Abstract
Poly(ADP-ribose) polymerase (PARP) knockout mice are resistant to murine models of human diseases such as cerebral and myocardial ischemia, traumatic brain injury, diabetes, Parkinsonism, endotoxic shock and arthritis, implicating PARP in the pathogenesis of these diseases. Potent selective PARP inhibitors are therefore being evaluated as novel therapeutic agents in the treatment of these diseases. Inhibition or depletion of PARP, however, increases genomic instability in cells exposed to genotoxic agents. We recently demonstrated the presence of a genomically unstable tetraploid population in PARP–/– fibroblasts and its loss after stable transfection with PARP cDNA. To elucidate whether the genomic instability is attributable to PARP deficiency or lack of PARP activity, we investigated the effects of PARP inhibition on development of tetraploidy. Immortalized wild-type and PARP–/– fibroblasts were exposed for 3 weeks to 20 µM GPI 6150 (1,11b-dihydro-[2H]benzopyrano[4,3,2-de]isoquinolin-3-one), a novel small molecule specific competitive inhibitor of PARP (Ki = 60 nM) and one of the most potent PARP inhibitors to date (IC50 = 0.15 µM). Although GPI 6150 initially decreased cell growth in wild-type cells, there was no effect on cell growth or viability after 24 h. GPI 6150 inhibited endogenous PARP activity in wild-type cells by ∼91%, to about the residual levels in PARP–/– cells. Flow cytometric analysis of unsynchronized wild-type cells exposed for 3 weeks to GPI 6150 did not induce the development of tetraploidy, suggesting that, aside from its catalytic function, PARP may play other essential roles in the maintenance of genomic stability.
INTRODUCTION
PARP–/– mice, with a homozygous disruption of the poly(ADP-ribose) polymerase (PARP) gene, do not express any PARP detectable by immunoblot analysis (1,2). They suffer far less tissue injury in murine models of a number of human diseases, including focal cerebral ischemia (3,4), traumatic brain injury (5), myocardial ischemia (6,7), streptozocin-induced diabetes (8–10), 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced Parkinsonism (11), endotoxic shock (12,13) and peroxynitrite- and dextran sulfate-induced inflammation (14,15), suggesting that PARP activation, triggered by oxidative or nitric oxide-induced stress, plays a role in the pathophysiology of these diseases. Thus, potent and specific inhibitors of PARP are now being developed as novel therapeutic agents in the treatment of these diseases. For acute treatment, PARP inhibitors have been shown to diminish brain damage after cerebral focal ischemia (4,16–21), protect cardial myocytes in regional heart ischemia (6,7,22), reduce renal ischemia–reperfusion injury (23) and decrease neuronal injury in a head trauma model (24). The onset of some diseases, such as the development of hyperglycemia in streptozocin-induced type I diabetes, is delayed in PARP+/– mice (8), suggesting that even partial inhibition of PARP can provide therapeutic benefits and that treatment with potent PARP inhibitors at the time of diagnosis may, thus, prevent or slow disease progression. Even weak PARP inhibitors, such as nicotinamide, have shown some limited beneficial effects in clinical trials for the treatment of human diabetes (25,26). Most PARP inhibitors, including the benzamide and isoquinoline families, however, exhibit low potency and specificity; thus, in the search for more specific, potent, small molecule PARP inhibitors, we have identified GPI 6150 (1,11b-dihydro-[2H]benzopyrano[4,3,2-de]isoquinolin-3-one), which has recently demonstrated remarkable efficacy in reducing tissue damage in animal models of human diseases (7,20,24,27) such as cerebral and myocardial ischemia, traumatic brain injury, arthritis, septic shock, type I diabetes and Parkinsonism. GPI 6150 is a specific competitive inhibitor of PARP (Ki = 60 nM), as revealed by enzyme kinetic analysis, and has no effect on other nicotinamide adenine dinucleotide (NAD)-utilizing enzymes (28).
On the other hand, PARP is also implicated in the maintenance of genomic stability and inhibition of PARP by chemical inhibitors or by dominant negative mutants, or depletion of PARP by antisense RNA expression have been shown to increase the frequency of recombination, gene amplification, sister chromatid exchanges (SCE) and micronucleus formation, markers of genomic instability, in cells exposed to genotoxic agents (29–35). Although exhibiting varying phenotypes, two strains of PARP knockout mice developed by different laboratories both exhibit increased genomic instability, as indicated by elevated frequencies of SCE and micronucleus formation after treatment with DNA-damaging agents, providing further support for a role for PARP in the maintenance of genomic integrity (1,2,36). Development of tetraploidy or aneuploidy—markers of genomic instability in cells—is typical of many tumors and is associated with progression to malignancy or metastasis (37). By flow cytometric (FACS) analysis, we recently demonstrated the presence of an unstable tetraploid population among immortalized fibroblasts derived from PARP–/– mice (38). Comparative genomic hybridization further detected partial chromosomal gains in 4C5-ter, 5F-ter and 14A1-C1 in PARP–/– mice and immortalized PARP–/– fibroblasts. These chromosomal gains and the tetraploid population were not seen in PARP–/– cells stably transfected with PARP cDNA [PARP–/–(+PARP)], indicating negative selection of cells with these genetic aberrations after reintroduction of PARP cDNA. PARP deficiency has also been associated with an increased frequency of chromosome fusions and aneuploidy (39). However, it remains to be clarified whether the genomic instability resulting from disruption of the PARP gene may be attributed to the deficiency in PARP protein itself or to a lack of significant PARP activity. Thus, it is of interest, particularly to laboratories developing potent PARP inhibitors as therapeutic agents, to test whether pharmacological inhibition of PARP, at least during the duration of acute treatment intended for initial clinical use, could increase genomic instability, as indicated for example by development of tetraploidy. Accordingly, in the present study immortalized fibroblasts derived from PARP+/+ and PARP–/– mice were continuously grown and subcultured in the presence of 20 µM GPI 6150 or with vehicle alone (dimethyl sulfoxide, DMSO) for a period of 3 weeks. At the end of weeks 1, 2 and 3, FACS analysis was performed to detect the presence of mixed ploidy or a genomically unstable tetraploid population in GPI 6150-treated PARP+/+ cells.
MATERIALS AND METHODS
GPI 6150
After a combination of high-throughput screening of chemical libraries and structure-based drug design to identify novel small molecule PARP inhibitors, GPI 6150 or 1,11b-dihydro-[2H]benzopyrano[4,3,2-de]isoquinolin-3-one (Guilford Pharmaceuticals Inc., Baltimore, MD) was synthesized and evaluated. One of the tetracyclic compounds in a series of PARP inhibitors, GPI 6150, is a specific competitive inhibitor of PARP (Ki = 60 nM) and has recently provided protection from tissue damage in several animal models of human diseases (7,20,24,27) such as cerebral and myocardial ischemia, traumatic brain injury, arthritis, septic shock, type I diabetes and Parkinsonism. GPI 6150, with a molecular mass of 237, was solubilized in 100% DMSO and added to the cells at a final concentration of 20 µM.
Cell lines
Homozygous PARP–/– mice, generated by disrupting exon 2 of the PARP gene by homologous recombination (1), and wild-type (PARP+/+) littermates (strain 129/Sv × C57BL/6, female) were used to derive wild-type (PARP+/+ clone A19) and PARP–/– (clone A1) fibroblasts, which were immortalized spontaneously by a standard 3T3 protocol (1) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml).
Immunoblot analysis
SDS–PAGE and transfer of proteins to nitrocelluose membranes were performed according to standard procedures. Membranes were stained with Ponceau S (0.1%) to confirm equal loading and transfer of proteins and then incubated with antibodies to PARP (1:2000 dilution; BioMol) and poly(ADP-ribose) (PAR) (1:250 dilution; 10HA). Immune complexes were detected by enhanced chemiluminescence (Pierce) after incu-bation with appropriate horseradish peroxidase-conjugated antibodies to mouse or rabbit IgG (1:3000 dilution).
Reverse transcription–polymerase chain reaction (RT–PCR)
Unique oligonucleotide primer pairs for mouse PARP (mPARP) were designed and prepared. Total RNA, purified from cell pellets with an RNA extraction kit (Pharmacia Biotech), was subjected to RT–PCR using oligonucleotide primer pairs for mPARP and a Perkin Elmer Gene Amp EZ tTh RNA PCR kit. The reaction mix (50 µl) contained 300 µM of each dGTP, dATP, dTTP and dCTP, 0.45 µM of each primer, 1 µg total RNA and 5 U rTth DNA polymerase. RNA was transcribed at 65°C for 40 min and DNA was amplified by an initial incubation at 95°C for 2 min, followed by 40 cycles at 95°C for 1 min, 60°C for 1.5 min and 65°C for 0.5 min, with a final extension at 70°C for 22 min. The PCR products were then separated by electrophoresis in a 1.5% agarose gel and visualized by ethidium bromide staining.
PARP activity assays
PARP+/+ fibroblasts were harvested and washed with ice-cold phosphate-buffered saline (PBS). Cell extracts were then derived and subjected to enzyme assays to measure PARP activity in the presence of various concentrations of GPI 6150 in order to determine the IC50 for this potent PARP inhibitor. For PARP activity assays incorporation of [32P]NAD in vitro into acid-insoluble acceptors was measured at 25°C for 1 min, with 30 µg protein per determination and triplicate determinations per treatment, as described previously (40). PARP+/+ and PARP–/– fibroblasts, grown continuously in the presence of 20 µM GPI 6150 or with vehicle alone, were harvested at days 1, 2, 3 and 4 (prior to passaging) and washed extensively with ice-cold PBS. Equal amounts of cell extracts were then derived and subjected to PARP activity assays to confirm inhibition of endogenous PARP activity in the GPI 6150-treated cells. Given that GPI 6150, a reversible inhibitor, forms a stable enzyme–inhibitor complex with PARP that lasts up to 2 h (28), the cell extracts were prepared and subjected to PARP activity assays in less than 30 min, during which ∼90% of the GPI 6150 is expected to be in the enzyme–inhibitor complex.
Flow cytometry
Nuclei were prepared for FACS analysis as described previously (41). Cells were exposed to trypsin and resuspended in 100 µl of a solution containing 250 mM sucrose, 40 mM sodium citrate (pH 7.6) and 5% (v/v) DMSO. The cells were lysed for 10 min in a solution containing 3.4 mM sodium citrate, 0.1% (v/v) NP-40, 1.5 mM spermine tetrahydrochloride and 0.5 mM Tris–HCl (pH 7.6). After incubation of lysates for 10 min with RNase A (0.1 mg/ml), nuclei were stained for 15 min with propidium iodide (0.42 mg/ml), filtered through a 37 µm nylon mesh and analyzed with a dual laser flow cytometer (FACScan; Becton Dickinson).
RESULTS
GPI 6150 is a potent PARP inhibitor with an IC50 of 0.15 µM
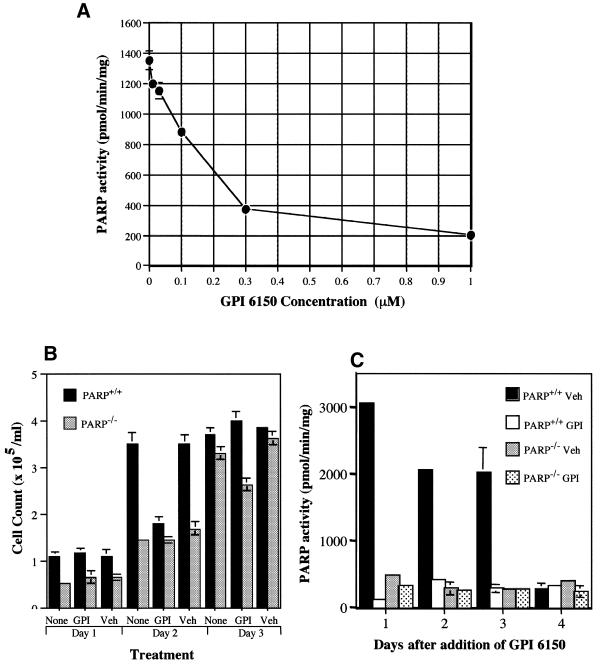
We first confirmed the lack of immunoreactive PARP in immortalized fibroblasts derived from PARP knockout mice (clone A1) and its presence and activity in wild-type (PARP+/+) cells (clone A19) by immunoblot analysis with antibodies to PARP and PAR (Fig. 1A). As expected, RT–PCR analysis detected mPARP transcripts in wild-type but not in PARP–/– cells (Fig. 1B). To determine the IC50 for GPI 6150 under our laboratory conditions, PARP+/+ fibroblasts were harvested, washed with ice-cold PBS and cell extracts were derived and subjected to enzyme assays to measure PARP activity in the presence of various concentrations of GPI 6150. Although the basal levels of PARP activity in the PARP+/+ fibroblasts were not induced by any DNA-damaging agent exogenously applied to the cells, the in vitro reaction included equal amounts of nicked DNA (activated calf thymus DNA) needed to activate PARP. PARP activity assays, performed by measurement of [32P]NAD incorporation in vitro into acid-insoluble acceptors at 25°C for 1 min, showed that GPI 6150 inhibited PARP activity by 50% at a concentration of 0.15 µM (IC50) (Fig. 2A). Thus, GPI 6150 is more potent than most PARP inhibitors, including 4-amino-1,8-napthalimide, phenanthridinones and dihydroxyisoquinoline, which have reported IC50 values of 0.18, 0.35 and 0.30 µM, respectively (42). Its 50% inhibitory concentration is about two orders of magnitude lower than the most commonly used PARP inhibitors 3-aminobenzamide (33 µM) and benzamide (22 µM) (42).
Figure 1.

PARP expression in immortalized wild-type and PARP–/– fibroblasts. (A) Cell extracts of wild-type and PARP–/– fibroblasts (30 µg protein) were subjected to immunoblot analysis with antibodies to PARP (upper) and PAR (middle). The blot was stained with Ponceau S to verify equal loading and transfer of proteins in both lanes (lower). (B) RT–PCR was performed with specific primers for the mPARP mRNA. The positions of PARP and PAR are indicated.
Figure 2.
Determination of the IC50 for GPI 6150 (A), effects of GPI 6150 on cell growth (B) and endogenous PARP activity (C) of wild-type and PARP–/– fibroblasts. (A) PARP+/+ cells were washed with ice-cold PBS, cell extracts were derived and equal amounts of protein (30 µg) were subjected to PARP activity assays in the presence of various concentrations of GPI 6150. PARP activity assays were performed by measurement of [32P]NAD incorporation in vitro into acid-insoluble acceptors at 25°C for 1 min, with triplicate determinations per treatment. (B) Cells were grown in the presence of 20 µM GPI 6150 or with vehicle alone (DMSO) for 3 weeks, during which GPI 6150 was replenished when the culture medium was changed every other day and when the cells were passaged once a week. During the first week, effects of GPI 6150 on cell viability and cell growth were determined by cell counts with Trypan blue staining every 24 h for the 3 days of exposure to GPI 6150 and prior to passaging the cells on day 4. (C) Inhibition of endogenous PARP activity in the wild-type cells exposed to 20 µM GPI 6150 at the indicated times was also verified by PARP activity assays. Equal amounts of total cellular protein (30 µg) were subjected to PARP activity assays in triplicate determinations. All the data are presented as means ± SD of three replicates of a representative experiment; essentially the same results were obtained in three independent experiments.
Inhibition of endogenous PARP activity by GPI 6150 and effects on cell growth and viability in immortalized wild-type and PARP–/– cells
To investigate whether pharmacological inhibition of PARP by GPI 6150, at least for the duration of treatment intended for initial acute clinical indication, could suffice to induce development of tetraploidy in cells, immortalized wild-type and PARP–/– fibroblasts were continuously grown and subcultured in the presence of 20 µM GPI 6150 or with vehicle alone (DMSO) for 3 weeks. Given the stable chemical structure and relatively long half-life of GPI 6150, the PARP inhibitor was replenished when the culture medium was changed every other day and when the cells were passaged once a week.
To first confirm that treatment of wild-type cells with GPI 6150 (20 µM) does not affect cell viability or cell growth during the period of treatment, cell counts were performed after Trypan blue staining every 24 h for the first 3 days of exposure to GPI 6150, prior to passaging at day 4. Figure 2B is a representative experiment showing cell counts in the presence of GPI 6150 or vehicle during the first 3 days (of the total 3 week period), prior to the first passaging. The cell counts for the next 3 days after passaging at day 4 and every 3 days thereafter were essentially the same as those shown in Figure 2B. As expected, the growth rate (rate of cell doubling) of PARP–/– cells was significantly lower than that of wild-type cells (Fig. 2B). This is consistent with previous studies showing that fibroblasts derived from PARP knockout mice exhibit proliferation deficiencies in culture (1,43). Furthermore, we have also shown that PARP is a core component of the multiprotein DNA replication complex (44) and plays an essential role in DNA replication associated with re-entry into the cell cycle after release from serum starvation or aphidicolin block (45,46). Although treatment of wild-type cells with GPI 6150 for 24 h resulted in an initial decrease in the rate of cell doubling to ∼50% of that in vehicle-treated cells, there was no difference in the rate of cell growth between the GPI 6150-treated cells and those treated with vehicle alone at 48 h and thereafter (Fig. 2B). There was also no significant increase in cell death, as indicated by increased internalization of Trypan blue, in the GPI 6150-treated PARP–/– or wild-type cells (data not shown).
To further verify inhibition of endogenous PARP activity in wild-type cells exposed to 20 µM GPI 6150 during the 3 week period of treatment, PARP activity assays were performed every day for the first 3 days of exposure to GPI 6150 and at day 4, when the cells were passaged. Cells were harvested and extensively washed with ice-cold PBS to remove traces of extracellular GPI 6150. Cell extracts were then derived and equal amounts of total cellular protein (30 µg) were subjected to PARP activity assays by incorporation of [32P]NAD into acid-insoluble acceptors as described in Materials and Methods. As expected, exposure to GPI 6150 resulted in ∼91% inhibition of endogenous PARP activity in the wild-type cells every day during the first 3 day exposure to the drug prior to passaging at day 4 (Fig. 2C). At day 4 endogenous PARP activity is expectedly low even in wild-type cells, presumably because the cells are reaching confluency. Consistent with previous reports of PAR-synthesizing activity in PARP–/– mice and cells, presumably due to PARP homologs such as PARP-2 (47,48) or sPARP (49), such a residual activity was also noted in the PARP–/– cells, which is ∼9–15% of that in wild-type cells (Fig. 2C). GPI 6150 also slightly inhibited this residual activity. This residual activity in PARP–/– cells has not been shown to modify proteins aside from itself, thus, it may not fully compensate for PARP depletion (47,48). Nevertheless, GPI 6150 inhibited endogenous PARP activity in wild-type cells to approximately the basal levels noted in PARP–/– cells.
Complete inhibition of PARP activity by GPI 6150 was also confirmed in intact cells, aside from cell extracts, by induction of Fas-mediated apoptosis in PARP+/+ fibroblasts in the presence of 20 µM GPI 6150 or with vehicle alone. Transient poly(ADP-ribosyl)ation of nuclear proteins occurs early during apoptosis induced by serum deprivation, camptothecin or antibodies to Fas in different cell lines (50–53). Immunoblot analysis with anti-PAR of cell extracts derived at various times after induction of apoptosis revealed that in the presence of vehicle alone a transient peak of poly(ADP-ribosyl)ation occurred 8 h after induction, which subsequently declined (Fig. 3), concomitant with the onset of caspase 3-mediated PARP cleavage (data not shown). In contrast, in the presence of GPI 6150 this early PARP activation was completely blocked, verifying complete inhibition of PARP activity in vivo by GPI 6150 (Fig. 3). The specificity of the anti-PAR antibody used in these experiments has been confirmed in experiments showing that removal of PAR from immunoblots by phosphodiesterase treatment eliminates the polymer signal (44).
Figure 3.

Complete inhibition of transient poly(ADP-ribosyl)ation of nuclear proteins during Fas-mediated apoptosis in wild-type fibroblasts by GPI 6150. Immortalized wild-type fibroblasts were exposed to anti-Fas (50 ng/ml) and cycloheximide (10 µg/ml) for the indicated times in the presence of 20 µM GPI 6150 or with vehicle alone. Equal amounts of total cellular protein (30 µg) were then subjected to immunoblot analysis with monoclonal antibodies to PAR. The positions of molecular size standards (in kDa) are indicated.
Short-term inhibition of PARP activity by GPI 6150 does not suffice to induce tetraploidy in immortalized PARP+/+ cells
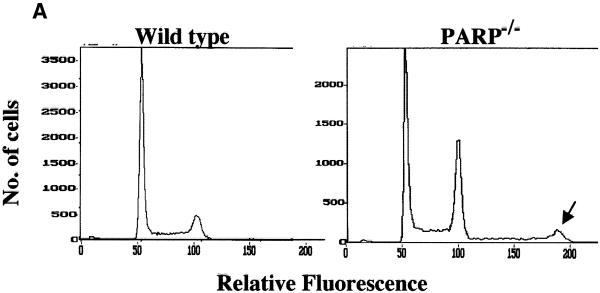
Tetraploidy is caused by the absence of either chromosome segregation or cytokinesis when cells exit from mitosis; such cells are genetically unstable and become aneuploid at subsequent mitoses (54). FACS analysis of unsynchronized immortalized PARP–/– (clone A1) cells revealed an unstable tetraploid population of cells (Fig. 4A). Whereas DNA histograms of wild-type cells (clone A19) (left panel) showed two major peaks of nuclei in the G0/G1 (haploid) and G2/M (diploid) phases of the cell cycle, unsynchronized PARP–/– cells (clone A1) (right panel) showed a third peak corresponding to the G2/M peak of an unstable tetraploid cell population in these cells, in addition to the two major peaks. This tetraploid population was also observed when PARP–/– cells were synchronized by either serum deprivation or aphidicolin and released into S phase (38). Similar results were also obtained with primary fibroblasts derived from PARP–/– and wild-type mice (55). Thus, the loss of PARP in diploid cells results in formation of unstable tetraploid cells predisposed to chromosome segregation abnormalities
Figure 4.
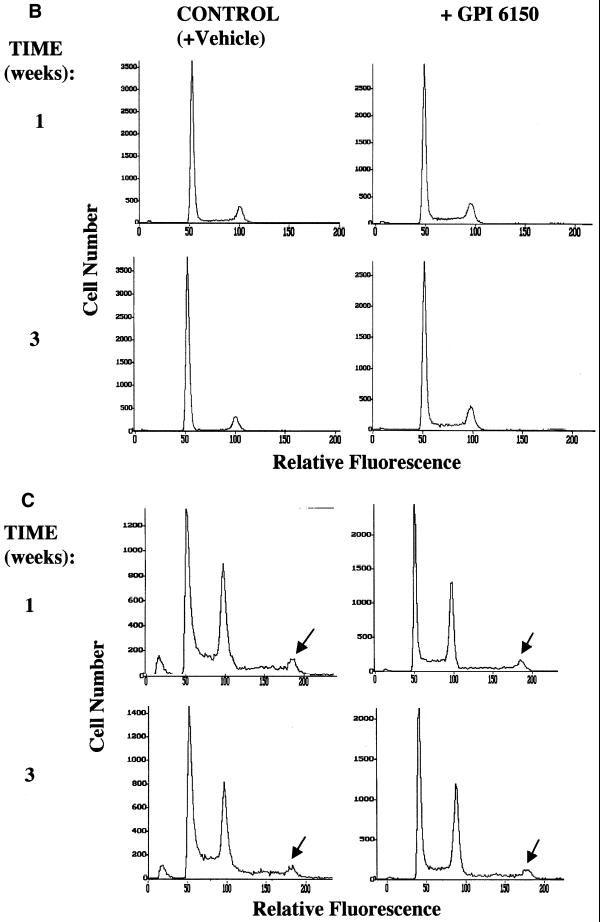
(Above and opposite) Effects of inhibition of PARP activity by GPI 6150 on the development of tetraploidy in immortalized wild-type and PARP–/– fibroblasts. FACS analysis was performed using unsynchronized immortalized wild-type (left) and PARP–/– (right) fibroblasts (A). Cells were harvested and nuclei were prepared and stained with propidium iodide for FACS analysis. In addition to the two major peaks of nuclei at G0/G1 and G2/M apparent in the DNA histograms of unsynchronized wild-type cells, the DNA histograms of PARP–/– cells exhibit a third peak corresponding to the G2/M peak of an unstable tetraploid cell population (arrows). FACS analysis was performed after continuous exposure of wild-type (B) or PARP–/– (C) fibroblasts to GPI 6150 (20 µM) or to vehicle (DMSO) alone for 1, 2 or 3 weeks to detect the presence of an unstable tetraploid population in the GPI 6150-treated wild-type cells. The G2/M peak of the unstable tetraploid cell population in the DNA histograms of PARP–/– cells is indicated by arrows. Essentially identical results were obtained in three independent experiments.
Previous studies have shown that under certain conditions, such as during DMBA-induced carcinogenesis in hamster keratinocytes (56), a significant increase in karyotypes showing tetraploidy or near-tetraploidy occurs after 2 weeks treatment. We next investigated whether development of the unstable population of tetraploid cells in immortalized PARP–/– fibroblasts can be induced by a lack of PARP activity for a 3 week period in wild-type cells. FACS analysis was performed after continuous exposure of cells to GPI 6150 for 1, 2 and 3 weeks to detect the presence of mixed ploidy or a tetraploid population in the GPI 6150-treated wild-type cells. FACS analysis of wild-type cells grown for 1, 2 or 3 weeks in the presence of 20 µM GPI 6150 or with vehicle (DMSO) alone did not show induced development of any tetraploidy in the cells, which retained a typical pattern of two major peaks of nuclei in the G0/G1 (haploid) and G2/M (diploid) phases of the cell cycle after 3 weeks (Fig. 4B). As expected, exposure of immortalized PARP–/– fibroblasts to 20 µM GPI 6150 or to vehicle alone for the same time period had no effect on the presence of the unstable tetraploid population in these cells; DNA histograms of unsynchronized PARP–/– cells retained the third peak corresponding to the G2/M peak of an unstable tetraploid cell population after 3 weeks exposure to GPI 6150 (Fig. 4C). Thus, short-term inhibition of PARP activity by GPI 6150 in PARP+/+ cells is not sufficient to induce the development of tetraploidy.
DISCUSSION
The present study demonstrates that the short-term pharmacological inhibition of PARP by GPI 6150, a novel potent competitive inhibitor of PARP, at least during the period of acute treatment intended for initial clinical use, does not suffice to induce genetic instability, as indicated by development of tetraploidy in cells exposed to this potent PARP inhibitor. It is possible that inhibition of PARP activity with GPI 6150 for a longer period of time may be necessary to induce the genetic alterations necessary for development of tetraploidy in wild-type cells, as in the case of life-time depletion of PARP in PARP knockout mice. Alternatively, these results suggest that PARP may play other essential roles in the maintenance of genomic stability, aside from its catalytic function. For example, immunoprecipitation experiments have shown that PARP physically associates with different nuclear proteins, such as DNA polymerase α (40,57,58), topoisomerase I (59) and p53 (60). The association of PARP and DNA polymerase α requires the PARP DNA-binding domain and occurs during the S and G2 phases of the cell cycle (58). Physical association with other proteins may represent a mechanism by which PARP can recruit certain proteins, such as PCNA and topoisomerase I, into the DNA replication/repair complex (45). PARP also physically associates with the base excision repair (BER) protein XRCC1 via the central XRCC domain, which contains a BRCT module, and XRCC1 interacts with ligase III via this BRCT module (61). PARP is therefore suggested to be a member of the BER multiprotein complex and to play a role in recruitment of XRCC1 and its partners to DNA breaks. This may explain why immortalized PARP–/– cells show severe defects in DNA BER, as indicated by a delay in rejoining of DNA strand breaks after exposure to genotoxic agents (43).
PARP may confer genetic stability via its putative role in p53 induction, accumulation and stabilization. p53 is implicated in the maintenance of diploidy as a component of the spindle checkpoint (62) and by regulating centrosome duplication (63). Given that the loss of p53 from diploid cells promotes the survival of cells with severe DNA damage and the development of tetraploidy (62,64,65), the presence of a tetraploid population among the immortalized PARP–/– cells is consistent with a lack of immunoreactive p53 in these cells (38). Interestingly, loss of the tetraploid population in PARP–/– cells stably transfected with PARP cDNA further correlates with partial restoration of p53 expression in these cells (38). Both PARP activity and p53 accumulation are induced by DNA damage and both proteins have been implicated as sensors of such damage. A functional association of PARP and p53 has been suggested by immunoprecipitation experiments (60); however, whether this interaction between the two proteins plays a role in p53 function or in p53 accumulation and stabilization remains to be clarified.
Accumulating evidence has also recently shown that PARP and poly(ADP-ribosyl)ation play dual roles in different nuclear processes, depending on the levels of the substrate NAD and the presence of PARP-activating DNA breaks. Thus, physical interaction of PARP with DNA polymerase α in the absence of NAD activates polymerase α (57), while addition of NAD to the DNA replication complex inhibits polymerase α catalytic activity (45). Similarly, in the absence of NAD, PARP interacts with different transcription factors to enhance activator-dependent transcription, while the presence of NAD and consequent PARP activation represses transcription, presumably by poly(ADP-ribosyl)ation of certain transcription factors (66). PARP in the absence of NAD enhances activator-dependent transcription by interacting with RNA polymerase II-associated factors (66). Similarly, PARP binds transcription enhancer factor 1 to enhance muscle-specific gene transcription (67), as well as transcription factor AP-2 to co-activate AP-2-mediated transcription (68). PARP-dependent silencing of transcription involves poly(ADP-ribosyl)ation of a number of transcription factors, which prevents the formation of active transcription complexes; modification of these proteins prevents binding to their respective DNA consensus sequences (69). Further work is clearly necessary to elucidate other molecular mechanisms by which PARP, aside from its catalytic activity, plays a role in the maintenance of genomic integrity.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Z. Q. Wang (IARC, France) for the PARP+/– and PARP–/– fibroblasts, and Dr Owen Blair (Cell Cycle Core Facility Cancer Center) for help with the FACS analysis. This work was supported in part by a gift from Guilford Pharmaceuticals Inc., grants CA25344 and 1P01 CA74175 from the National Cancer Institute, by the US Air Force Office of Scientific Research (grant AFOSR-89-0053) and by the US Army Medical Research and Development Command (contract DAMD17-90-C-0053 to M.E.S. and DAMD 17-96-C-6065 to D.S.R).
References
- 1.Wang Z.Q., Auer,B., Stingl,L., Berghammer,H., Haidacher,D., Schweiger,M. and Wagner,E.F. (1995) Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev., 9, 509–520. [DOI] [PubMed] [Google Scholar]
- 2.de Murcia J., Niedergang,C., Trucco,C., Ricoul,M., Dutrillaux,B., Mark,M., Oliver,J., Masson,M., Dierich,A., LeMeur,M., Waltzinger,C., Chambon,P. and de Murcia,G. (1997) Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and men. Proc. Natl Acad. Sci. USA, 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eliasson M., Sampei,K., Mandir,A., Hurn,P., Traystman,R., Bao,J., Pieper,A., Wang,Z.Q., Dawson,T., Snyder,S. and Dawson,V. (1997) Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nature Med., 3, 1089–1095. [DOI] [PubMed] [Google Scholar]
- 4.Endres M., Wang,Z.Q., Namura,S., Waeber,C. and Moskowitz,M. (1997) Ischemic brain injury is mediated by the activation of poly(ADP-ribose) polymerase. J. Cereb. Blood Flow Metab., 17, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 5.Whalen M., Clark,R., Dixon,C., Robichaud,P., Marion,D., Vagni,V., Graham,S., Virag,L., Hasko,G., Stachlewitz,R., Szabo,C. and Kochanek,P. (1999) Reduction of cognitive and motor deficits after traumatic brain injury in mice deficient in poly(ADP-ribose) polymerase. J. Cereb. Blood Flow Metab., 19, 835–842. [DOI] [PubMed] [Google Scholar]
- 6.Zingarelli B., Salzman,A. and Szabo,C. (1998) Genetic disruption of poly(ADP-ribose) synthetase inhibits the expression of P-selectin and intercellular adhesion molecule-1 in myocardial ischemia/reperfusion injury. Circ. Res., 83, 85–94. [DOI] [PubMed] [Google Scholar]
- 7.Walles T., Wang,P., Pieper,A., Li,J.-H., Zhang,J., Snyder,S. and Zweier,J. (1998) Demonstration that poly(ADP-ribose) accumulation occurs in the postischemic heart and is associated with myocardial necrosis. Circulation, Suppl. 98, 260. [Google Scholar]
- 8.Pieper A., Brat,D., Krug,D., Watkin,C., Gupta,S., Blackshaw,S., Verma,A., Wang,Z.Q. and Snyder,S. (1999) Poly(ADP-ribose) polymerase-deficient mice are protected from streptozocin-induced diabetes. Proc. Natl Acad. Sci. USA, 96, 3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkart V., Wang,Z.,Q., Radons,J., Heller,B., Herceg,Z., Stingl,L., Wagner,E. and Kolb,H. (1999) Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nature Med., 5, 314–319. [DOI] [PubMed] [Google Scholar]
- 10.Masutani M., Suzuki,H., Kamada,N., Watanabe,M., Ueda,O., Nozaki,T., Jishage,K., Watanabe,T., Sugimoto,T., Nakagama,H., Ochiya,T. and Sugimura,T. (1999) Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozocin-induced diabetes. Proc. Natl Acad. Sci. USA, 96, 2301–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandir S., Przedboeski,S., Jackson-Lewis,V., Wang,Z.Q., Simbulan-Rosenthal,C., Smulson,M., Hoffman,B., Guastella,D., Dawson,V. and Dawson,T. (1999) Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc. Natl Acad. Sci. USA, 96, 5774–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver F., Mennisier-de Murcia,J., Nacci,C., Decker,P., Andriantsitohaina,R., Muller,S., de la Rubia,G., Stoclet,J. and de Murcia,G. (1999) Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly(ADP-ribose) polymerase-1 deficient mice. EMBO J., 18, 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhule S., Nicotera,P., Wendel,A. and Leist,M. (1999) Prevention of endotoxin-induced lethality, but not of liver apoptosis in poly(ADP-ribose) polymerase-deficient mice. Biochem. Biophys. Res. Commun., 263, 433–438. [DOI] [PubMed] [Google Scholar]
- 14.Szabo C., Virag,L., Cuzzocrea,S., Scott,G., Hake,P., O’Connor,M., Zingarelli,B., Salzman,A. and Kun,E. (1998) Protection against peroxynitrite-induced fibroblast injury and arthritis development by inhibition of poly(ADP-ribose) synthase. Proc. Natl Acad. Sci. USA, 95, 3867–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zingarelli B., Szabo,C. and Salzman,A. (1999) Blockade of poly(ADP-ribose) synthetase inhibits neutrophil recruitment, oxidant generation and mucosal injury in murine colitis. Gastroenterology, 116, 335–345. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K., Greenberg,J., Jackson,P., Maclin,K. and Zhang,J. (1997) Neuroprotective effects of inhibiting poly(ADP-ribose) synthetase on focal cerebral ischemia in rats. J. Cereb. Blood Flow Metab., 17, 1137–1142. [DOI] [PubMed] [Google Scholar]
- 17.Tokime T., Nozaki,K., Sugino,T., Kikuchi,H., Hashimoto,N. and Ueda,K. (1998) Enhanced poly(ADP-ribosyl)ation after focal ischemia in rat brain. J. Cereb. Blood Flow Metab., 18, 991–997. [DOI] [PubMed] [Google Scholar]
- 18.Sun A. and Cheng,J. (1998) Neuroprotective effects of poly(ADP-ribose) polymerase inhibitors in transient focal cerebral ischemia of rats. Acta Pharmacol. Sin., 19, 104–108. [PubMed] [Google Scholar]
- 19.Ayoub I., Lee,E., Ogilvy,C., Beal,M. and Maynard,K. (1999) Nicotinamide reduces infarction up to two hours after the onset of permanent focal cerebral ischemia in Wistar rats. Neurosci. Lett., 259, 21–24. [DOI] [PubMed] [Google Scholar]
- 20.Lautar S., Pieper,A., Verma,A., Xi,L., Tang,Z., Liu,W., Snyder,S. and Zhang,J. (1998) Post ischemia treatment with GPI 6150 diminishes poly(ADP-ribose) accumulation during ischemia-reperfusion injury in rat brain. Soc. Neurosci. Abstr., 24, 1226. [Google Scholar]
- 21.Endres M., Scott,G., Salzman,A., Kun,E., Moskowitz,M. and Szabo,C. (1998) Protective effects of 5-iodo-6-amino-1,2-benzopyrone, an inhibitor of poly(ADP-ribose) synthetase against peroxynitrite-induced glial damage and stroke development. Eur. J. Pharmacol., 351, 377–382. [DOI] [PubMed] [Google Scholar]
- 22.Thiemermann C., Bowes,J., Myint,F. and Vane,J. (1997) Inhibition of the activity of poly(ADP-ribose) synthetase reduces ischemia-reperfusion injury in the heart and skeletal muscle. Proc. Natl Acad. Sci. USA, 94, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaterjee P., Zacharowski,K., Cuzzocrea,S., Otto,M. and Thiemermann,C. (2000) Inhibitors of poly(ADP-ribose) synthetase reduce renal ischemia-reperfusion injury in anesthetized rats in vivo. FASEB J., 14, 641–651. [DOI] [PubMed] [Google Scholar]
- 24.LaPlaca M., Zhang,J., Li,J.-H., Raghupathi,R., Smith,F., Bareye,F., Graham,D. and McIntosh,T.J. (1999) Pharmacologic inhibition of poly (ADP-ribose) polymerase is neuroprotective following traumatic brain injury in rats. Neurotrauma, 16, 976. [DOI] [PubMed] [Google Scholar]
- 25.Mandrup-Poulson T., Remiers,J., Andersen,H., Pociot,F., Karlsen,A., Bjerre,U. and Nerup,J. (1993) Nicotinamide treatment in the prevention of insulin-dependent diabetes mellitus. Diabetes Metab. Rev., 9, 295–309. [DOI] [PubMed] [Google Scholar]
- 26.Vague P., Picq,R., Bernal,M., Lassman-Vague,V. and Vialettes,B. (1989) Effect of nicotinamide treatment on the residual insulin secretion in type 1 (insulin-dependent) diabetic patients. Diabetologia, 32, 316–321. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J., Li,J.-H., Lautar,S., Mooney,M., Zhou,Y. and Williams,P. (1999) GPI 6150, a PARP inhibitor, exhibits strong anti-inflammatory effect in rat models of arthritis. Acta Physiol. Scand., Suppl. 167, 90. [Google Scholar]
- 28.Zhang J., Lautar,S., Huang,S., Ramsey,C., Cheung,A. and Li,J.-H. (2000) GPI 6150 prevents H2O2 cytotoxicity by inhibiting poly(ADP-ribose) polymerase. Biochem. Biophys. Res. Commun., 278, 590–598. [DOI] [PubMed] [Google Scholar]
- 29.Morgan W. and Cleaver,J. (1982) 3-Aminobenzamide synergistically increases sister-chromatid exchanges in cells exposed to methyl methanesulfonate but not to ultraviolet light. Mutat. Res., 104, 361–366. [DOI] [PubMed] [Google Scholar]
- 30.Burkle A., Heilbronn,R. and Zur,H.H. (1990) Potentiation of carcinogen-induced methotrexate resistance and dihydrofolate reductase gene amplification by inhibitors of poly(adenosinediphosphate-ribose) polymerase. Cancer Res., 50, 5756–5760. [PubMed] [Google Scholar]
- 31.Waldman A. and Waldman,B. (1991) Stimulation of intrachromosomal homologous recombination in mammalian cells by an inhibitor of poly(ADP-ribosyl)ation. Nucleic Acids Res., 19, 5943–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber V., Hunting,D., Trucco,C., Gowans,B., Grunwald,P., de Murcia,G. and de Murcia,J. (1995) A dominant negative mutant of human PARP affects cell recovery, apoptosis and sister chromatid exchange following DNA damage. Proc. Natl Acad. Sci. USA, 92, 4753–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kupper J., Muller,M. and Burkle,A. (1996) Trans-dominant inhibition of poly(ADP-ribosyl)ation potentiates carcinogen-induced gene amplification in SV40-transformed Chinese hamster cells. Cancer Res., 56, 2715–2717. [PubMed] [Google Scholar]
- 34.Ding R., Pommier,Y., Kang,V.H. and Smulson,M. (1992) Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J. Biol. Chem., 267, 12804–12812. [PubMed] [Google Scholar]
- 35.Ding R. and Smulson,M. (1994) Depletion of nuclear poly(ADP-ribose) polymerase by antisense RNA expression: influences on genomic stability, chromatin organization and carcinogen cytotoxicity. Cancer Res., 54, 4627–4634. [PubMed] [Google Scholar]
- 36.Wang Z., Stingl,L., Morrison,C., Jantsch,M., Los,M., Schulze-Osthoff,K. and Wagner,E. (1997) PARP is important for genomic stability but dispensable in apoptosis. Genes Dev., 11, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson J., Rademaker,A., Goolsby,C., Traczyk,T. and Zoladz,C. (1996) DNA ploidy in nonmelanoma skin cancer. Cancer, 77, 284–291. [DOI] [PubMed] [Google Scholar]
- 38.Simbulan-Rosenthal C., Haddad,B., Rosenthal,D., Weaver,Z., Coleman,A., Luo,R., Young,H., Wang,Z.Q., Ried,T. and Smulson,M. (1999) Chromosomal aberrations in PARP–/– mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc. Natl Acad. Sci. USA, 96, 13191–13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.d’Adda di Fagagna F., Hande,M., Tong,W., Landsorp,P., Wang,Z.Q. and Jackson,S. (1999) Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nature Genet., 23, 76–80. [DOI] [PubMed] [Google Scholar]
- 40.Smulson M.E., Kang,V.H., Ntambi,J.M., Rosenthal,D.S., Ding,R. and Simbulan,C.M.G. (1995) Requirement for the expression of poly(ADP-ribose) polymerase during the early stages of differentiation of 3T3-L1 preadipocytes, as studied by antisense RNA induction. J. Biol. Chem., 270, 119–127. [DOI] [PubMed] [Google Scholar]
- 41.Vindelov L.L., Christensen,I.J., Jensen,G. and Nissen,N.I. (1983) Limits of detection of nuclear DNA abnormalities by flow cytometric DNA analysis. Results obtained by a set of methods for sample-storage, staining and internal standardization. Cytometry, 3, 332–339. [DOI] [PubMed] [Google Scholar]
- 42.Banasik M. and Ueda,K. (1994) Inhibitors and activators of ADP-ribosylation reactions. Mol. Cell. Biochem., 138, 185–197. [DOI] [PubMed] [Google Scholar]
- 43.Trucco C., Oliver,F., de Murcia,G. and de Murcia,J. (1998) DNA repair defect in PARP-deficient cell lines. Nucleic Acids Res., 26, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simbulan-Rosenthal C.M.G., Rosenthal,D.S., Hilz,H., Hickey,R., Malkas,L., Applegren,N., Wu,Y., Bers,G. and Smulson,M. (1996) The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that this enzyme is a component of the multiprotein DNA replication complex. Biochemistry, 35, 11622–11633. [DOI] [PubMed] [Google Scholar]
- 45.Simbulan-Rosenthal C.M., Rosenthal,D.S., Boulares,A.H., Hickey,R.J., Malkas,L.H., Coll,J.M. and Smulson,M.E. (1998) Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry, 37, 9363–9370. [DOI] [PubMed] [Google Scholar]
- 46.Simbulan-Rosenthal C., Rosenthal,D., Luo,R. and Smulson,M.E. (1999) Poly(ADP-ribose) polymerase upregulates E2F-1 promoter activity and DNA pol a expression during early S-phase. Oncogene, 18, 5015–5023. [DOI] [PubMed] [Google Scholar]
- 47.Shieh W.M., Ame,J.C., Wilson,M., Wang,Z.Q., Koh,D., Jacobson,M. and Jacobson,E. (1998) Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem., 273, 30069–30072. [DOI] [PubMed] [Google Scholar]
- 48.Ame J., Rolli,V., Schreiber,V., Niedergang,C., Apiou,F., Decker,P., Muller,S., Hoger,T., de Murcia,J. and de Murcia,G. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem., 274, 17860–17868. [DOI] [PubMed] [Google Scholar]
- 49.Sallmann F., Vodenicharov,M., Wang,Z.Q. and Poirier,G. (2000) Characterization of sPARP-1. An alternative product of PARP-1 gene with poly(ADP-ribose) polymerase activity independent of DNA strand breaks. J. Biol. Chem., 275, 15504–15511. [DOI] [PubMed] [Google Scholar]
- 50.Rosenthal D.S., Ding,R., Simbulan-Rosenthal,C.M.G., Vaillancourt,J.P., Nicholson,D.W. and Smulson,M.E. (1997) Intact cell evidence for the early synthesis and subsequent late apopain-mediated suppression of poly(ADP-ribose) during apoptosis. Exp. Cell Res., 232, 313–321. [DOI] [PubMed] [Google Scholar]
- 51.Simbulan-Rosenthal C.M., Rosenthal,D.S., Iyer,S., Boulares,A.H. and Smulson,M.E. (1998) Transient poly(ADP-ribosyl)ation of nuclear proteins and role for poly(ADP-ribose) polymerase in the early stages of apoptosis. J. Biol. Chem., 273, 13703–13712. [DOI] [PubMed] [Google Scholar]
- 52.Simbulan-Rosenthal C.M., Rosenthal,D.S., Iyer,S., Boulares,H. and Smulson,M.E. (1999) Involvement of PARP and poly(ADP-ribosyl)ation in the early stages of apoptosis and DNA replication. Mol. Cell. Biochem., 193, 137–148. [PubMed] [Google Scholar]
- 53.Simbulan-Rosenthal C.M., Rosenthal,D.S., Luo,R. and Smulson,M.E. (1999) Poly(ADP-ribosyl)ation of p53 during apoptosis in human osteosarcoma cells. Cancer Res., 59, 2190–2194. [PubMed] [Google Scholar]
- 54.Andreassen P., Martineau,S. and Margolis,R. (1996) Chemical induction of mitotic checkpoint override in mammalian cells results in aneuploidy following a transient tetraploid state. Mutat. Res., 372, 181–194. [DOI] [PubMed] [Google Scholar]
- 55.Simbulan-Rosenthal C.M., Ly,D.H., Rosenthal,D.S., Konopka,G., Luo,R., Wang,Z.-Q., Schultz,P. and Smulson,M. (2000) Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc. Natl Acad. Sci. USA, 97, 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin M., Hsieh,S., Li,S.H.S., Chiang,T., McBride,J.R.T., Chou,L., Chou,M. and Wong,D. (1994) Sequential cytogenetic alterations in hamster oral keratinocytes during DMBA-induced oral carcinogenesis. Eur. J. Cancer B Oral Oncol., 30, 252–264. [DOI] [PubMed] [Google Scholar]
- 57.Simbulan C., Suzuki,M., Izuta,S., Sakurai,T., Savoysky,E., Kojima,K., Miyahara,K., Shizuta,Y. and Yoshida,S. (1993) Poly(ADP-ribose) polymerase stimulates DNA polymerase alpha. J. Biol. Chem., 268, 93–99. [PubMed] [Google Scholar]
- 58.Dantzer F., Nasheuer,H., Vonesch,J., de Murcia,G. and de Murcia,J. (1998) Functional association of poly(ADP-ribose) polymerase with DNA polymerase α-primase complex: a link between DNA strand-break detection and DNA replication, Nucleic Acids Res., 26, 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferro A.M., Higgins,N.P. and Olivera,B.M. (1983) Poly(ADP-ribosylation) of a DNA topoisomerase. J. Biol. Chem., 258, 6000–6003. [PubMed] [Google Scholar]
- 60.Vaziri H., West,M., Allsop,R., Davison,T., Wu,Y., Arrowsmith,C., Poirier,G. and Benchimol,S. (1997) ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J., 16, 6018–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cross S., Sanchez,C., Morgan,C., Schimke,M., Ramel,S., Idzerda,R., Raskind,W. and Reid,B. (1995) A p53-dependent mouse spindle checkpoint. Science, 267, 1353–1356. [DOI] [PubMed] [Google Scholar]
- 63.Fukasawa K., Choi,T., Kuriyama,R., Rulong,S. and Vande Woude,G. (1996) Abnormal centrosome amplification in the absence of p53. Science, 271, 1744–1747. [DOI] [PubMed] [Google Scholar]
- 64.Ramel S., Sanchez,C., Schimke,M., Neshat,K., Cross,S., Raskind,W. and Reid,B. (1995) Inactivation of p53 and the development of tetraploidy in the elastase-SV40 T antigen transgenic mouse pancreas. Pancreas, 11, 213–222. [DOI] [PubMed] [Google Scholar]
- 65.Yin X., Grove,L., Datta,N., Long,M. and Prochownik,E. (1999) C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene, 18, 1177–1184. [DOI] [PubMed] [Google Scholar]
- 66.Meisterernst M., Stelzer,G. and Roeder,R. (1997) Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc. Natl Acad. Sci. USA, 94, 2261–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butler A. and Ordahl,C. (1999) Poly(ADP-ribose) polymerase binds with transcription factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol. Cell. Biol., 19, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kannan P., Yu,Y., Wankhade,S. and Tainsky,M. (1999) Poly(ADP-ribose) polymerase is a coactivator for AP-2-mediated transcriptional activation. Nucleic Acids Res., 27, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oei S., Griesenbeck,J., Schweiger,M. and Ziegler,M. (1998) Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem., 273, 31644–31647. [DOI] [PubMed] [Google Scholar]