Abstract
Carbonyl-modified proteins are considered markers of oxidative damage caused by oxidative stress, aging, and disease. Here we use a previously developed capillary electrophoretic method for detecting femtomole (10−15 mole) carbonyl levels in mitochondrial proteins that are size separated and profiled. For protein labeling, carbonyls were tagged with Alexa 488 hydrazine and amine groups in proteins with 3-(2-furoyl)quinoline-2-carboxaldehyde. Total mitochondrial protein carbonyl levels were statistically higher in fast- than in slow-twitch muscle of young Fischer 344 rats, and statistically higher in old than in young slow-twitch muscle. Even when some statistical comparisons of the total protein carbonyl levels would not reveal differences, principal component analysis (PCA) classified the carbonyl profiles into four distinct sample groups of different age and muscle types. In addition, PCA was used to predict that most age-related or muscle-type-related changes in carbonyl levels occur in proteins with a molecular weight between 9.8 and 11.7 kD.
Keywords: Mitochondria, Carbonyl, Principal component analysis, Fluorescence, Capillary electrophoresis
Carbonyl-Modified proteins are considered markers of oxidative damage in aged tissue and diseases such as Parkinson's, diabetes, emphysema, and atherosclerosis (1,2). Multiple reports describe age-related increased levels of carbonyl-modified proteins in cultured human fibroblasts, human brain tissue, human eye lens proteins, and rat hepatocytes (3). Skeletal muscle is composed of two major types of muscle fibers: slow- and fast-twitch fibers. The two major fiber types use different metabolic processes for energy production. The fast-twitch fibers mainly produce adenosine-5′-triphosphate (ATP) via anaerobic catabolism, whereas the slow-twitch muscle uses aerobic pathways. The decline in skeletal muscle performance associated with aging is muscle fiber type-specific, with fast-twitch muscles declining more rapidly during aging than slow-twitch muscles (4–6).
Frequently total carbonyl levels in proteins are determined spectrophotometrically following derivatization with 2,4-dinitrophenyl hydrazine (DNPH) (7–15). We recently reported the detection and quantification of carbonylated proteins using capillary-sieving electrophoresis with post-column laser-induced fluorescence detection (CSE-LIF). Two fluorescent labels were used: Carbonyl groups were labeled with Alexa 488 hydrazide, whereas primary amines in proteins were labeled with 3-(2-furoyl) quinoline-2-carboxaldehyde (FQ). The dual-channel fluorescence detection made it possible to identify proteins that are carbonylated due to co-detection of the two fluorescent labels. Because the labeled proteins are separated in a capillary containing a sieving matrix, the migration time is inversely related to the protein molecular weight (MW), thereby producing a protein profile with an x-axis associated with either migration time or MW; the y-axis (i.e., Alexa 488 fluorescence intensity) in the profile corresponds to the abundance of carbonyls. Lastly, this method allows for detection of femtomole (fmole) amounts of carbonyls in proteins, which makes it compatible with the analysis of small samples, such as single cells or tissue microbiopsies.
Chemometric techniques have been previously used to extract relevant analytical information from the large amount of data generated by capillary electrophoresis (CE). Principal component analysis (PCA) is one of the most common techniques used to bring out data patterns that are not easily identified in electrophoretic profiles (16–19). It does so by reducing the number of dimensions (e.g., number of data points in an electropherogram) in multidimensional data sets to fewer dimensions (i.e., principal components [PCs]), while retaining the data set's variability (20,21). In PCA, each sample has scores associated with each PC. The most important PCs can be used to define a coordinate system, and their score values represent each sample in the score plot (22). In a score plot, the closer the distance between samples, the more similar they are with respect to the features represented by the PCs selected for the score plot (16).
In this study, the overall goal was to characterize the complex carbonyl profiles of skeletal muscle mitochondrial proteins associated with aging and muscle type. CSE-LIF electropherograms were used as fingerprints of protein carbonylation in the analyzed samples. We hypothesized that PCA is able to identify unique protein carbonyl patterns in the electropherograms of four sample groups, even though statistical analysis would not reveal differences among the groups being compared. The four sample groups included young slow-twitch, young fast-twitch, old slow-twitch, and old fast-twitch muscles. Our findings indicate that this technique (CSE-LIF), in conjunction with PCA, is capable of detecting muscle-type and age-dependent changes in protein carbonyl profiles in rat skeletal muscle mitochondria, and that the four distinct sample groups are identified in score plots. These results demonstrate that the combination of CSE-LIF and PCA is a powerful tool that may predict the age and muscle type of an unknown sample, characterize key features in the sample, and identify the MW range in which proteins susceptible to oxidative damage (i.e., carbonylation) are found.
Materials and Methods
Reagents
Alexa 488 hydrazide, FQ, and potassium cyanide were obtained from Invitrogen (Carlsbad, CA). Sodium cyanoborohydride, sodium borohydride, sodium dodecyl sulfate (SDS), sodium acetate, Tris, Tricine, bovine serum albumin (BSA), sucrose, potassium chloride, potassium phosphate monobasic, ethylene glycol tetraacetic acid (EGTA), 2,4-dinitrophenylhydrazine (DNPH), HEPES, ascorbate, guanidine, dextran (425∼575 kD), trypsin inhibitor (20.1 kD), trypsinogen (24 kD), carbonic anhydrase (29 kD), glyceraldehyde-3-phosphate dehydrogenase (36 kD), and albumin (66 kD) were purchased from Sigma-Aldrich (St. Louis, MO). Bicinichoninic acid (BCA) assay and nagarse were purchased from Pierce (Rockford, IL). Amicon Ultra-4 Centrifugal Filter Units were purchased from Millipore (Billerica, MA). UltraTrol LN was obtained from Target Discovery (Palo Alto, CA). The mitochondrial isolation buffer was composed of 100 mM sucrose, 100 mM KCl, 50 mM Tris-HCl, 1 mM KH2PO4, 100 μM EGTA, and 0.2% BSA at pH 7.4. The Alexa 488 hydrazide labeling buffer consisted of 100 mM sodium acetate and 20 mM NaCl, 1% SDS at pH 5.5. The separation buffer contained 8% dextran, 20 mM Tris, 20 mM tricine, and 0.5% SDS (pH 8).
Animal and Tissue Preparation
Skeletal muscle samples were obtained from female Fischer 344 rats with a mean life span of 29 months (23). The rats were divided into two groups based on their age: six young adult rats (12 months old, 100% survival rate) and six old rats (26 months old, ∼25% survival rate). They were maintained under specific pathogen-free conditions in our animal facility (Minneapolis VA Medical Center). In our study, we selected the soleus muscle, which is composed mainly of myosin heavy chain type 1 fibers (slow-twitch), and the semimembranosus, plantaris, extensor digitorum longus, and tibialis anterior muscles, which are mainly composed of myosin heavy chain type 2 fibers (fast-twitch).
Mitochondria were isolated by tissue homogenization and differential centrifugation as described by Madsen and colleagues (24). Briefly, muscle pieces were minced initially with scissors and suspended in the mitochondrial isolation buffer (10 mL/g tissue). Then nagarse at 0.2 mg/mL was added to the minced muscle suspension. After 1 minute of incubation, the minced tissue was homogenized and the homogenate was diluted fourfold with the isolation buffer. The homogenate was centrifuged at 700 × g for 10 minutes. The supernatant was then centrifuged at 10,000 × g for 10 minutes to obtain an enriched mitochondrial pellet. After cleaning up the fluffy layer at the top of the mitochondrial pellet, the mitochondria were resuspended in the isolation buffer (without BSA) and centrifuged at 7000 × g for 10 minutes. The resulting pellet was solubilized in the Alexa 488 hydrazide labeling buffer. Insoluble materials were removed by centrifugation at 13,000 × g for 10 minutes and discarded. The protein concentration was determined using the bicinichoninic acid (BCA) assay.
Labeling of Mitochondrial Proteins with Alexa 488 Hydrazide
The labeling reaction of Alexa 488 hydrazide with carbonyl groups has been described previously (25,26). Briefly, for labeling mitochondrial samples with Alexa 488 hydrazide, each sample (20 μL, 1.5 mg/mL) was incubated with 2 μL of 3.5 mM Alexa 488 hydrazide at 37°C for 2 hours. After incubation, 22 μL of 30 mM sodium cyanoborohydride in phosphate buffer (pH 6.0) was added to stabilize the reaction product. To remove the unreacted Alexa 488 hydrazide, an Amicon Ultra-4 centrifugal filter unit (5 kD cutoff) was used. Briefly, 3 mL of washing buffer (20 mM Tris, 20 mM tricine, and 0.1% SDS at pH 8.0) was added. Then the sample was centrifuged at 7500 × g for 20 minutes, which led to retention of the species with MW above 5 kD and filtration of other species (e.g., unreacted dye). Upon repeating the washing step 5 times, the unreacted dye peaks were reduced significantly (see Supplemental Material, Supplemental Figure 2). After removal of the excessive dye, 5 μL of 20% SDS in separation buffer was added to the sample to denature proteins and then the sample was taken to a final volume of 100 μL.
FQ Labeling of Protein Standards
To determine the MW of the detected mitochondrial proteins, we used 3-(2-furoyl) quinoline-2-carboxaldehyde (FQ) to label protein standards that were co-injected with the sample. Briefly, 50 μL of the protein standards solution containing trypsin inhibitor (20.1 kD), trypsinogen (24 kD), carbonic anhydrase (29 kD), glyceraldehyde-3-phosphate dehydrogenase (36 kD), and albumin (66 kD) was mixed with 35 μL of 10 mM KCN in a 500-μL microcentrifuge tube, which contained 500 nmole of dry FQ prepared in a Speedvac (27–30). The reaction mixture was incubated for 5 minutes at 65°C. Then 5 μL of these FQ-labeled protein standards were added to 50 μL of Alexa 488 hydrazide-labeled mitochondrial proteins and analyzed by CSE-LIF. The addition of FQ-labeled protein standards does not affect the detection of Alexa 488 fluorescence (i.e., carbonyls) because these two fluorophores emit at different spectral ranges and are detected in separate channels.
Capillary Electrophoresis
A commercial capillary electrophoresis instrument, Beckman Coulter P/ACE MDQ system (Fullerton, CA) was used for the analysis of protein carbonyls. For excitation, the LIF detector used an argon-ion laser (488 nm line, 3 MW) that was directed to a detector window in the capillary using a fiber optic; this wavelength excites both Alexa 488 and FQ-labeled proteins. Fluorescence from proteins was directed to a long-pass dichroic mirror (XF2013; Omega Optical, Brattleboro, VT) placed at 45° relative to the direction of the incoming fluorescence; transmission and reflection occur above and below 540 nm, respectively. Transmitted light (i.e., FQ-labeled protein fluorescence) and reflected light (i.e., Alexa 488 fluorescence) were further filtered with a 635DF55 bandpass filter (∼608–662 nm) and a 520DF20 bandpass filter (∼510– 530 nm), respectively, before reaching separate photomultiplier tubes (PMTs). The PMT output signals were sampled at 4 Hz.
Separations were performed using a 50-μm inner diameter (i.d.), 365-μm outer diameter (o.d.) fused-silica capillary. UltraTrol LN, a class of linear polyacrylamide made of N-substituted acrylamide copolymers, was used for precoating the capillary walls. This procedure decreases the electroosmotic flow and inhibits protein binding to the inner wall of the capillary (31). The total capillary length and the length to the detection window were 30 and 20 cm, respectively. The sample was injected hydrodynamically at 11 kPa for 4 seconds into the capillary containing the separation buffer. The total protein content for each CSE analysis was calculated to be 1.65 ng. Separations were performed at −570 V/cm. The capillary was reconditioned between consecutive runs with sequential pressure-driven flushes (15 psi for 3 minutes each) of 0.5 M sodium hydroxide, H2O, the dynamic coating reagent UltraTrol LN, separation buffer, and the running buffer (32).
Calibration of CSE-LIF With Oxidized BSA Standards
BSA (10 mg/mL) was oxidized with a hydroxyl radical-generating system consisting of ascorbate/Fe(III)/O2 as described previously by Requena and colleagues (33) and was used as a carbonylated protein standard. The carbonyl content of the oxidized BSA was calculated from the absorbance of the complex with DNPH at 375 nm (molar absorption coefficient, 22,000 M/cm) to be 12.3 nmole carbonyl/mg protein (34). This oxidized BSA was then diluted to 1, 0.5, 0.25, 0.1, and 0.04 of the original concentration with labeling buffer. Each of these five preparations of oxidized BSA (5 μL each) were labeled with 2 μL of 3.5 mM Alexa 488 hydrazide at 37°C for 2 hours. The following procedure for labeling of carbonyls was essentially the same as that used for labeling mitochondrial proteins, except that the volumes of 30 mM sodium cyanoborohydride and the final volume of each labeled BSA standards were 7 μL and 300 μL, respectively. Each of the Alexa 488 hydrazide-labeled oxidized BSA standards was injected hydrodynamically at 11 kPa for 3 seconds (with an injection volume of 0.8 nL) into the capillary containing the separation buffer. The standards were analyzed in triplicate, and the areas of the peaks detected in the Alexa 488 channel (y) was plotted versus the different amounts (moles) of carbonyl groups (x) to obtain the calibration curve:
| (1) |
MW Calibration
The second detector of the dual-channel fluorescence detection scheme was used for detection of protein standards labeled with FQ. Supplemental Figure 1A displays five conspicuous peaks representing FQ labeling standard proteins with MWs from 20,000 to 66,000. Supplemental Figure 1B shows the relationship between the logarithm of molecular mass and mobility for the five standard proteins indicating that the dextran separation is size based. Hence, using this relationship, the migration time could be transformed to protein MW.
Computational Data Analysis
Igor Pro software (Wavemetrics, Lake Oswego, OR) was used to analyze electropherograms. This program provides intensity values at each migration time point. The fluorescence intensity values detected in the Alexa 488 channel were used to calculate the area associated with carbonyl amounts. The in-house-written Igor procedure, Wide Peak Analysis, was used for this purpose (26).
Statistical Analysis
Statistical analysis was performed with the QuickCalcs program (GraphPad, San Diego, CA). For comparison of carbonyl levels, the triplicates of a given sample were averaged and the sample variance was determined. Samples with a high sample variance were excluded from subsequent data analysis (i.e., one from the slow- and one from the fast-twitch muscle [young groups]). A Grubbs test was then used to confirm that there were no outliers in the remaining samples within each group. Comparison between groups was done using one-tailed t tests. Results were converted to nmole carbonyl/mg protein using Equation 1 and reported as mean ± standard error of the mean.
Principal Component Analysis
PCA was used to analyze the electropherograms of Alexa 488 hydrazide-labeled mitochondrial protein samples described above. As mentioned in the introduction, PCA transforms data sets to extract PCs that contribute the most to data variance, thereby eliminating redundant, orthogonal, and interdependent variables and reducing the data set complexity. The PCA-processed data set is transformed into a new coordinate system, such that the greatest variance lies on the first coordinate (first PC), the second greatest variance on the second coordinate (second PC), and so forth. The position of a sample in an N-dimensional space of N PCs is then defined by its respective score vector (cf. Figure 3). The contribution of all variables to the variance of a PC is represented by a loading vector, which defines how each of the variables contributed to the total variance explained by a PC (cf. Figure 4).
Figure 3.
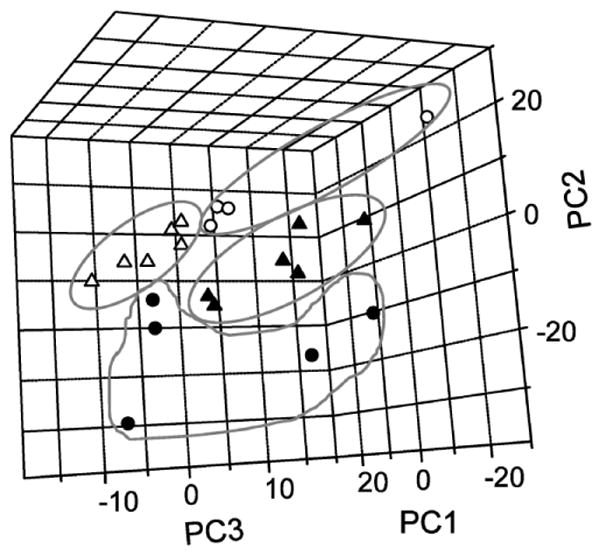
Principal component analysis (PCA) of mitochondrial protein electropherograms. Score plot of PCA-analyzed samples using the first three PCs (filled triangle, young fast-twitch, filled circle, old fast-twitch; open triangle, young slow-twitch; open circle, old slow-twitch). A series of two-dimensional panels of the score plot are shown in Supplemental Figure 3.
Figure 4.
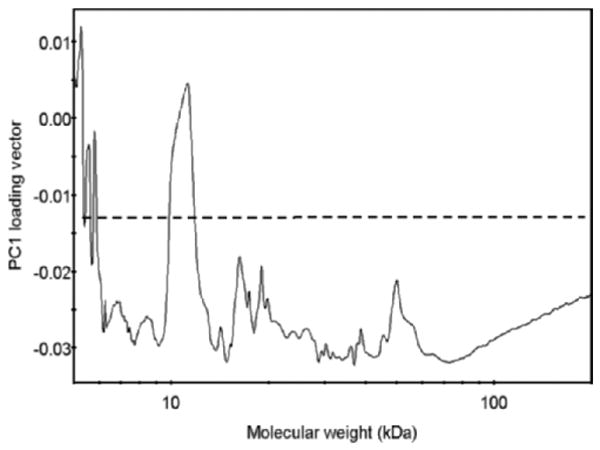
Loading vector of the first principal component (PC) in the principal component analysis (PCA) model. The loading vector represents how variables (i.e., Alexa 488 fluorescence values corresponding to proteins in a particular molecular weight [MW] range) contribute to the variance explained by the first PC. The variables with the most contribution fall above the set threshold (dashed line).
Here, every electropherogram had 3600 data points (i.e., variables representing 3600 migration time points with their corresponding fluorescence intensity values), each of which was treated as an independent variable. To classify samples based on their CE profiles, individual electropherograms in each of the four sample groups were arbitrarily divided into the calibration and validation data sets, representing 75% and 25% of the total number of available sample measurements, respectively. Four calibration data sets, one for each of the four sample groups, were used to build four disjoint PCA models in SIMCA-P (Umetrics, Umea, Sweden). These disjoint PCA models allow for pattern recognition based on modeling each of the categories by a separate PCA model (35). The four built disjoint models were then cross-validated using the samples in the validation data sets. Each model gave rise to a distribution of individual samples in an N-dimensional space of the PC scores, N being the number of PCs used. A PC was considered significant if its normalized eigenvalue was >2 and the contribution to the total explained variance was >1%. For each sample, a probability of membership was calculated, which represents the probability that the measurement belongs to one of the four disjoint models (36).
To identify the group of proteins that were carbonylated the most, the time points in each electropherogram were transformed into MW values using parameters from the MW calibration line (cf. Supplemental Figure 1); each sample was analyzed in triplicate, and the triplicate runs were aligned and averaged, resulting in a single average electropherogram for each sample.
To align and average replicate CSE-LIF measurements, migration times of 5 MW standards (detected only in the red spectral channel; Supplemental Figure 1A) were used to construct a calibration curve (Supplemental Figure 1B). The calibration curve was then applied to data obtained with the green detector channel (c.f. Figure 1) to determine the respective carbonyl levels, and the migration time values were transformed to MW values in kilodaltons (kDs). The data points corresponding to a particular sample value were then linearly interpolated to match the exact MW values of all samples. This alignment process yielded data sets, each containing exactly 1432 data points spanning the 5- to 200-kD range, which were subsequently averaged for each sample type and subjected to PCA. Prior to PCA, the data set was normalized and centered between −1 and 1 using the “mapminmax” function in Matlab. PCA was performed using the “princomp” function in Matlab 7.4 (MathWorks, Natick, MA). Samples with the Mahalanobis distance from the center of the score plot greater than the threshold (probability of 0.95) were flagged as outliers and were excluded from the data set.
Figure 1.
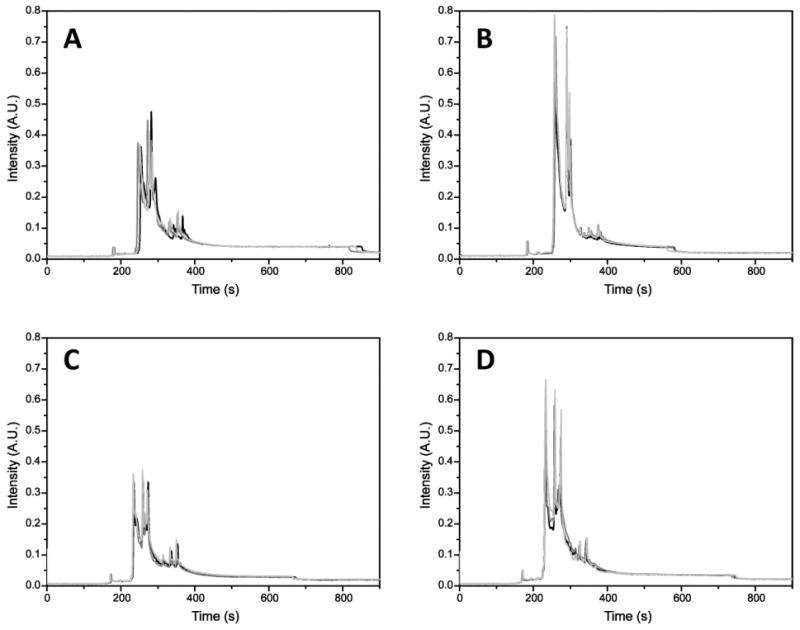
Electropherograms of mitochondrial proteins labeled with Alexa 488 hydrazide. A, Young, fast-twitch. B, Old, fast-twitch. C, Young, slow-twitch. D, Old, slow-twitch. Separation, −570 V/cm; hydrodynamic injection, 11 kPa, 4 seconds; sieving matrix, 20 mM Tris, 20 mM tricine, 8% dextran (462 kD), 0.5% sodium dodecyl sulfate, pH 8. The samples were analyzed in triplicate. Capillary conditioning, fluorescence labeling, and detection are described in Materials and Methods. A.U. = arbitrary units.
The processed data from all samples in the four groups defined above were pooled to construct a variance PCA model that defines the variables (i.e., MWs) associated with variations among these groups. As described above, the loading vector of the first PC represents the contribution of a particular MW value to the model variance captured by the first PC. High values of the loading vector (i.e., those above the preset threshold of −0.014 representing 50% of the maximum value) represent the MW values contributing the most to the total model variance (cf. Figure 4).
Results
Carbonyl Abundance in Protein Profiles
We previously introduced a CSE-LIF method for the detection and quantification of protein carbonyls in mitochondria samples containing femtomole levels of carbonyls. This method also made it possible to obtain an MW profile of the carbonylated proteins (26). Here, we used this analytical method for the comparison of carbonyl levels and profiles in mitochondrial preparations obtained from slow- and fast-twitch muscles isolated from six 12- and six 26-month old Fischer 344 rats. Each one of the 24 samples was analyzed in triplicate. Losses attributed to the multiple washes in the Amicon filter units were negligible as demonstrated for the recovery of oxidized BSA (see Supplemental Material, Figure 2 for details). The sample variances of the peak areas for each sample analyzed in triplicate ranged from 0.07 (1% relative standard deviation, [RSD]) to 13 (26% RSD), except for one young (variance 45, 40% RSD) and one old (variance 39, 26% RSD) fast-twitch samples that were eliminated in subsequent t test comparisons. Within a given group, none of the remaining samples were outliers (Grubbs test). The total carbonyl content was then calculated based on the total average area of each peak sample (c.f. Equation 1). Fast-twitch and slow-twitch muscle in the young adult group had different carbonyl levels (0.92 ± 0.11 vs 0.54 ± 0.07 nmole carbonyl/mg protein, respectively, p =.006, n = 5 and 6 samples in each group, respectively). As shown in Figure 2, in fast-twitch muscle, we did not detect a significant increase of carbonyl levels in proteins of old (26-month-old) versus young (12-month-old) rats (i.e., 1.34 ± 0.31 vs 0.92 ± 0.11 nmole carbonyl/mg protein, respectively; p =.110, n = 5 samples in each group). Only in slow-twitch muscle was there a significant difference in protein carbonyl levels between old and young adult rats (i.e., 1.23 ± 0.32 vs 0.54 ± 0.07 nmole carbonyl/mg protein, respectively; p =.051, n = 6 samples in each group).
Figure 2.
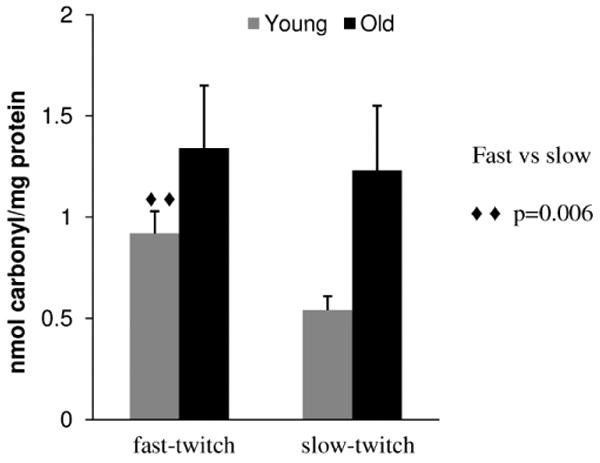
Protein carbonylation levels in Fischer 344 rat skeletal muscle mitochondria. Values reported are in nanomoles carbonyl per milligram protein and are reported as the mean ± standard error of the mean.
Sample Categorization by PCA
As noted above, the electropherograms (i.e., carbonylated profiles) of the four sample types contain a number of distinct features, but the differences between the samples are difficult to capture visually (cf. Figure 1). The overlapping peaks are mainly caused by the large number of carbonylated proteins present in the sample and the wide dynamic range of their abundance. For instance, in a previous proteomic study, 94 carbonylated proteins were identified in the skeletal muscle mitochondria of Fischer 344 rats (37). However, not all of these proteins necessarily have changes in carbonyl levels with aging. To extract valuable information from the electropherograms from such complex samples, PCA was used. The principles of PCA have been described in detail and can be found elsewhere (21,38,39). To classify samples into one of the four groups based on their electropherogram profiles, we constructed four disjoint PCA models used for pattern recognition and sample classification (35). Particular attention was paid to the number of PCs, as this often affects the outcome of PCA models—too few PCs will not adequately model the data set, whereas too many will overfit the data and lead to a less robust model.
Each disjoint model used two to four PCs to explain up to 87.5% of total variance (see Supplemental Table 1 for details). These four disjoint models were then used to analyze the samples in the validation data set (i.e., the remaining 25% of observations). For each sample in the validation set, classification to one of the four categories was based on its probability of membership value (Table 1), which represents the probability that an observation belongs to the model based on the normalized distance of a validation observation to the model. Because a probability > .05 (95% confidence interval) is commonly considered statistically significant, the highest probability value indicates that a sample belongs to a particular disjoint model. Table 1 shows that, of 20 samples, there are 17 correct assignments at 95% confidence (17 out of 19 cells with bold text); the other three samples (cell with text in italics) are correctly assigned but with a smaller confidence level (samples Y5fc, 05Sa, and Y2Sa). These results corroborate the fact that PCA is a powerful tool for multivariate analysis of complex data matrices, such as electropherograms describing carbonyl profiles.
Table 1. Probability of Membership in the Four Age/Muscle-Type Groups for Samples in the Validation Data Set.
| Sample | Young Fast-Twitch | Old Fast-Twitch | Young Slow-Twitch | Old Slow-Twitch |
|---|---|---|---|---|
| Y1Fa | .173 | 0 | 0 | 0 |
| Y2Fa | .058 | 1.08 × 10−22 | 3.90 × 10−31 | 5.56 × 10−29 |
| Y3Fb | 1.000 | 1.39 × 10−13 | 2.11 × 10−24 | 9.26 × 10−25 |
| Y4Fc | .567 | 2.34 × 10−31 | 1.53 × 10−37 | 1.08 × 10−34 |
| Y5Fc | .020 | 1.16 × 10−26 | 1.10 × 10−33 | 2.75 × 10−31 |
| O1Fb | 1.61 × 10−5 | .384 | 2.66 × 10−17 | 1.06 × 10−17 |
| O1Fc | .071 | .837 | 7.07 × 10−19 | 5.02 × 10−18 |
| 03Fb | 0 | .555 | 1.68 × 10−21 | 2.80 × 10−19 |
| 04Fb | 1.06 × 10−24 | .424 | 1.74 × 10−31 | 3.40 × 10−25 |
| 06Fa | 3.81 × 10−23 | .063 | 0 | 1.01 × 10−36 |
| Y1Sb | 8.79 × 10−32 | 6.43 × 10−21 | .655 | 4.42 × 10−13 |
| Y2Sa | 0 | 1.39 × 10−21 | .005 | 4.69 × 10−11 |
| Y4Sb | 3.45 × 10−30 | 2.11 × 10−20 | .158 | 2.57 × 10−5 |
| Y4Sc | 3.33 × 10−29 | 2.00 × 10−20 | .143 | 2.41 × 10−6 |
| Y6Sb | 2.75 × 10−37 | 1.72 × 10−20 | .764 | .225 |
| O1Sc | 0 | 2.30 × 10−20 | .001 | .685 |
| 02Sa | 0 | 4.56 × 10−24 | 3.79 × 10−28 | .986 |
| 02Sc | 0 | 2.32 × 10−21 | 0 | .489 |
| 04Sb | 0 | 2.12 × 10−21 | .002 | .414 |
| 05Sa | 8.62 × 10−20 | 7.37 × 10−22 | 7.83 × 10−25 | .037 |
Notes: The probability of membership values >.05 (95% confidence interval) are shown in boldface; values between .01 and .05 are shown in italics.
The number between the two letters identifies the animal; the last character is a notation describing the run number in the triplicate analysis. Example: 05Sa=old rat #5, slow-twitch muscle, run a).
YF = young fast-twitch; OF = old fast-twitch; YS = young slow-twitch; OS = old slow-twitch.
Prediction of MW
PCA can be used to evaluate the “importance” of variables (i.e., migration times transformed to MW values) by examining the loading vectors of PCs. To do this, a new PCA model was built using averaged replicate electropherograms for all the samples transformed to MW values. Because of its complexity, this model required six PCs to explain 94.7% of total variance. In score plots using the first three PCs (Figure 3), which explain ∼87% of the total variance, the four sample types are reasonably well separated, indicating that the model allows for identification of a sample using its PC scores (i.e., score plot coordinates). As it can sometimes be difficult to appreciate the degree to which individual groups are separated in a three-dimensional plot, we have also prepared a series of two-dimensional score plots shown in Supplemental Figure 3.
By inspecting the loading vector of the first PC (Figure 4), which explains 64% of total variance, the contribution of protein MWs to the PCA model can be evaluated. Here, the loading vector is a measure of contribution of a variable (i.e., a given MW value) to the variance: The higher the loading vector value, the bigger the contribution of a given MW value to the total variance of the model. Arbitrarily, 50% of the loading vector maximum was set as a threshold (−0.014) (dashed line in Figure 4), above which all values were considered significant contributors to the model variance.
As seen in Figure 4, the main MW regions contributing to the model variance are at 5–6 kD and 9.8–11.7 kD. The first region is associated with small, nonprotein fluorescent species present in the samples. The second interval represents a MW range in which most age-related or muscle-type-related changes in the protein carbonyl levels occur.
Discussion
This work demonstrated that CSE-LIF can be used to investigate muscle-type and age-dependent changes in protein carbonyl in rat skeletal muscle mitochondria. It also showed that PCA is useful to successfully classify protein carbonyl profiles and to predict MW ranges in which most age-related or muscle type-related protein carbonyl changes occur. In this work, we focused on carbonyls of mitochondrial proteins, because (i) mitochondria is one of the major sources of reactive oxygen species (ROS) in the cell (40) and (ii) proteins are believed to be a major targets of ROS, which results in oxidative molecular damage (e.g., carbonyl formation) (40).
The effects of variations in MW, caused by different forms of oxidative modifications or different degrees of labeling with FQ, on the separation profiles deserves some attention as they could potentially affect the PCA results. The resolution (i.e., based on the full width at half maximum of a typical protein peak) in the CSE separation is ∼2.5 kD for proteins with a MW of 22 kD. This resolution is not sufficient to detect small variations caused by the modification or labeling procedures described earlier. For example, glutamic semialdehyde, being one of the main products of metal-catalyzed oxidation, changes the MW of proline and arginine by 16 and −43 Da, respectively (33). A given protein would need to have ∼60 oxidized arginine residues to display a peak different of that of the nonoxidized protein. Thus, the CSE separations provide complex profiles that will not be affected by minor modifications in MW. These profiles can only be compared using chemometric approaches such as PCA.
Age-related increase in the protein carbonyl content has been previously reported in rat hepatocytes (41), gerbil brain (42), houseflies (43), human skin fibroblasts (44), and rat kidney cells (45). Fano and colleagues (46) reported that protein carbonyl levels showed a significant increase from young to old in human skeletal muscle (i.e., 2.0–2.9 nmole carbonyl/mg protein, respectively). Overall, our slow-twitch data are in agreement with reports that state that skeletal muscle accumulates carbonyls with aging (47,48), which may be a consequence of decreased protein degradation or increased ROS generation (15).
Other studies reported no increase in protein carbonyl contents with aging of rat skeletal muscle (49), rat liver (50), gerbil brain cortex (50), and Fischer 344 rat brain, liver, lung, and heart (51) with DNPH spectrophotometric assays. A particularly relevant study to the results presented here determined that the carbonyl content was 0.7 nmole carbonyl/mg protein in gastrocnemius muscle from both 10- and 30-month-old Fischer 344 rats (15). This muscle is one of the components of the fast-twitch muscles investigated in our study. The carbonyl levels at young ages determined in our study are comparable (i.e., 0.9 nmole carbonyl/mg protein) and do not statistically significantly increase with aging.
The differences in the protein carbonyl levels between slow- and fast-twitch muscle types reported here underline different metabolic mechanisms. Fast-twitch muscle had more carbonyls than did slow-twitch muscle in young adult (12-month-old) Fischer 344 rats. The fact that oxidative damage is much more evident in fast-twitch than in slow-twitch muscle suggests (i) increased ROS levels, and consequently ROS-associated oxidative damage, or (ii) less effective degradation of oxidized proteins and greater antioxidant capacity in slow-twitch muscle. These observations are in agreement with a report stating that superoxide produced in both mitochondrial complexes I and III is higher in fast-twitch than in slow-twitch muscle and that superoxide scavenging capacity is lower in fast-twitch than in slow-twitch muscle (52). Thus, it appears that carbonyl levels in both muscle types may be associated with ROS produced by the electron transport chain in mitochondria, even when fast-twitch muscle relies primarily on glycolysis for ATP production. In contrast, it is possible that other sources of ROS in the cytoplasm of fast-twitch muscle fibers (e.g., cytoplasmic xanthine oxidase) contribute to oxidative damage in the respective muscle type (53,54).
The relationship between oxidative stress (especially total protein carbonyls) and normal aging has been studied extensively in other tissues (41–44,46,47,49,51,55), but this is the first time that PCA has been used to classify mitochondrial protein carbonyl profiles obtained by CSE-LIF according to age and muscle type. As shown in Figure 3 (and Supplemental Figure 3), the four sample types cluster differently, indicating the distinct oxidative stress characteristics of these sample types. Because these plots are landmarks representing the variations in carbonyl levels of mitochondrial proteins from fast- or slow-twitch skeletal muscle of young or old Fischer 344 rats, they may be used for cataloguing unknown samples. In such studies, it would be beneficial to include more samples to calibrate the PCA model, which would in turn improve its robustness. More reproducible results (in terms of fluorescence intensity as well as migration times) would further improve the accuracy of the model. Lastly, there are chemometric methods, (e.g., artificial neural network) that are more sophisticated than PCA and that could be powerful alternatives to characterize unknown samples based on their carbonyl profiles (20), but their examination is beyond the scope of this report.
By analyzing the electropherograms shown in Figure 1, it can be seen that most carbonylated proteins lie between 220 and 400 seconds that correspond to a MW range from 4.2 to 51.8 kD, whereas the useful range extends up to 800 seconds that corresponds to 240 kD. The absence of significant levels of carbonyls in proteins ranging from 51.8 to 240 kD may be caused by (a) low carbonyl levels in mitochondrial proteins with high MW and (b) low solubility of these proteins during solubilization of mitochondria. However, the causes of these losses were not further investigated because the most dramatic changes were expected in the 10- to 30-kD range, which contains the most abundant mitochondrial proteins (56). In fact, the range 9.8–11.7 kD, determined from the loading vector analysis, ought to include the subset of proteins that contributes the most to the variations associated with age and muscle type. This approach allows us to identify six carbonylated proteins (Table 2) of 94 that have been previously reported as carbonylated (37,57).
Table 2. Identified Carbonylated Proteins with Molecular Weight (MW) Intervals of 9.8–11.7 kd.
| N | Accession | Protein Name | MW (kd) |
|---|---|---|---|
| Carbonylated proteins identified from both proteomics studies (37,57) | |||
| 1 | P26772 | 10 kd heat shock protein | 10.9 |
| Carbonylated proteins identified from first proteomics study (37) | |||
| 2 | Q5M9I5 | Cytochrome b-c1 complex subunit 6 | 10.4 |
| 3 | P62898 | Cytochrome c | 11.6 |
| 4 | IPI00358015 | 1 NADH dehydrogenase (ubiquinone) 1β, 3 | 11.2 |
| Carbonylated proteins identified from second proteomics study (59) | |||
| 5 | IPI00191103 | 1 NADH dehydrogenase (ubiquinone) lα | 10.8 |
| 6 | Q2XTA8 | NADH dehydrogenase 1β 4 [Fragment] | 9.9 |
Note: Swiss-Prot accession numbers are given for each protein. For those proteins without a Swiss-Prot accession number, IPI accession numbers are given instead.
The majority of the proteins listed in Table 2 are mitochondrial inner membrane complexes I–V from the oxidative phosphorylation (OXPHOS) machinery, known to be targets of ROS (58,59). In addition, thioredoxin and 10-kD heat shock protein identified as carbonylated belong to the 9.8- to 11.7-kD MW range. At the present time, the effect of carbonylation on thioredoxin and heat shock protein function is unknown, and one hypothesis to investigate further is that carbonylation decreases their function and leads to age-related losses of antioxidant defense and apoptosis regulation.
Clearly, fast profiling by CSE-LIF followed by PCA could be a prescreening tool to identify the MW range of interest, which could in turn tremendously reduce the complexity of subsequent proteomic studies. The value of such a prescreening method would be even greater when working with precious muscle samples. For example, we are conducting a longitudinal study in which muscle biopsies are taken from the same muscle of the same animal at different time points over the life span of the animal. This methodology may also be useful to elicit correlations between protein oxidative damage and muscle function and shed light on how much damage the system can withstand before function failure.
Conclusion
In this article, a strategy for quantitation and profiling of carbonylated proteins by CSE-LIF, down to femtomole carbonyl levels, was combined with PCA. Even when statistical comparisons would not reveal differences between muscle types and age, the PCA correctly identified four different sample types. In addition, the PCA of the carbonyl profiles resulting from the CSE-LIF measurements suggests that the most important variations in carbonylation associated with age and muscle type occur in proteins with masses in the 9.8- to 11.7-kD MW range. This range includes 10-kD heat shock protein and thioredoxin, which previously have been associated with oxidative stress defense systems. This range also included proteins involved in oxidative phosphorylation that may be associated with different roles that mitochondria play in fast- and slow-twitch muscle types. These results highlight a new approach to fast classification and quantification of carbonyls. This approach has potential application in investigating oxidative stress found in aging, disease, exercise and dietary regimens, or anti-aging treatments.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (AG025371). E.A.A. acknowledges the support of the NIH by a Career Award (1K02-AG21453).
We also thank Dr. Dmitry Andreyev for writing an Igor procedure for peak analysis and Kate O'Connor for isolating the rat skeletal muscle tissue used in this study.
References
- 1.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 2.Bautista J, Mateos-Nevado MD. Immunological detection and quantification of oxidized proteins by labelling with digoxigenin. Biosci Biotechnol Biochem. 1998;62:419–423. doi: 10.1271/bbb.62.419. [DOI] [PubMed] [Google Scholar]
- 3.Stadtman ER, Levine RL. Protein oxidation. Ann N Y Acad Sci. 2000;899:191–208. doi: 10.1111/j.1749-6632.2000.tb06187.x. [DOI] [PubMed] [Google Scholar]
- 4.Sandmann ME, Shoeman JA, Thompson LV. The fiber-type-specific effect of inactivity and intermittent weight-bearing on the gastrocnemius muscle of 30-month-old rats. Arch Phys Med Rehabil. 1998;79:658–662. doi: 10.1016/s0003-9993(98)90040-5. [DOI] [PubMed] [Google Scholar]
- 5.Thompson LV. Skeletal muscle adaptations with age, inactivity, and therapeutic exercise. J Orthop Sports Phys Ther. 2002;32:44–57. doi: 10.2519/jospt.2002.32.2.44. [DOI] [PubMed] [Google Scholar]
- 6.Holloszy JO, Chen M, Cartee GD, Young JC. Skeletal muscle atrophy in old rats: differential changes in the three fiber types. Mech Ageing Dev. 1991;60:199–213. doi: 10.1016/0047-6374(91)90131-i. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn AC, Doe WF, Buffinton GD. Protein carbonyl formation on mucosal proteins in vitro and in dextran sulfate-induced colitis. Free Radic Biol Med. 1999;27:262–270. doi: 10.1016/s0891-5849(99)00065-9. [DOI] [PubMed] [Google Scholar]
- 8.Chevion M, Berenshtein E, Stadtman ER. Human studies related to protein oxidation: protein carbonyl content as a marker of damage. Free Radic Res. 2000 33:S99–S108. [PubMed] [Google Scholar]
- 9.Shacter E, Williams JA, Lim M, Levine RL. Differential susceptibility of plasma proteins to oxidative modification: examination by western blot immunoassay. Free Radic Biol Med. 1994;17:429–437. doi: 10.1016/0891-5849(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 10.Bizzozero OA, Ziegler JL, De Jesus G, Bolognani F. Acute depletion of reduced glutathione causes extensive carbonylation of rat brain proteins. J Neurosci Res. 2006;83:656–667. doi: 10.1002/jnr.20771. [DOI] [PubMed] [Google Scholar]
- 11.Mosoni L, Breuille D, Buffiere C, Obled C, Mirand PP. Age-related changes in glutathione availability and skeletal muscle carbonyl content in healthy rats. Exp Gerontol. 2004;39:203–210. doi: 10.1016/j.exger.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Pansarasa O, Bertorelli L, Vecchiet J, Felzani G, Marzatico F. Age-dependent changes of antioxidant activities and markers of free radical damage in human skeletal muscle. Free Radic Biol Med. 1999;27:617–622. doi: 10.1016/s0891-5849(99)00108-2. [DOI] [PubMed] [Google Scholar]
- 13.Pansarasa O, Castagna L, Colombi B, Vecchiet J, Felzani G, Marzatico F. Age and sex differences in human skeletal muscle: role of reactive oxygen species. Free Radic Res. 2000;33:287–293. doi: 10.1080/10715760000301451. [DOI] [PubMed] [Google Scholar]
- 14.Radak Z, Sasvari M, Nyakas C, et al. Regular training modulates the accumulation of reactive carbonyl derivatives in mitochondrial and cytosolic fractions of rat skeletal muscle. Arch Biochem Biophys. 2000;383:114–118. doi: 10.1006/abbi.2000.2042. [DOI] [PubMed] [Google Scholar]
- 15.Radak Z, Takahashi R, Kumiyama A, et al. Effect of aging and late onset dietary restriction on antioxidant enzymes and proteasome activities, and protein carbonylation of rat skeletal muscle and tendon. Exp Gerontol. 2002;37:1423–1430. [Google Scholar]
- 16.Johannesson N, Olsson L, Backstrom D, Wetterhall M, Danielsson R, Bergquist J. Screening for biomarkers in plasma from patients with gangrenous and phlegmonous appendicitis using CE and CEC in combination with MS. Electrophoresis. 2007;28:1435–1443. doi: 10.1002/elps.200600606. [DOI] [PubMed] [Google Scholar]
- 17.Szymanska E, Markuszewski MJ, Capron X, et al. Increasing conclusiveness of metabonomic studies by chem-informatic preprocessing of capillary electrophoretic data on urinary nucleoside profiles. J Pharm Biomed Anal. 2007;43:413–420. doi: 10.1016/j.jpba.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Mohana M, Reddy K, Jayshanker G, Suresh V, Sarin RK, Sashidhar RB. Principal opium alkaloids as possible biochemical markers for the source identification of Indian opium. J Sep Sci. 2005;28:1558–1565. doi: 10.1002/jssc.200500078. [DOI] [PubMed] [Google Scholar]
- 19.Scampicchio M, Mannino S, Zima J, Wang J. Chemometrics on microchips: towards the classification of wines. Electroanalysis. 2005;17:1215–1221. [Google Scholar]
- 20.Sentellas S, Saurina J. Chemometrics in capillary electrophoresis. Part B. Methods for data analysis. J Sep Sci. 2003;26:1395–1402. [Google Scholar]
- 21.Jolliffe IT. Principal component analysis. 2nd. New York: Springer; 2002. [Google Scholar]
- 22.Rodriguez-Nogales JM, Vivar-Quintana AM, Revilla I. Influence of somatic cell count and breed on capillary electrophoretic protein profiles of ewes' milk: a chemometric study. J Dairy Sci. 2007;90:3187–3196. doi: 10.3168/jds.2006-589. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi R, Goto S. Age-associated accumulation of heat-labile aminoacyl-tRNA synthetases in mice and rats. Arch Gerontol Geriatr. 1987;6:73–82. doi: 10.1016/0167-4943(87)90040-9. [DOI] [PubMed] [Google Scholar]
- 24.Madsen K, Ertbjerg P, Djurhuus MS, Pedersen PK. Calcium content and respiratory control index of skeletal muscle mitochondria during exercise and recovery. Am J Physiol. 1996;271:E1044–E1050. doi: 10.1152/ajpendo.1996.271.6.E1044. [DOI] [PubMed] [Google Scholar]
- 25.Triantafilou K, Triantafilou M, Fernandez N. Lipopolysaccharide (LPS) labeled with Alexa 488 hydrazide as a novel probe for LPS binding studies. Cytometry. 2000;41:316–320. [PubMed] [Google Scholar]
- 26.Feng J. Quantification of carbonylated proteins in rat skeletal muscle mitochondria using capillary sieving electrophoresis with laser-induced fluorescence detection. Electrophoresis. 2008;29:475–482. doi: 10.1002/elps.200700262. [DOI] [PubMed] [Google Scholar]
- 27.Presley AD, Fuller KM, Arriaga EA. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;793:141–150. doi: 10.1016/s1570-0232(03)00371-4. [DOI] [PubMed] [Google Scholar]
- 28.Stoyanov AV, Ahmadzadeh H, Krylov SN. Heterogeneity of protein labeling with a fluorogenic reagent, 3-(2-furoyl)quinoline-2-carbox-aldehyde. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;780:283–287. doi: 10.1016/s1570-0232(02)00535-4. [DOI] [PubMed] [Google Scholar]
- 29.Chen Z, Wu J, Baker GB, Parent M, Dovichi NJ. Application of capillary electrophoresis with laser-induced fluorescence detection to the determination of biogenic amines and amino acids in brain microdialysate and homogenate samples. J Chromatogr A. 2001;914:293–298. doi: 10.1016/s0021-9673(01)00539-8. [DOI] [PubMed] [Google Scholar]
- 30.Pinto D, Arriaga EA, Schoenherr RM, Chou SS, Dovichi NJ. Kinetics and apparent activation energy of the reaction of the fluorogenic reagent 5-furoylquinoline-3-carboxaldehyde with ovalbumin. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;793:107–114. doi: 10.1016/s1570-0232(03)00368-4. [DOI] [PubMed] [Google Scholar]
- 31.Adhihetty PJ, Irrcher I, Joseph AM, Ljubicic V, Hood DA. Plasticity of skeletal muscle mitochondria in response to contractile activity. Exp Physiol. 2003;88:99–107. doi: 10.1113/eph8802505. [DOI] [PubMed] [Google Scholar]
- 32.Kraly JR, Jones MR, Gomez DG, et al. Reproducible two-dimensional capillary electrophoresis analysis of Barrett's esophagus tissues. Anal Chem. 2006;78:5977–5986. doi: 10.1021/ac061029+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Requena JR, Chao CC, Levine RL, Stadtman ER. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci U S A. 2001;98:69–74. doi: 10.1073/pnas.011526698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine RL, Garland D, Oliver CN, et al. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 35.Wold S. Pattern recognition by means of disjoint principal components models. Pattern Recognit. 1976;8:127–139. [Google Scholar]
- 36.Krzanowski WJ. Principles of multivariate analysis: a user's perspective. New York: Oxford University Press; 2000. Rev. ed. [Google Scholar]
- 37.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 38.Navratil M, Cimander C, Mandenius CF. On-line multisensor monitoring of yogurt and filmjolk fermentations on production scale. J Agric Food Chem. 2004;52:415–420. doi: 10.1021/jf0304876. [DOI] [PubMed] [Google Scholar]
- 39.Kourti T. The Process Analytical Technology initiative and multivariate process analysis, monitoring and control. Anal Bioanal Chem. 2006;384:1043–1048. doi: 10.1007/s00216-006-0303-y. [DOI] [PubMed] [Google Scholar]
- 40.Reid MB. Nitric oxide, reactive oxygen species, and skeletal muscle contraction. Med Sci Sports Exerc. 2001;33:371–376. doi: 10.1097/00005768-200103000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Starke PE, Oliver CN, Stadtman ER. Modification of hepatic proteins in rats exposed to high oxygen concentration. FASEB J. 1987;1:36–39. doi: 10.1096/fasebj.1.1.2886388. [DOI] [PubMed] [Google Scholar]
- 42.Sohal RS, Agarwal S, Sohal BH. Oxidative stress and aging in the Mongolian gerbil (Meriones unguiculatus) Mech Ageing Dev. 1995;81:15–25. doi: 10.1016/0047-6374(94)01578-a. [DOI] [PubMed] [Google Scholar]
- 43.Sohal RS, Agarwal S, Dubey A, Orr WC. Protein oxidative damage is associated with life expectancy of houseflies. Proc Natl Acad Sci U S A. 1993;90:7255–7259. doi: 10.1073/pnas.90.15.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver CN, Ahn BW, Moerman EJ, Goldstein S, Stadtman ER. Age-related changes in oxidized proteins. J Biol Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 45.Goto S, Nakamura A, Radak Z, et al. Carbonylated proteins in aging and exercise: immunoblot approaches. Mech Ageing Dev. 1999;107:245–253. doi: 10.1016/s0047-6374(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 46.Fano G, Mecocci P, Vecchiet J, et al. Age and sex influence on oxidative damage and functional status in human skeletal muscle. J Muscle Res Cell Motil. 2001;22:345–351. doi: 10.1023/a:1013122805060. [DOI] [PubMed] [Google Scholar]
- 47.Mecocci P, Fano G, Fulle S, et al. Age-dependent increases in oxidative damage to DNA, lipids, and proteins in human skeletal muscle. Free Radic Biol Med. 1999;26:303–308. doi: 10.1016/s0891-5849(98)00208-1. [DOI] [PubMed] [Google Scholar]
- 48.Sato A, Huang MZ, Watanabe S, et al. Protein carbonyl content roughly reflects the unsaturation of lipids in muscle but not in other tissues of stroke-prone spontaneously hypertensive strain (SHRSP) rats fed different fats and oils. Biol Pharm Bull. 1998;21:1271–1276. doi: 10.1248/bpb.21.1271. [DOI] [PubMed] [Google Scholar]
- 49.Bejma J, Ji LL. Aging and acute exercise enhance free radical generation in rat skeletal muscle. J Appl Physiol. 1999;87:465–470. doi: 10.1152/jappl.1999.87.1.465. [DOI] [PubMed] [Google Scholar]
- 50.Cao G, Cutler RG. Protein oxidation and aging. I. Difficulties in measuring reactive protein carbonyls in tissues using 2,4-dinitrophenylhydrazine. Arch Biochem Biophys. 1995;320:106–114. doi: 10.1006/abbi.1995.1347. [DOI] [PubMed] [Google Scholar]
- 51.Goto S, Nakamura A, Radak Z, et al. Carbonylated proteins in aging and exercise: immunoblot approaches. Mech Ageing Dev. 1999;107:245–253. doi: 10.1016/s0047-6374(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 52.Anderson EJ, Neufer PD. Type II skeletal myofibers possess unique properties that potentiate mitochondrial H(2)O(2) generation. Am J Physiol Cell Physiol. 2006;290:C844–C851. doi: 10.1152/ajpcell.00402.2005. [DOI] [PubMed] [Google Scholar]
- 53.Reid MB, Haack KE, Franchek KM, Valberg PA, Kobzik L, West MS. Reactive oxygen in skeletal muscle. I. Intracellular oxidant kinetics and fatigue in vitro. J Appl Physiol. 1992;73:1797–1804. doi: 10.1152/jappl.1992.73.5.1797. [DOI] [PubMed] [Google Scholar]
- 54.Meydani M, Evans WJ, Handelman G, et al. Protective effect of vitamin E on exercise-induced oxidative damage in young and older adults. Am J Physiol. 1993;264:R992–R998. doi: 10.1152/ajpregu.1993.264.5.R992. [DOI] [PubMed] [Google Scholar]
- 55.Ohira Y, Yoshinaga T, Ohara M, et al. Myonuclear domain and myosin phenotype in human soleus after bed rest with or without loading. J Appl Physiol. 1999;87:1776–1785. doi: 10.1152/jappl.1999.87.5.1776. [DOI] [PubMed] [Google Scholar]
- 56.Taylor SW, Fahy E, Zhang B, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 57.Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ. Quantitative proteomic profiling of muscle-type and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63:1137–1152. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 59.Kudin AP, Bimpong-Buta NY, Vielhaber S, Elger CE, Kunz WS. Characterization of superoxide-producing sites in isolated brain mitochondria. J Biol Chem. 2004;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


