., Eiden L. E., Bonner T. I., Usdin T. B. (1994) J. Biol. Chem. 269, 21929–21932 [PubMed] [Google Scholar]
- 37. Berrard S., Varoqui H., Cervini R., Israël M., Mallet J., Diebler M. F. (1995) J. Neurochem. 65, 939–942 [DOI] [PubMed] [Google Scholar]
- 38. Berse B., Blusztajn J. K. (1995) J. Biol. Chem. 270, 22101–22104 [DOI] [PubMed] [Google Scholar]
- 39. Berse B., Lopez-Coviella I., Blusztajn J. K. (1999) Biochem. J. 342, 301–308 [PMC free article] [PubMed] [Google Scholar]
- 40. López-Coviella I., Berse B., Krauss R., Thies R. S., Blusztajn J. K. (2000) Science 289, 313–316 [DOI] [PubMed] [Google Scholar]
- 41. Lecomte M. J., De Gois S., Guerci A., Ravassard P., Faucon Biguet N., Mallet J., Berrard S. (2005) Mol. Cell. Neurosci. 28, 303–313 [DOI] [PubMed] [Google Scholar]
- 42. Madziar B., Lopez-Coviella I., Zemelko V., Berse B. (2005) J. Neurochem. 92, 767–779 [DOI] [PubMed] [Google Scholar]
- 43. Madziar B., Shah S., Brock M., Burke R., Lopez-Coviella I., Nickel A. C., Cakal E. B., Blusztajn J. K., Berse B. (2008) J. Neurochem. 107, 1284–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brock M., Nickel A. C., Madziar B., Blusztajn J. K., Berse B. (2007) Brain Res. 1145, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brandon E. P., Mellott T., Pizzo D. P., Coufal N., D'Amour K. A., Gobeske K., Lortie M., López-Coviella I., Berse B., Thal L. J., Gage F. H., Blusztajn J. K. (2004) J. Neurosci. 24, 5459–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujii T., Okuda T., Haga T., Kawashima K. (2003) Life Sci. 72, 2131–2134 [DOI] [PubMed] [Google Scholar]
- 47. Haberberger R. V., Pfeil U., Lips K. S., Kummer W. (2002) J. Invest. Dermatol. 119, 943–948 [DOI] [PubMed] [Google Scholar]
- 48. Hanna-Mitchell A. T., Beckel J. M., Barbadora S., Kanai A. J., de Groat W. C., Birder L. A. (2007) Life Sci. 80, 2298–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lips K. S., Pfeil U., Haberberger R. V., Kummer W. (2002) Cell Tissue Res. 307, 275–280 [DOI] [PubMed] [Google Scholar]
- 50. Lips K. S., Pfeil U., Reiners K., Rimasch C., Kuchelmeister K., Braun-Dullaeus R. C., Haberberger R. V., Schmidt R., Kummer W. (2003) J. Histochem. Cytochem. 51, 1645–1654 [DOI] [PubMed] [Google Scholar]
- 51. Pfeil U., Haberberger R. V., Lips K. S., Eberling L., Grau V., Kummer W. (2003) Life Sci. 72, 2087–2090 [DOI] [PubMed] [Google Scholar]
- 52. Lind G. J., Cavanagh H. D. (1993) Invest. Ophthalmol. Vis. Sci. 34, 2943–2952 [PubMed] [Google Scholar]
- 53. Lind G. J., Cavanagh H. D. (1995) Invest. Ophthalmol. Vis. Sci. 36, 1492–1507 [PubMed] [Google Scholar]
- 54. Gobeil F., Fortier A., Zhu T., Bossolasco M., Leduc M., Grandbois M., Heveker N., Bkaily G., Chemtob S., Barbaz D. (2006) Can. J. Physiol. Pharmacol. 84, 287–297 [DOI] [PubMed] [Google Scholar]
- 55. Okuda H., Shioda S., Nakai Y., Nakayama H., Okamoto M., Nakashima T. (1993) Brain Res. 625, 145–151 [DOI] [PubMed] [Google Scholar]
- 56. Jacob M. H., Lindstrom J. M., Berg D. K. (1986) J. Cell Biol. 103, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cai Y., Cronin C. N., Engel A. G., Ohno K., Hersh L. B., Rodgers D. W. (2004) EMBO J. 23, 2047–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Augusti-Tocco G., Biagioni S., Tata A. M. (2006) J. Mol. Neurosci. 30, 45–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.