Abstract
Previous work has shown that the α-tocopherol transfer protein (α-TTP) can bind to vesicular or immobilized phospholipid membranes. Understanding the molecular mechanisms by which α-TTP associates with membranes, is thought to be critical to understand its function and role in the secretion of tocopherol from hepatocytes into the circulation. Calculations presented in the Orientations of Proteins in Membranes (OPM) database have provided a testable model for the spatial arrangement of α-TTP and other CRAL-TRIO-family proteins with respect to the lipid bilayer. These calculations predicted that a hydrophobic surface mediates the interaction of α-TTP with lipid membranes. To test the validity of these predictions, we employed site-directed mutagenesis and examined the substituted mutants with regards to intermembrane ligand transfer, association with lipid layers, abd biological activity in cultures hepatocytes. Substitution of residues in helices A8 (F165A and F169A) and A10 (I202A, V206A and M209A) decreased the rate of intermembrane ligand transfer as well as protein adsorption to phospholipid bilayers. The largest impairment was observed upon mutation of residues that are predicted to be fully immersed in the lipid bilayer in both apo- (open) and holo- (closed) conformations such as Phe165 and Phe169. Mutation F169A, and especially F169D, significantly impaired α-TTP-assisted secretion of α-tocopherol outside cultured hepatocytes. Mutation of selected basic residues (R192H, K211A, and K217A) had little effect on transfer rates indicating no significant involvement of non-specific electrostatic interactions with membranes.
Keywords: α-tocopherol, tocopherol transfer protein, lipid transfer protein, immobilized bilayer, hydrophobic effect, free energy of binding
Introduction
The α-tocopherol transfer protein (α-TTP) is a soluble protein mainly expressed in liver in mammals that is responsible for the selective retention of α-tocopherol over other forms of vitamin E ingested in the diet. It binds α-tocopherol with high affinity (Kd ~ 25 nM 1) and catalyzes the intermembrane transfer of α-tocopherol.2–4 We have recently shown5,6 that α-TTP expressed in cultured hepatocytes is localized to the late endocytic compartment and that α-TTP co-localizes with a fluorescent tracer of tocopherol (R)-2,5,7,8-tetramethyl-chroman-2-[9-(7-nitro-benzo[1,2,5]oxadiazol-4-yl amino)-nonyl]-chroman-6-ol, (NBD-Toc) shortly after uptake.7,8 That α-TTP localizes to endocytic vesicles raises the question of how this protein finds its way to that particular membrane surface. In vitro studies suggest that this may in part be a function of the propensity of endosomes to display highly curved membrane surfaces since α-TTP delivers NBD-Toc much faster to small highly curved vesicles than to larger vesicles of lower curvature.9
Human α-TTP belongs to the CRAL-TRIO protein family, that includes a number of lipid transfer proteins including the prototypical member of this family, Sec14p, a yeast phosphatidyl inositol/phosphatidyl choline transfer protein. Given the structural similarity of α-TTP with Sec14p, we were interested in confirming whether α-TTP also functions by the proposed “bulldozer” mechanism of membrane recognition and ligand delivery.10 To begin addressing this question we wished to explore what face of the protein was likely to interact with membranes and whether any specific amino acid residue(s) dominate the favorable free energy changes of this interaction.
Most lipid transfer proteins have the capacity for both ligand and membrane recognition. The molecular forces involved in protein-membrane recognition include electrostatics, hydrogen bonding, and hydrophobic effects. The latter come into play when proteins penetrate through the phospholipid headgroups into the hydrophobic interior of the phospholipid bilayer. The mechanism and forces involved in protein-membrane interactions has become an active area of research including description of proteins that have generic (charge) or explicit (i.e. PS) phospholipid dependence for binding, the ability to sense or induce membrane curvature, or recognize proteins already resident on membrane surfaces.
The role of hydrophobicity in protein-membrane interactions is well documented and databases exist that list these interactions and describe calculated changes in free-energy (ΔG) of protein insertion into the membrane. The Orientation of Proteins in Membranes (OPM) database11–13 has over 850 entries, including α-TTP (pdb code: 1oiz from the x-ray structure of Meir et al. 14 and 1r5l from Min et al.15). We have used the calculated orientation of α-TTP with respect to a hydrophobic surface as a model to probe the role of selected amino acid residues on this hydrophobic face. We have compared the membrane binding ability and ligand transfer rates of wild-type α-TTP with mutated variants of the protein in which hydrophobicity at residues F165, F169, V201, I202, and M209 was reduced or eliminated. We find that loss of these hydrophobic residues drastically alters the ability of α-TTP to associate with immobilized phospholipid bilayers and to deliver NBD-tocopherol to acceptor vesicles. Furthermore, the reduction in hydrophobicity at these sites all but abolished the protein s activity in cultured cells.
Results
Membrane binding modes of Sec14-like proteins
Human α-TTP belongs to the CRAL-TRIO protein family (PFAM: PF00650) of soluble proteins thought to participate in the transport of hydrophobic ligands. The three-dimensional structure of human α-TTP has been solved both with14,15 and without14 bound α-tocopherol (1OIP, 1R5L and 1OIZ, respectively). Structures of other members of the family have been reported for yeast phosphatidylinositol-transfer protein Sec14p (1AUA)10, its close homologue Sfh1 (3B7A, 3B7N, 3B7Q, 3B7Z)16,17 and human supernatant protein factor SPF (1O6U, 1OLM)18,19, as well as a more distant SpoIIAA-like bacterial proteins with still unknown function (2Q3L, 2OOK).20 The three-dimensional structures of the open and closed conformations of two SpoIIAA-like proteins (YP_749275.1 and YP_001095227.1) provide insights into membrane association and ligand binding.20
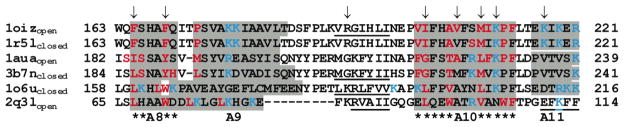
The characteristic feature of Sec14p-like proteins is the presence of two domains (CRAL-TRIO) first identified in the retinal-binding protein (CRALBP) and the Trio guanine nucleotide exchange factor, namely: an N-terminal all-helical domain and a C-terminal domain composed of βαβ units that form a hydrophobic lipid-binding cavity. Several members of this family are now considered to be lipid transfer proteins and the highly homologous lipid-binding domains of these proteins may be superimposed with a RMSD of less than 2.5 for core residues by the SSM server (http://www.ebi.ac.uk/msd-srv/ssm/). Comparison of these structures shows a significant conformational change in the C-terminal domain upon binding of natural ligands. The major conformational rearrangement involves the movement of a helical segment (residues 201–213 of α-TTP) proposed to regulate the entry and exit of ligands from the hydrophobic binding cavity. In the “open” (apo) conformation the distance between two “gating” helices located at the mouth of the binding cavity (between α-carbons of F213 and K177) is 15.6 A in human α-TTP14 and as much as 25 Å (from homologous residues F231 to R195) in yeast Sec14p 10,21 whereas in the closed (holo) state the distance is 6.2 Å for α-TTP15 and ~10.9Å in Sec14p.21 In most CRAL-TRIO domains the movable lid helix is characterized by the highly hydrophobic solvent exposed surface and by the presence of a conserved KPFL motif (Figure 1). Therefore, it was proposed that this “gating” helix is critical for the ability of Sec14 to bind membranes and to exchange lipophilic ligands between the hydrophobic pocket and the lipid bilayer. 10,21
Figure 1.
Sequence alignment of membrane-binding regions of proteins from Sec14 family. Helical fragments are shadowed, β-strands are underlined, spatially fixed and mobile “gating” helices are marked by asterisks. Residues that penetrate the hydrocarbon core of the lipid bilayer are colored in red, basic residues in the proximity to the membrane boundary are colored in blue. α-TTP residues mutated in this work are indicated by arrows. Helical numbering is based on that of Min et al.15
In order to understand the role of non-polar residues from “gating” helices we combined the theoretical calculations of membrane binding modes of Sec14-like proteins with the subsequent mutagenesis of predicted membrane-penetrating residues of α-TTP. Our previously developed PPM1.0 method 11 allows us to define spatial positions and penetration depths of proteins into the hydrocarbon core of the lipid bilayer. Moreover, PPM1.0 method provides rough estimations of the transfer free energies of peripheral proteins from water to the lipid bilayer, which were in agreement with the experimental membrane binding affinities of the proteins in the absence of specific lipid binding or significant conformational transitions.11 We have recently developed an improved version of the method, PPM2.0, to properly account for the electrostatic component of transfer energy and interactions with head groups of lipids. PPM2.0 allows a more accurate energy calculation. Results of these calculations were deposited in the OPM database 13 and shown in Figures 2, 3, Supplementary Figure 1 and Table 2 and Supplementary Table 1.
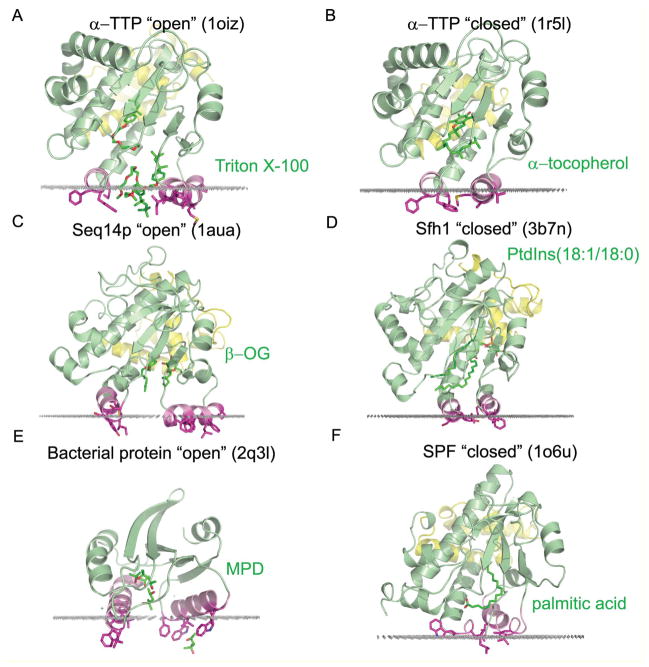
Figure 2.
Calculated interactions between the lipid-binding domains of proteins from Sec14 family with the hydrocarbon core of the lipid bilayer: α-TTP in “open” (1OIZ, A) and “closed” (1R5L, B) conformations, yeast phosphatidylinositol transfer protein Sec14p in “open” (1AUA, C) and its homolog Sfh1 in “closed” (3B7N, D) conformation, the remotely related SpoIIAA-like bacterial protein in “open” conformation (2Q3L, E), and supernatant protein factor, SPF (1O6U, F) in “closed” conformation The loosely packed jelly-roll GOLD domain of SPF that blocks the entrance to the binding cavity and occludes membrane binding of Sec14-like domain was removed from the structure as it presumably rearrange or dissociates during “opening” of the closed SPF structure determined by X-ray crystallography18. Protein backbone is shown in a cartoon representation with helices colored in green, N-terminal α-helical domain is colored in yellow, and lid helices (fixed and mobile) enclosing the binding cavity are colored in pink. Residues that penetrate into the acyl chain region of the lipid bilayer are colored purple. Molecules of detergents bound to “open” conformations (Triton X-100 in 1oiz, β-octylglucoside (β-OG) in 1aua, and methylpentanediol (MPD) in 2q3l) and hydrophobic ligands bound to “closed” conformations (α-tocopherol in 1r5l, phosphatidylinositol in 3b7n, and palmitic acid in 1o6u) are shown by sticks colored in dark green. Calculated boundaries between lipid head groups and acyl chain region are shown by grey dots.
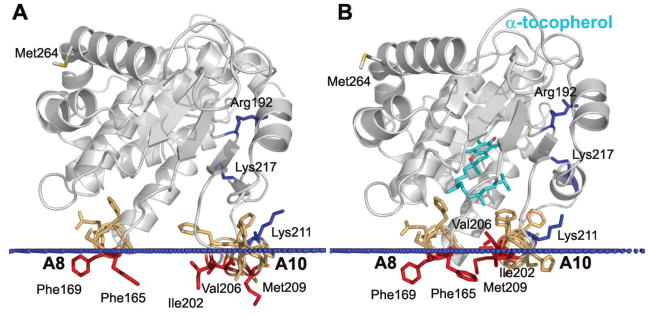
Figure 3.
Comparison of the predicted association between the “open” (A) and “closed” (B) conformations of the α-TTP and the lipid bilayer. “Open” and “closed” conformations correspond to 1oiz and 1r5l PDB entries, respectively. Cartoon representations of α-TTP structures are colored grey. αTocopherol (cyan), mutated residues (red for hydrophobic and blue for basic), and residues from “gating” helices A8 and A10 (orange) are shown by sticks. Calculated membrane boundaries are shown by blue dots.
Table 2. Association of structures of Sec14-like proteins with lipid bilayers.
Shown are results of PPM 2.0 calculations of spatial positions of CRAL-TRIO proteins in a model lipid bilayer (9:1 DOPC:DOPS) are presented by the following parameters: predicted lipid-penetration residues, calculated transfer free energies (ΔG, kcal/mol), and maximal penetration depth (D, )
| Protein | PDB code | State | Predicted residues penetrating into lipid acyl chain region |
Transfer free energies ΔG (kcal/mol) | Maximal penetratio n depth D (Å) | |
|---|---|---|---|---|---|---|
| in fixed lid helix | in movable lid helix | |||||
| α-TTP | 1OIZ | open | F165, F169, P173 | V201, I202, A205, V206, M209, I210, P212, F213 | −19.8 | 7.9 ± 0.1 |
| α-TTP | 1R5L | closed | F165, F169, P173 | V201, I202, H204, A205, S208, M209, P212 | −13.6 | 4.9 ± 1.0 |
| Sec14p | 1AUA | open | I184, Y188, M191 | F219, G220, T223, A224, L227, F228, P230, F231 | −14.1 | 5.1 ± 1.8 |
| Sfh1 | 3B7N | closed | L186, S187, Y190 | F221, G222, T225, M229 | −5.5 | 3.0 ± 1.8 |
| SPF | 1O6U | closed | L160, L163, W164 | L197, V200 | −5.3 | 3.6 ± 1.5 |
| SpoIIAA -like bacterial protein | 2Q3L | open | L67, W71, L74, L78 | L95, W98, V102, W104, F105 | −12.1 | 6.0 ± 0.8 |
All analyzed structures of Sec14-like proteins associate to the membrane through the same pair of “gating” helices (colored pink in Figure 2A–E) and several homologous solvent-accessible hydrophobic residues (Figure 1, 2 and Table 2). Calculated transfer free energies ranged from −5 to −20 kcal/mol (Table 2). It appeared that in “open” conformations “gating” helices penetrate deeper into the hydrophobic membrane core (D from 5.1± 1.8 to 7.9±0.1 Å), which results in more negative transfer free energy ( Gtransf from −12.1 to −19.8 kcal/mol), as compared with “closed” conformations (D from 3.0±1.8 to 4.9±1.0 Å; Gtransf from −5.5 to −13.6 kcal/mol).
More detailed computational analyses of spatial orientations of α-TTP relative to the lipid bilayer (Figure 3) demonstrates that the protein binds to the membrane via immersion into the hydrocarbon region of hydrophobic residues from two lid helices, A8 and A10, that enclose the ligand binding cavity in the “open” (1OIZ) and “closed” (1R5L) conformations. In the ligand-free “open” conformation large patches of exposed hydrophobic residues from the spatially fixed helix A8 (F165, F169) and the movable “gating” helix A10 (V201, I202, A205, V206, M209, I210, P212, F213) of α-TTP penetrate deep into the lipid bilayer. Overall, the depth of penetration of the “open” conformation of α-TTP into the hydrocarbon core was estimated to be 7.9 ± 0.1 A with calculated ΔGtransfer of −19.8 kcal/mol. Binding of α-tocopherol stabilizes the “closed” conformation, where the lid helix A10 moves closer to helix A8 and side chains of V206, M209 and F213 reorient toward the ligand binding cavity and become involved in hydrophobic contacts between two “gating” helices. The conformational changes in α-TTP during the “open” to “closed” transition lead to a slight reorientation of the protein relative to the membrane. The membrane penetration depth decreases to 4.5 ± 1.0 Å and ΔGtransf becomes −13.6 kcal/mol.
Role of hydrophobic residues in α-TTP binding to membranes
Calculated structural models of α-TTP bound to a lipid bilayer were used to predict which non-polar residues that penetrate deep into the hydrophobic phase should be mutated to alanine and the acidic residue aspartate. The latter substitution was expected to produce much larger effects on the free energy of transfer given the cost of submerging these ionizable residues into the membrane. Our previous studies have determined that α-TTP does bind membranes and that this is occurring when α-TTP delivers or picks up tocopherol from membranes.9,22 Mutant forms of α-TTP were generated and assessed for three aspects of α-TTP function: 1) transfer of the protein-bound ligand to membrane vesicles, 2) protein adsorption to immobilized bilayers and 3) protein binding to unilamellar vesicles. The residues chosen for mutagenesis are shown in red and gold in Figure 3.
Rates of Ligand Transfer by Mutated α-TTP
Using a fluorescent analogue of α-tocopherol, (R)-2,5,7,8-tetramethyl-chroman-2-[9-(7-nitro-benzo[1,2,5]oxadiazol-4-yl amino)-nonyl]-chroman-6-ol, (NBD-Toc), we measured the rates of ligand transfer of α-TTP-bound NBD-Toc to small unilamellar vesicles (SUVs) that contain TRITC-PE as a FRET acceptor.7 We have recently shown9 that transfer to SUVs (mean diameter ~ 60 nm) is about eight to ten times faster than to LUVs (mean diameter ~ 150 nm) at equimolar phospholipid concentration. The α-TTP/NBD-Toc complex is rapidly mixed with vesicles and the decrease in fluorescence is monitored over time as NBD-Toc is transferred to the vesicle and quenched. The results of these measurements are shown in Figure 4 and Table 3.
Figure 4.
Rates of transfer of NBD-Toc from α-TTP variants to SUVs (A) and LUVs (B). All trials were repeated in triplicate and errors shown are standard deviations. See Materials and Methods for details of protein and lipid concentrations. Transfer rates of F169D to LUVs were zero, and consequently do not have a bar in the histogram. Bars with an asterisk (*) indicate values significantly different from wild-type (unpaired two-tailed T-test, P<0.05)
Table 3.
Rates of NBD-Toc delivery to PC vesicles (LUV and SUV) by wild type and mutant forms of α-TTP. KDs for binding of NBD-Toc to α-TTP variants are averages of duplicate measurements; estimated errors are ± 15%
| TTP mutant | NBD-Toc KD (nM) | Rate of NBD-Toc transfer to | |||
|---|---|---|---|---|---|
| SUV | LUV | ||||
| Rate (s−1) | Ratemut/Ratewt % | Rate (s−1) | Ratemut/Ratewt % | ||
| WT | 19 | 0.127 ± 0.012 | 100 | 0.016 ± 0.002 | 100 |
| F165D | 17 | 0.013 ± 0.001 | 10 | 0.009 ± 0.005 | 56 |
| F169D | 79 | 0.015 ± 0.002 | 12 | 0 | 0 |
| I202D | 26 | 0.028 ± 0.002 | 22 | 0.005 ± 0.002 | 31 |
| M209D | 12 | 0.047 ± 0.004 | 37 | 0.013 ± 0.001 | 81 |
| I202D/M209D | 25 | 0.014 ± 0.001 | 11 | 0.008 ± 0.003 | 50 |
| F165D/M209D | 29 | 0.034 ± 0.003 | 27 | 0.010 ± 0.001 | 62 |
| M264D | 29 | 0.116 ± 0.013 | 91 | 0.008 ± 0.001 | 50 |
| F165A | 52 | 0.037 ± 0.002 | 29 | 0.005 ± 0.002 | 34 |
| F169A | 70 | 0.067 ± 0.006 | 53 | 0.008 ± 0.002 | 48 |
| V206A | 23 | 0.154 ± 0.012 | 121 | 0.016 ± 0.002 | 100 |
| M209A | 90 | 0.140 ± 0.012 | 110 | 0.025 ± 0.005 | 156 |
| R192H | 22 | 0.106 ± 0.008 | 83 | 0.012 ± 0.003 | 75 |
| K211A | 36 | 0.105 ± 0.008 | 83 | 0.011 ± 0.001 | 69 |
| K217A | 32 | 0.130 ± 0.011 | 102 | 0.010 ± 0.004 | 62 |
The complex of wild-type α-TTP with ligand transfers NBD-Toc to SUVs composed of bovine liver PC with a rate constant of 0.127 ± 0.012 s−1. This rate is reduced 8-fold if the assay is performed with larger vesicles (LUVs), supporting our previous observations of the importance of membrane curvature for the TTP-membrane interaction. It is worth noting that none of the mutations reported here have disturbed this preference for SUV over LUVs. The rate of ligand transfer is significantly reduced if residues predicted to insert into the hydrophobic domain of a bilayer are changed to aspartate. The rate of transfer to SUVs by the F165D mutant is reduced by 90%, F169D by 88%, I202D by 78% and M209D by 63%. The more conservative mutations to alanine also showed substantial reduction in the rate of ligand transfer: F165A (71 % lower) and F169A (47 % lower). A similar conservative mutation of M209 (M209A) had essentially no effect on the rate of transfer. The double mutants F165D/M209D and I202D/M209D demonstrated rates of NBD-Toc transfer to SUVs similar to those obtained for single F165D and I202D mutants. These data point out the small contribution of non-polar interaction of M209 to the transfer free energy of α-TTP from aqueous to the lipid phase. The mutants V206D and I202A were not assayed due to difficulties in expression and purification. We also prepared the M264D mutant in which a surface accessible hydrophobic methionine is replaced with aspartate, in a region predicted to be distant from the membrane plane. The rate of ligand deposition in SUVs for this mutant was reduced by only 9%, but 50% for LUVs. The reason(s) for the larger rate reduction for transfer to LUVs is not obvious, but is likely at least in part due to the much lower rates generally observed for transfer to LUVs. To inquire whether ligand transfer rate variations were caused by differing abilities of α-TTP variants to associate with membranes we employed dual polarization interferometry (DPI) to measure protein adsorption to an immobilized lipid bilayer.22 These results are shown in Figure 5 and Table 4.
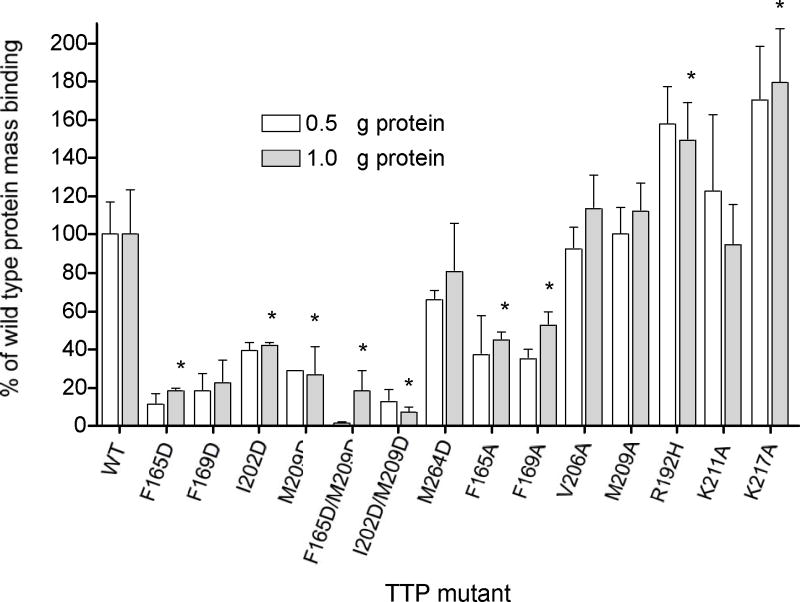
Figure 5.
Comparison of protein binding to immobilized phospholipids bilayers (9:1 PC:PS) determined by dual polarization interferometry. All proteins were tested at both 0.5 (open bars) and 1.0 μM (grayed bars) that approach the saturation values for wild type α-TTP under the conditions tested. Shown are averages of quadruplicate measurements (two channels are measured for each injection) with associated standard deviations. Bars with an asterisk (*) indicate values significantly different from wild-type (unpaired two-tailed T-test, P<0.05)
Table 4.
Affinity of wild type and mutant forms of α-TTP for immobilized phospholipid bilayers (9:1 PC/PS) measured by dual polarization interferometry. Kd values were determined by fitting a saturation binding curve produced by injecting concentrations of proteins that spanned low masses of adsorbed protein to saturation, and were fitted to a one site saturation binding model. nd = not determined. KD values without listed errors are averages of duplicates with an estimated error of ± 20%
| TTP Mutant | Kd (μM) | Fold change in affinity | Adsmut/Adswt (0.50 μM injection) | Adsmut/Adswt (1.0 μM injection) |
|---|---|---|---|---|
| Wild Type | 0.410 ± 0.08 | 0 | 1 | 1 |
| F165D | 4.994 ± 1.9 | 12.2 | 0.11 | 0.19 |
| F169D | 5.85 ± 0.54 | 14.3 | 0.18 | 0.23 |
| I202D | 2.05 | 5 | 0.29 | 0.27 |
| M209D | 0.488 | 1.2 | 0.39 | 0.42 |
| F165D/M209D | nd | - | 0.01 | 0.18 |
| I202D/M209D | 4.49 ± 2.1 | 10.9 | 0.13 | 0.07 |
| M264D | nd | - | 0.66 | 0.81 |
| F165A | 2.11 ± 0.61 | 5.1 | 0.37 | 0.45 |
| F169A | nd | - | 0.33 | 0.51 |
| V206A | nd | - | 0.93 | 1.14 |
| M209A | nd | - | 1.01 | 1.12 |
| R192H | nd | - | 1.58 | 1.50 |
| K211A | nd | - | 1.23 | 0.95 |
| K217A | nd | - | 1.71 | 1.80 |
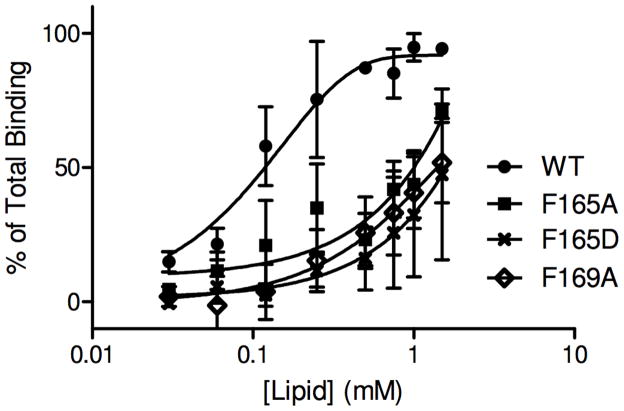
We have shown that α-TTP binds more efficiently to smaller vesicles of higher curvature than to larger vesicles9 so variations in membrane binding as measured with flat bilayers deposited on the DPI sensor chip should be compared to membranes that can exhibit curvature. We therefore performed vesicle-binding assays22,23 using sonicated SUVs and extruded 100 nm LUVs. The results for wild-type α-TTP is shown in Figure 6 and the binding curves for other mutants in Figure 7. Our previous observation of higher rates of α-TTP-dependent ligand transfer to SUVs are supported by the vesicle binding assays which show weaker association with LUVs (Figure 6). The F165A, F165D, and F169A mutants that had the greatest impact on the transfer rate of NBD-Toc also exhibited diminished vesicle binding (Figure 7).
Figure 6.
Binding of α-TTP to sonicated small and large unilamellar vesicles (SUVs and LUVs). Binding was assayed using a filtration-based separation of lipid-bound protein from free protein as described previously22.
Figure 7.
Binding of α-TTP variants to small unilamellar vesicles (SUVs) as determinend by a filtration vesicle binding assay. Values plotted are averages ± standard deviation (n>3).
Comparison of calculated membrane binding energies with functional characteristics of α-TTP mutants
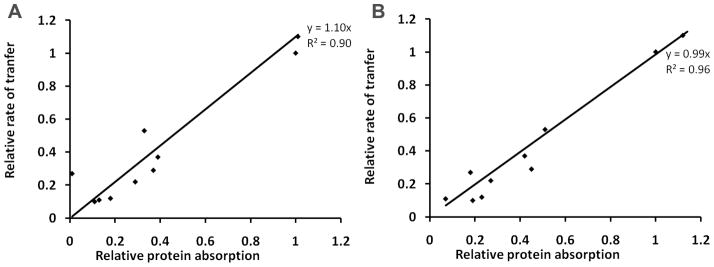
The pattern of protein adsorption to immobilized membranes (Figure 5 and Table 4) closely parallels the variation in rates using the FRET transfer assay (Figure 4 and Table 3). For instance, the F165D mutant adsorbs only 11% of the mass of wild type α-TTP and the rate of ligand transfer to SUV was reduced to a similar extent (10 %). Good correlation (0.94 > R2 >0.88) was observed between rates of TTP-catalyzed NBD-Toc transfer to SUVs and protein adsorption to phospholipid bilayers among different TTP mutants relative to the wild-type protein (Figure 8). This linear correlation may indicate that protein binding to phospholipid membranes followed by the conformational transition from a “closed” to an “open” conformation may represents a rate-limiting step in FRET kinetics.
Figure 8.
Correlation of rates of TTP-catalyzed NBD-Toc transfer to SUV and protein adsorption to immobilized phospholipid bilayers (9:1 PC:PS) at 0.5 μM (A) and 1 μM (B) protein concentrations.
Changes in protein adsorption of TTP mutants relative to the wild type proteins were compared with calculated changes in binding energy of these mutants modeled using both “open” and “closed” protein conformations (Supplementary Figure 1, Supplementary Table 1). For both conformations calculated transfer free energies from water to the DOPC/DOPS (9:1) bilayer correlated equally well with experimental values of protein adsorption to immobilized phospholipid bilayers with similar composition (Supplementary Figure 1). R-values for the “closed” and “open” conformations were 0.89 and 0.83, respectively, when compared with protein adsorption data measured at 0.5 μM protein injection; and 0.72 and 0.84, when compared at 1.0 μM protein injection. It s noteworthy that correlations significantly improved for calculation performed by the advanced PPM2.0 method, which better accounts for electrostatic energy and interactions with lipid head groups than PPM1.0. For example, correlation coefficient between protein adsorption data and transfer energies of the “open” conformation increased from 0.58 to 0.84, when estimated by PPM1.0 or PPM2.0, respectively.
The role of Phe169 in a-TTP-facilitated secretion of α-tocopherol from intact cells
As described above, two solvent-exposed hydrophobic residues from helix A8, Phe165 and Phe169, are theoretically predicted to be critical for the interaction of α-TTP with membranes. This prediction is borne out in experiments where the association between α-TTP and artificial lipid bilayers was measured (Figure 5). To examine the physiological relevance of these findings, we tested the F169A/D variants of α-TTP in the only known activity of the protein in vivo: stimulation of vitamin E secretion from cultured hepatocytes.6,24,25 We followed a modified version of the original assay, in which we employ fluorescence microscopy to visualize the amount of NBD-Toc that accumulates in cells under different conditions26. HepG2 cells were transfected with the various constructs, and incubated with serum-complexed NBD-Toc, which is taken up through endocytosis.6,24 After 16 hrs incubation with NBD-Toc, mock-transfected cells accumulate significant amount of the vitamin, visible as intense fluorescence throughout the cytoplasm (leftmost panel in Figure 9B). Expression of α-TTP stimulates secretion of the vitamin, such that the amount of NBD-Toc is reduced by >80% (Figure 9B, C). Cells that expressed TTP(F169A) or TTP(F169D) displayed markedly enhanced NBD-Toc fluorescence than those expressing the wild-type protein, indicating compromised biological activity of the mutated proteins (Figure 9B). Specifically, the F169A and F169D substitutions lead to a 2-fold and 5-fold inhibition of tocopherol secretion, respectively, as compared to the wild-type TTP (Figure 9C), despite similar expression levels of the different proteins (Figure 9A). The stronger inhibition elicited by the F169D substitution as compared to that observed with F169A (Figure 9) is consistent with the impairments caused by these mutations to association with lipid bilayers in vitro (Figure 5 & 7). These data demonstrate that membrane binding is an important/obligatory component of α-TTP s physiological activity.
Figure 9.
Cultured HepG2 cells were transiently transfected with the indicated constructs and loaded with NBD-tocopherol as described in Materials and Methods. NBD-Toc fluorescence intensity was measured in > 14 fields from each slide, averaged, and normalized to cell number. A. Expression levels of the various α-TTP variants as detected by Western blotting. B. Representative fluorescence micrographs. C. Averages and standard deviations of relative NBD-Toc fluorescence intensity measured in the indicated transfectants. Representative of two independent experiments are presented.
Role of charged residues in the interaction of α-TTP with membranes
In addition to the significant contribution of hydrophobic residues in the binding energy of peripheral proteins to the lipid bilayer11, an important role is often suggested for basic residues, as they may interact electrostatically with negatively charged head groups of phospholipids, and with some signaling lipids, such as phosphoinositides. Analysis of structural models of α-TTP oriented relative to the lipid bilayer (Figure 2) demonstrated that at least seven positively charged surface lysines (K75, K125, K177, K178, K211, K217, and K219) are good candidates for such electrostatic interactions. Among these residues K211 is highly conserved in all Sec14-like proteins as a part of KPFL motif (homologous to K229 in Sec14p).21 Some other basic residues, located more distantly from the predicted lipid boundary (R59, R68, K190, R192, R221) together with K75 and K177 form a cluster of positively charged residues, which may be involved in a specific binding of phosphoinositides. To evaluate the possible roles of ionic interactions in the binding of α-TTP to lipid bilayers, we mutated two surface lysines (Lys211, K217; located in helices A10 and A11, respectively) into alanines. In addition, we generated the naturally-occurring α-TTP R192H mutant that is associated with heritable mild vitamin E deficiency in humans.27,28
The effects of K211A and R192H substitution mutations on α-TTP-facilitated transfer of NBD-Toc to SUV and the affinity of α-TTP binding to immobilized PC/PS bilayers was very small (Figures 4, 5, Table 3). α-TTP(K211A) exhibited only a minor impairment in NBD-Toc delivery to SUVs and LUVs (83% and 69% of the transfer observed with the wild-type protein, respectively). Two other mutants, α-TTP(K217A) and α-TTP(R192H), showed similar affinity to NBD-Toc as the wild type TTP (32 nM and 22 nM versus 19 nM), and essentially equivalent ligand transfer to SUVs (102% and 83% of WT, respectively). Curiously, however, both K217A and R192H bound to immobilized bilayers considerably better than wild type TTP: 171% and 158% of WT for SUVs; 180% and 150% for LUVs, respectively. The apparent tighter binding of these mutants to immobilized membranes, despite equal rates of ligand transfer, prompted us to test the binding of α-TTP(K217A) in a filtration vesicle binding assay using SUVs (Figure 10). These experiment showed that the mutant binding to SUV composed of PC is essentially indistinguishable from the binding of the wild type protein. The reason for this discrepancy is not clear, but could reflect a change in the equilibrium favoring the open form of the protein that has a greater exposed hydrophobic surface than the closed form.
Figure 10.
Vesicle binding assay of wild type α-TTP and its K217A mutant to PC SUVs. Shown are the averages of four determinations ± the standard deviation for wild-type protein and average of two measurements for K217A.
These results indicate that the suggested involvement of basic residues in non-specific ionic interactions of the protein with lipid head-groups is not required for successful membrane binding and ligand transfer. That the R192H and K217A mutants bound better to immobilized membranes might be due to an improved stability of these mutants compared to the wild type protein. The stability of the R192H and K217A mutants might be enhanced due to introduction of residues with increased secondary structure propensities (His in β-sheet and Ala in α-helix29 or by elimination of electrostatic repulsions between spatially close R192 and K217 residues.
Discussion
Our theoretical analyses of the membrane binding modes of Sec14-like proteins using the PPM method 11 underline the essential role of hydrophobic residues of two “gating” helices located at the mouth of the ligand binding pocket in recruitment of these proteins to membranes. The subsequent mutagenesis studies support the role of hydrophobic residues in membrane binding of α-TTP and subsequent transfer of NBD-Toc from α-TTP into the lipid bilayer. Indeed, mutations of residues from both lid helices (F165, F169, I202, V206 and M209) to either Ala or Asp significantly affected the rate of NDB-Toc transfer to SUVs (Figure 4, Table 3) and protein absorption to phospholipid bilayers as measured by DPI (Figure 5, Table 4) or vesicle filtration experiments (Figures 6, 7). The largest effect on the protein absorption was obtained when replacements involved residues that are predicted to be fully immersed in the lipid bilayer. This included Phe165 and Phe169 from helix A8 that we predict anchors α-TTP to membranes in both “open” and “closed” states. On the other hand, mutations of Ile202, and Met209, Val206 residues in the flexible A10 helix, predicted to be fully lipid-exposed only in the “open” state, exhibited either normal membrane binding behavior (V206A, M209A) or moderate functional impairment (I202D, V206D, M209D). In all experiments with the artificial lipid bilayers it appeared that mutations of Phe165 and Phe169 affect ligand transfer and membrane binding the most. Likewise, α-TTP(F169D) was more defective in assisting tocopherol secretion from cultured hepatocytes than the F169A mutant (Figures 10). The larger effect of substitution to negatively charged Asp as compared to Ala substitution can be explained by a deionization penalty of the charged Asp residues when penetrating into the nonpolar membrane interior.
Our data supported and extend our previous observation that α-TTP binds tighter and exchanges the α-tocopherol at a higher rate with highly-curved SUVs, as compared to more flat LUVs. Accordingly, all observed effects of residue substitutions were more pronounced with SUVs (Figures 4, 6, Table 3). This effect may be related to the decreased lateral pressure in outer leaflets of curved SUV membranes, as compared to the flatter membranes of LUVs, which is known to facilitate insertion of proteins and peptides into membranes.30,31 However, we also observed a five-to-ten–fold loss of membrane binding affinity for F165A, F169D, and F165D mutants in the DPI experiments, which presumably employ “flat” membranes immobilized on the sensor chip. The fact that the extent of protein-membrane binding assessed by DPI paralleled results obtained by vesicle binding assays is likely due to the extreme sensitivity of the DPI assay. Thus, we found the DPI technique more advantageous, because it is capable of reproducibly detecting small changes in an already small response. Vesicle binding assays that use filtration-based separation of lipid-bound protein appeared to be less sensitive and showed considerably more variability.
In this work we theoretically estimated the transfer free energy of peripheral membrane proteins from water to the lipid bilayer of a defined lipid composition using an improved PPM method that accounts for the hydrophobic, H-bonding and electrostatic interactions of proteins with membranes. The calculated membrane binding affinity (ΔGtransf) may be overestimated because it does not include the entropy of protein immobilization in membrane12 and the lipid bilayer deformation that is due to the lateral pressure12. The contribution of lateral pressure is expected to be smaller or negative for curved membrane surfaces32, facilitating protein binding.30 To eliminate these potential errors, we operated with transfer energy differences of mutants and wild-type protein (ΔΔGtransf) instead of the absolute values. This allowed us to obtain good correlations (R2 in the range 0.8 – 0.9) between predicted transfer free energy from aqueous to the lipid phase and relative protein adsorption measured by DPI for α-TTP and its mutants (Supplementary Figure 1, Supplementary Table 1). This result indicates that our theoretical method is adequate for quantitative analysis of protein-membrane binding energetics of different protein conformations.
The theoretical analysis supports the previously proposed three-step process of inter-membrane transfer of α-tocopherol by α-TTP.22 However, it must be modified to include the transition between protein conformations within the lipid bilayer. The modified scheme of α-tocopherol extraction includes the following steps that are shown in Figure 11: (1) binding of the ligand-free α-TTP to the membrane (A) followed by opening of the protein (B) in the membrane; (2) ligand binding in membrane with simultaneous closing of the structure (C); and (3) dissociation from the membrane of ligand-loaded α-TTP (D). During step 2 the equilibrium between protein states (B and C) depends on the concentration of α-tocopherol. Additional steps 4 and 5 correspond to the transitions between different protein states in water, such as “closed” ligand-loaded conformations (D) or ligand-free state (A), and a metastable “open” conformation of the ligand-free α-TTP (E). The free energy changes during transitions from water to membrane of 1R5L-like and 1OIZ-like conformations (transitions 3 and 6, respectively) were estimated by the PPM.
Figure 11.
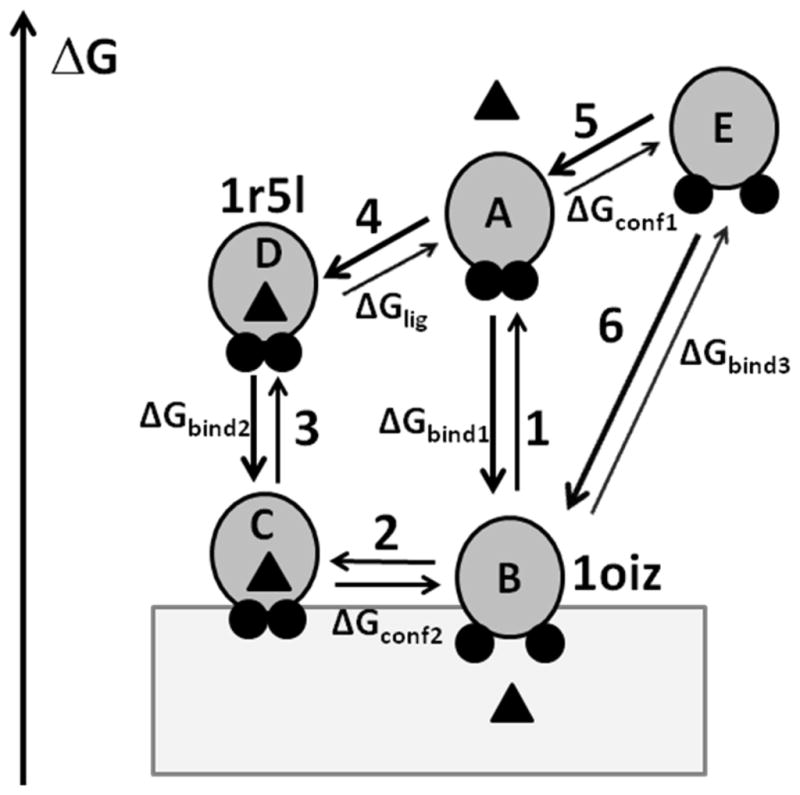
Transitions between different states of α-TTP. α-TTP is shown by gray circles with A8 and A10 helices shown by small black circles, α-tocopharol is shown by a triangle, bilayer is shown by a grey rectangle. α-TTP can adopt a “closed” or an “open” conformations in water and in the membrane-bound state. Two lid helices are tightly packed in the “closed” conformation, but move apart in the “open” conformation. Left axis shows relative free energy of different states. In the aqueous solution, the ligand-bond closed conformation (D) has a lower energy than ligand-free open conformation (E) because of the increased exposure of non-polar residues to water during the opening of the structure and ligand dissociation. However, the corresponding conformations C and B are expected to have similar energies in membrane because the nonpolar residues remain buried in the lipid environment. Note, that the energy gain for membrane binding of ligand-free state (transition 1, ΔGbind1) is larger than for ligand-loaded state (transition 3, ΔGbind2). Calculations of transfer energy with PPM 2.0 for 1r5l and 1oiz structures provide estimations of ΔGbind1 (transition 3) and ΔGbind3 (transition 6), respectively.
As follows from our calculations, there is a significant free energy gain associated with the membrane binding of the α-TTP, which is smaller for the ligand-loaded “closed” conformation (ΔGbind2 = −13.6 kcal/mol for 1R5L) than for the ligand-free “open” conformation (ΔGbind3 = −19.8 kcal/mol for 1OIZ). The difference between these values (−6.2 kcal/mol) can be attributed to the energetic cost of the conformational transition (ΔGconf1) and the energy of ligand binding (ΔGlig) in water. Thus, the “closed” conformation loaded with the ligand should have a weaker membrane binding affinity than its apo-conformation (ΔGbind2 < ΔGbind1) and dissociates more easily from the membrane. This result agrees with our previous observation that the apparent affinity of α-TTP to lipid vesicles decreases in the presence of α-tocopherol.22 Analysis of crystal structures demonstrated that for all Sec14-like proteins have a common conformational transition from a “closed” to an “open” state, which involves the dissociation of helices homologous to A8 and A10 of α-TTP. This transition is expected to be energetically unfavorable in water (transition 5, ΔGconf1), because it leads to the exposure of hydrophobic residues of both helices and the ligand-binding cavity to water. A similar transition in the non-polar lipid environment (transition 2, ΔGconf2) may be of lower energy, thus enabling the reversibility of the α-tocopherol binding and transport. Energies of the “open” and “closed” states in membranes are likely similar because there is no penalty for exposure of non-polar residues in the lipid environment. Some energy may be lost due to dissociation of tocopherol, but the ligand has been partially replaced by lipids because of the opening and deeper penetration of the protein structure to membrane. The lower transfer energies from water to the lipid bilayer and the deeper immersion (by 2–3 Å) into the lipid core of “open” conformations, as compared to the “closed” ones, seems to be a common feature of all Sec14-like proteins (Table 2). As modeled in Figure 11, the processes observed in DPI or filtration-based essays can be described by transitions 1–2 for the ligand free TTP and by transitions 3–2 for the ligand-loaded TTP. The NBD-Toc transfer to phospholipid vesicles monitored in FRET experiments may be described by transitions 3–2, while the NBD-Toc extraction from the membranes likely corresponds to transitions 2-3-4.
The present work has emphasized the role of surface hydrophobic residues on the ability of the protein to bind and transfer ligand to lipid bilayers. However, the question remains about the possible functional role of multiple solvent-exposed basic residues. We have previously reported22 on the effect of mutations that cause ataxia with vitamin E deficiency (AVED), which include a number of charge replacement mutations. In this work we confirmed that R192H has near normal rates of ligand delivery (83% of wild type TTP) and binds immobilized bilayers with slightly greater efficiency (~150% of wild type TTP) as judge by DPI. The mutations of two basic residues, K211A and K217, located at or near movable A10 helix also have little effect on tocopherol delivery to membranes (83% and 102% of WT), but increased TTP binding in DPI assay (123% and 171% of WT, respectively). This result indicates that all basic residues tested do not contribute to the membrane binding of TTP via non-specific ionic interactions with lipid headgroups, but may be involved in intramolecular interactions that would stabilize the native structure of TTP or some of its conformations, thus resulting in the improved binding or facilitated conformational transition, which are essential for the efficient inter-membrane transfer of the vitamin E.
Our failure to see any change in NBD-transfer rates for wild type TTP when the receiving vesicles contain a variety of anionic lipids (PS, PI, PA, LBPA) also confirmed that non-specific electrostatic effects play only a minor role in binding of TTP to membranes.9 However, our results were obtained with bilayers devoid of other possible protein partners or targeting lipids such as phosphoinositides. It remains possible that PtdIns(3,5)P2, that is highly enriched on late endosomes33 may be essential for the recruitment of TTP to late endosomes, the major intracellular compartment to where the α-TTP has been originally found in cultured hepatocytes6. An interaction of TTP with unspecified phosphoinositides has been reported34 and we eagerly await further reports on this phenomenon.
In summary, we report the sensitivity of TTP binding to membranes to the loss of particular surface hydrophobic residues that can insert into the hydrophobic core of membranes, while the protein is insensitive to the presence of anionic lipids9. That such insertion/binding is more favorable in smaller unilamellar vesicles (i.e. SUVs versus LUVs) is a phenomenon matched by other membrane binding proteins and peptides.35,36
Materials and Methods
Enzymes, lipids and chemicals
Pfu Turbo DNA polymerase, XL-1 blue and BL21(DE3) E. coli competent cells were purchased from Stratagene (Cedar Creek, TX). Qiaprep Spin Miniprep kit was from Qiagen (Mississauga, ON). DpnI restriction enzyme was obtained from New England Biolabs (Pickering, ON). The reagents used for protein purification were same as published (9) except glutathione agarose, which was acquired from Sigma (St. Louis, MI). Polycarbonate membranes used for lipid extrusion were purchased from Avestin, Inc (Ottawa, ON). N-(6-tetramethylrhodaminethio-carbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanol-amine, triethylammonium salt (TRITC-DHPE) was from Molecular Probes (Invitrogen, OR). All phospholipids were obtained from Avanti Polar Lipids (Alabaster, AL): bovine liver L-α-phosphatidylcholine (liver PC), 1,2-dioleoyl-sn-glycerol-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS). NBD-Toc was previously synthesized and characterized in our laboratory.8
Spectroscopy and chromatography
Unmodified sensor chips utilized for the Dual Polarization Interferometric analysis of protein lipid binding interactions were acquired from the Farfield Sensors Ltd. (Manchester, England). Microcon YM-30 for size exclusion filtration assay was obtained from Fisher Scientifics (Ottawa, ON). Precast 10% Tris·HCl mini gels were from BioRad Laboratories (Mississauga, ON). ProtoBlue Safe Colloidal Coomassie G-250 Stain was obtained from National Diagnostics (Atlanta, GA). All other reagents were purchased from BioShop Canada Inc. (Burlington, ON).
Site directed mutagenesis
The pGEX fusion construct of human α-TTP 2 was used as a template to create the following mutant constructs: F169A/D, F169A, I202A/D, V206A/D, M209A/D, F165D/M209D, I202/M209 and M264D. PfuTurbo DNA polymerase was utilized to generate these mutations following the QuickChange Site-Directed mutagenesis protocol as per the manufacturer s instructions (Stratagene) with 10% DMSO in the reaction mixture. All primers listed in the Table 1 were designed according to the guidelines suggested by the QuickChange protocol and were purchased from Sigma-Genosys (Oakville, Ontario). Plasmid DNA purification was achieved using Qiaprep Spin Miniprep kit. The intended mutations were confirmed by Roberts Research Institute (London, ON) with a custom primer 5′-ATGGCAGAGGCGCGATCCCAGC-3′. The double mutant constructs such as F165D/M209D and I202D/M209D were created by introducing F165D or I202D to the template with confirmed M209D mutation.
Table 1.
Oligonucleotide primers used for site-directed mutagenesis
| Mutants | Sequences of mutagenic primersa | Tm (°C) |
|---|---|---|
| F165A | 5′-CTTTGATCTGGAAGGTTGGCAGGCTTCTCATGCTTTTCAAATC-3′ | 82.4 |
| F165D | 5′-CTTTGATCTGGAAGGTTGGCAGGATTCTCATGCTTTTCAAATCA-3′ | 82.4 |
| F169A | 5′-CAGTTTTCTCATGCTGATCAAATCACTCCATCC-3′ | 75.3 |
| I202A | 5′-GATAAATGAACCAGTAGCTTTCCATGCTGTCTTTTCC-3′ | 74.9 |
| I202D | 5′-GATAAATGAACCAGTAGATTTCCATGCTGTCTTTTCC-3′ | 73.6 |
| V206A | 5′-CAGTAATTTTCCATGCTGCCTTTTCCATGATCAAACC-3′ | 78.2 |
| V206D | 5′-CAGTAATTTTCCATGCTGACTTTTCCATGATCAAACC-3′ | 76.0 |
| M209A | 5′-CCATGCTGTCTTTTCCGCGATCAAACCATTCCTGAC-3′ | 82.8 |
| M209D | 5′-CCAGTAATTTTCCATGCTGTCTTTTCCGATATCAAACCATTCCT-3′ | 79.5 |
| M264D | 5′-GGAATGGACAAATTTTATAGATAAGTCTGAAGATTATCTC-3′ | 68.8 |
A complementary pair of primers was used to create each mutant, only the sense sequences are listed. The nucleotides to be changed are underlined and shown in boldface type.
Protein expression and purification
Wild type α-TTP and various mutant constructs were transformed into E. coli BL21 (DE3) competent cells for protein expression. Typically, E. coli cultures were grown in 1L baffled flasks at 37 °C until OD at 600 nm was between 0.4 and 0.6. The bacterial culture was cooled to room temperature before induction with 400 μM IPTG. To improve the yield of soluble protein, the culture was grown at room temperature for 16 hours, except for I202A and V206D. I202A was induced at 15 °C overnight. Most of V206D remained in the inclusion bodies, therefore the induction was carried out at 10 °C and 100 μM IPTG. The cells were harvested, frozen and thawed two times before the long-term storage at −80 °C.
The protein purification protocol was essentially the same as previously described 9. Briefly, the E. coli pellet was lysed in lysis buffer containing 4 mg/mL lysozyme. Lysate became less viscous after addition of DNase I and RNase. Lysate was further clarified after 30 seconds sonication (3 × 10 second pulses). The resulting lysate was centrifuged at 40,000g for 30 min at 4 °C. Supernatant was subjected to glutathione affinity purification followed by thrombin protease digestion while protein was still on the gel. α-TTP was eluted and a final concentration of 0.5 mM PMSF was added to prevent further thrombin degradation. Purified protein was stored at 4 °C and used within a few days after purification. The ligand binding affinity of α-TTP mutants was determined as previously described 8 by titrating 0.2 μM protein with increasing concentrations of fluorescent ligand NBD-Toc. The emission spectra were recorded at 535 nm with excitation wavelength set at 466 nm. The dissociation constants Kd values were determined after fitting the fluorescent data to a one site binding equation using GraphPad Prism (version 5).
Lipid Vesicle Preparation
Acceptor lipid vesicles for ligand transfer assays contained 97 mol% bovine liver PC and 3 mol% quencher TRITC-PE, unless otherwise specified. For the size exclusion and dual polarization interferometric (DPI) assessment of protein association, 90% DOPC and 10% DOPS phospholipids vesicles were employed. The inclusion of PS made bilayer formation on the sensor surface much more efficient. Lipid mixtures were first evaporated under reduced pressure (0.1 mm Hg) to completely remove chloroform. The lipid mixtures were rehydrated for at least 30 min at room temperature before liposome preparation as described previously 9. Large unilamellar vesicles (LUVs) of mean diameter 150 nm were prepared with a Liposofast mini-extruder (Avestin, Inc). Small unilamellar vesicles (SUVs) were prepared, following the procedure described by Schroeder and Thompson 37 with minor modifications. The sonicated liposomes were centrifuged at 100,000g for 1.5 hours at 4 °C to remove titanium particles. Due to the presence of high sucrose content in SET buffer, the standard phosphorus assay38 was not possible. Instead, the phosphorous assay was performed initially with SET buffer without sucrose to confirm there is no loss of phospholipids due to both extrusion and sonication procedures. Typically, the same rehydrated lipid mixture (multilamellar vesicles or MLVs) was used to prepare SUVs or LUVs of same stock concentration. Lipid samples were prepared freshly for each experiment.
Transfer of NBD-Toc to lipid vesicles investigated by FRET
The rate of NBD-Toc transfer from α-TTP mutants to lipid vesicles was investigated using a fluorescence resonance energy transfer (FRET) assay as described previously9. Experiments were performed with a QuantaMaster-QM-2001-4 fluorometer (Photon Technologies International, Inc.) equipped with SPF-17 stopped flow device. Standard transfer experiments were performed by incubating 0.45 μM NBD-Toc with 4 μM α-TTP wild type or mutant protein in SET buffer (250 mM Sucrose, 100 mM KCl, 50 mM Tris, 1 mM EDTA, pH 7.4) for 15 minutes prior to mixing with 200 μM acceptor vesicles using the stopped-flow device. The final concentrations after mixing were 0.225 μM NBD-Toc, 2 μM α-TTP and 100 μM vesicles. All experiments were performed at room temperature. The fluorescence quench was monitored over time and normalized to the starting fluorescent intensity of NBD-Toc bound to α-TTP as 100%. The rate of NBD-Toc transfer was best fitted with a single exponential decay equation provided by Prism GraphPad software version 5.
Size exclusion assessment of membrane binding by α-TTP wildtype and mutant proteins
Binding of α-TTP to lipid vesicles was performed by a method similar to that published by Huang et al. 23 with modifications. Wild type or mutant α-TTP (31.5 μg) was incubated with increasing concentrations of SUVs or LUVs in SET buffer for 30 minutes at room temperature in a total sample volume of 200 μl. At the end of incubation, samples were loaded onto pre-wetted YM-100 Microcon filters. These samples were centrifuged at 10,000g in a bench top microcentrifuge for 20 minutes. The filters were washed once with 100 μl SET buffer, the flow-through fractions were collected and corresponded to the free protein that failed to bind to lipid vesicles. The microcon filters were then inverted and washed three times with 100 ml SET buffer containing 150 mM Triton X-100. Each wash was completed by centrifugation at 3000g for 5 minutes. The flow-through collected represented the lipid-bound α-TTP. The samples were then separated on precast 10% Tris HCl mini gels and fixed for 45 minutes in a fixing solution (40% EtOH, 10% acetic acid and 50% MilliQ H2O). Gels were stained overnight with ProtoBlue Colloidal Coomassie G-250 working solution (10-fold dilution in absolute ethanol). Gels were destained with 5% MeOH, 7% acetic acid for a few hours or until the complete removal of background staining. The intensity of each protein band was quantified with Scion Image software (developed by Scion Corp.).
Dual polarization interferometric assessment of membrane binding by α-TTP wildtype and mutants
The Analight Bio 200 dual polarization interferometer (Farfield Sensors Ltd, Manchester, UK) was used to study the association of α-TTP wild type and mutants to a planer lipid bilayer containing 10% DOPS and 90% DOPC. This technique has been characterized in detail as a valuable quantitative tool to study the binding interaction between proteins or between protein and lipid membrane environments in vitro 39–41. Due to the hypersensitive detection response toward the immobilized phase on the sensor surface, all solutions were degassed for at least 30 minutes to remove any fine bubbles prior to use. Running buffer (10 mM potassium phosphate dibasic, 137 mM NaCl, pH 7.4) was used to dilute α-TTP and prepare LUVs α-TTP was dialyzed in running buffer for 1 hour at 4°C. All experiments were performed at 20°C. The instrument was first equilibrated with running buffer for 1 hour. A fresh unmodified silicon oxynitride sensor chip was then installed. The instrument was calibrated with 80% ethanol in water (v/v). Once a stable baseline was established, extruded DOPS/DOPC (1:9) vesicles were passed over the sensor chip at a flow rate of 25 μl/min for 8 minutes. Vesicles ruptured and a stable bilayer was attained after approximately 5 minutes. The bilayer was allowed to stabilize for an additional 5 minutes and during this time any extra lipid that was deposited on the top of lipid bilayer was removed. α-TTP at the desired concentration was then injected over the lipid bilayer at 25 μl/min for 8 minutes. At the end of the injection, the change in the protein layer was monitored for an additional few minutes. To regenerate the sensor chip for another run, existing lipid and protein layers were stripped with 2% SDS followed by 50% isopropyl alcohol (150 μl). This procedure was repeated until the entire set of data was collected. The data from the first 450 seconds of α-TTP injection were resolved and fitted to a one site binding equation to calculate the maximum amount of protein deposited onto the lipid bilayer at each protein concentration. The maximum binding data (Bmax) were further analyzed according to a single exponential association equation to determine the binding affinity (Kd) of α-TTP mutants to the lipid bilayer. Since the Kd represents a half maximal value of protein that can stick to the membrane, we can quickly compare the lipid-binding abilities of mutant α-TTP forms by comparison of one or two concentrations where we would expect to see significant changes in adsorbed specific mass (i.e. injections at or near the Kd value).
Orientation of proteins in membranes
Structures of α-TTP (1R5L, 1OIZ) and other proteins from the CRAL-TRIO family (1AUA, 3B7N, 1O6U, 2Q3L) were taken from the Protein Data Bank 42. Models of α-TTP mutants were generated using QUANTA (Accelrys Inc) by substituting the corresponding residues and optimizing their side chain conformers in the “closed” (1R5L) and “open” (1OIZ) structures. The spatial orientation of membrane-associated proteins with respect to the lipid bilayer, the maximal penetration depths of protein residues in the hydrocarbon core (D) and transfer energies of proteins from water to the membrane (ΔGtransf) were optimized using a new version (2.0) of our Positioning of Protein in Membranes (PPM) method 11. The new anisotropic solvent model of the lipid bilayer, PPM 2.0, includes the following free energy components: (a) surface area-dependent contribution that describes solute-solvent interactions in the first solvation shell (van der Waals, H-bonding, and entropy of solvent); (b) long-range electrostatic energy of protein dipoles and ionized groups in a media, and (c) deionization penalty for ionizable groups in nonpolar environment. The first-shell contribution was described by linear dependencies of the energy on solvent bulk dielectric constant (ε), hydrogen bond acidity (α) and basicity (β) parameters. The electrostatic term was described by a linear dependence of the energy on the solvatochromic polarizability parameter π* for dipoles and by a modified Born equation for ions. The solubility properties of the fluid lipid bilayer were defined by profiles of several polarity parameters (α, β, ε and π*) and concentration of water along the membrane normal. The profiles were calculated for DOPC bilayer based on the published distributions of volume fractions of lipid components obtained by X-ray and neutron scattering43. Parameters α and β were corrected to reflect the presence of ~10% of negatively charged DOPS. The preferential solvation of protein groups by water and electrostatic interactions with lipid phosphates were also included in the model.
Activity of α-TTP in cultured cells
α-TTP activity in cells was measured by quantifying the α-TTP-induced secretion of vitamin E, modified after previous methods6,25. Human hepatocytes (HepG2/C3A; ATCC CRL-10741) were cultured in Dulbeco modified Eagle s medium (DMEM) supplemented with 10% fetal bovine serum on collagen-coated glass cover slips. Indicated α-TTP variants were generated by site-directed mutagenesis (QuickChange, Invitrogen), using wild-type α-TTP in the pCDNA3.1 vector as a template. Transient transfections were carried out using Fugene6 according to manufacturer protocol (Roche). NBD-tocopherol was complexed to serum lipoproteins44 as described earlier6. Thirty hours after transfection, cells were incubated with 5 μM serum complexed NBD-tocopherol for 16–18 hrs in humidified 10% CO2 incubator at 37°C. After 4 washes, cells were incubated in normal media for 4 hrs, during which time α-TTP-facilitated secretion takes place (2). After 3 washes in phosphate buffered saline (PBS), cells were fixed in 3.7% paraformaldehyde/PBS for 20 min at room temperature, washed 4 times with PBS, and permeabilized with 0.1% triton X-100 for 5 minutes. Nuclei were stained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) in 0.2% BSA/PBS for 5 minutes at room temperature. Mounted slides were observed on a Leica DM 4000B fluorescence microscope at 40× magnification. Fluorescence intensity was quantified in digitized images using ScionImage software and normalized to cell number in each field.
Supplementary Material
Acknowledgments
This work was supported by an NSERC Discovery Grant (J.A.), and award DK067494 from the National Institutes of Health (D.M.) and NSF DBI award 0849713 (A.L. and I.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Panagabko C, Morley S, Hernandez M, Cassolato P, Gordon H, Parsons R, Manor D, Atkinson J. Ligand specificity in the CRAL-TRIO protein family. Biochemistry. 2003;42:6467–74. doi: 10.1021/bi034086v. [DOI] [PubMed] [Google Scholar]
- 2.Morley S, Panagabko C, Shineman D, Mani B, Stocker A, Atkinson J, Manor D. Molecular determinants of heritable vitamin E deficiency. Biochemistry. 2004;43:4143–9. doi: 10.1021/bi0363073. [DOI] [PubMed] [Google Scholar]
- 3.Morley S, Panagabko C, Stocker A, Atkinson J, Manor D. Structure-function relationship in the tocopherol transfer protein. Vitamin E And Health. 2004;1031:332–333. doi: 10.1196/annals.1331.035. [DOI] [PubMed] [Google Scholar]
- 4.Hosomi A, Arita M, Sato Y, Kiyose C, Ueda T, Igarashi O, Arai H, Inoue K. Affinity for alpha tocopherol transfer protein as a determinant of the biological activities of vitamin E analogs. FEBS Lett. 1997;409:105–108. doi: 10.1016/s0014-5793(97)00499-7. [DOI] [PubMed] [Google Scholar]
- 5.Qian J, Wilson K, Nava P, Morley S, Atkinson J, Manor D. Intracellular localization of alpha-tocopherol transfer protein and alpha-tocopherol. Vitamin E And Health. 2004;1031:330–331. doi: 10.1196/annals.1331.034. [DOI] [PubMed] [Google Scholar]
- 6.Qian J, Morley S, Wilson K, Nava P, Atkinson J, Manor D. Intracellular trafficking of vitamin E in hepatocytes: the role of tocopherol transfer protein. J Lipid Res. 2005;46:2072–82. doi: 10.1194/jlr.M500143-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Morley S, Cross V, Cecchini M, Nava P, Atkinson J, Manor D. Utility of a fluorescent vitamin E analogue as a probe for tocopherol transfer protein activity. Biochemistry. 2006;45:1075–1081. doi: 10.1021/bi052271y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nava P, Cecchini M, Chirico S, Gordon H, Morley S, Manor D, Atkinson J. Preparation of fluorescent tocopherols for use in protein binding and localization with the alpha-tocopherol transfer protein. Bioorg Med Chem. 2006;14:3721–36. doi: 10.1016/j.bmc.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 9.Zhang WX, Frahm G, Morley S, Manor D, Atkinson J. Effect of bilayer phospholipid composition and curvature on ligand transfer by the alpha-tocopherol transfer protein. Lipids. 2009;44:631–41. doi: 10.1007/s11745-009-3310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 11.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–33. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct Biol. 2007;7:44. doi: 10.1186/1472-6807-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–5. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 14.Meier R, Tomizaki T, Schulze-Briese C, Baumann U, Stocker A. The molecular basis of vitamin E retention: Structure of human alpha-tocopherol transfer protein. J Molec Biol. 2003;331:725–734. doi: 10.1016/s0022-2836(03)00724-1. [DOI] [PubMed] [Google Scholar]
- 15.Min KC, Kovall RA, Hendrickson WA. Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: implications for ataxia with vitamin E deficiency. Proc Natl Acad Sci U S A. 2003;100:14713–8. doi: 10.1073/pnas.2136684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaaf G, Betts L, Garrett TA, Raetz CR, Bankaitis VA. Crystallization and preliminary X-ray diffraction analysis of phospholipid-bound Sfh1p, a member of the Saccharomyces cerevisiae Sec14p-like phosphatidylinositol transfer protein family. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:1156–60. doi: 10.1107/S1744309106041728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stocker A, Tomizaki T, Schulze-Briese C, Baumann U. Crystal structure of the human supernatant protein factor. Structure. 2002;10:1533–40. doi: 10.1016/s0969-2126(02)00884-5. [DOI] [PubMed] [Google Scholar]
- 19.Stocker A, Baumann U. Supernatant protein factor in complex with RRR-alpha-tocopherylquinone: A link between oxidized vitamin E and cholesterol biosynthesis. Journal of Molecular Biology. 2003;332:759–765. doi: 10.1016/s0022-2836(03)00924-0. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Lomize A, Jin KK, Carlton D, Miller MD, Jaroszewski L, Abdubek P, Astakhova T, Axelrod HL, Chiu H-J, Clayton T, Das D, Deller MC, Duan L, Feuerhelm J, Grant JC, Grzechnik A, Han GW, Klock HE, Knuth MW, Kozbial P, Krishna SS, Marciano D, McMullan D, Morse AT, Nigoghossian E, Okach L, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Weekes D, Xu Q, Hodgson KO, Wooley J, Elsliger M-A, Deacon AM, Godzik A, Lesley SA, Wilson IA. Open and closed conformations of two SpoIIAA-like proteins ( YP_749275.1 and YP_001095227.1) provide insights into membrane association and ligand binding. Acta Crystallographica Section F. 2010;66:1245–1253. doi: 10.1107/S1744309109042481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan MM, Temple BR, Phillips SE, Bankaitis VA. Conformational dynamics of the major yeast phosphatidylinositol transfer protein sec14p: insight into the mechanisms of phospholipid exchange and diseases of sec14p-like protein deficiencies. Mol Biol Cell. 2007;18:1928–42. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morley S, Cecchini M, Zhang W, Virgulti A, Noy N, Atkinson J, Manor D. Mechanisms of ligand transfer by the hepatic tocopherol transfer protein. J Biol Chem. 2008;283:17797–804. doi: 10.1074/jbc.M800121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H, Ball JM, Billheimer JT, Schroeder F. Interaction of the N-terminus of sterol carrier protein 2 with membranes: role of membrane curvature. Biochem J. 1999;344(Pt 2):593–603. [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J, Atkinson J, Manor D. Biochemical Consequences of Heritable Mutations in the alpha-Tocopherol Transfer Protein. Biochemistry. 2006;45:8236–42. doi: 10.1021/bi060522c. [DOI] [PubMed] [Google Scholar]
- 25.Arita M, Nomura K, Arai H, Inoue K. a-Tocopherol transfer protein stimulates the secretion of a-tocopherol from a cultured liver cell line through a brefeldin A-insensitive pathway. Proc Natl Acad Sci U S A. 1997;94:12437–12441. doi: 10.1073/pnas.94.23.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morley S, Thakur V, Danielpour D, Parker R, Arai H, Atkinson J, Barnholtz-Sloan J, Klein E, Manor D. The tocopherol transfer protein sensitizes prostate cancer cells to vitamin E. J Biol Chem. 2010 doi: 10.1074/jbc.M110.169664. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Federico A. Ataxia with isolated vitamin E deficiency: a treatable neurologic disorder resembling Friedreich’s ataxia. Neurol Sci. 2004;25:119–121. doi: 10.1007/s10072-004-0245-0. [DOI] [PubMed] [Google Scholar]
- 28.Manor D, Morley S. The alpha-tocopherol transfer protein. Vitam Horm. 2007;76:45–65. doi: 10.1016/S0083-6729(07)76003-X. [DOI] [PubMed] [Google Scholar]
- 29.Lomize AL, Reibarkh MY, Pogozheva ID. Interatomic potentials and solvation parameters from protein engineering data for buried residues. Protein Sci. 2002;11:1984–2000. doi: 10.1110/ps.0307002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Brink-van der Laan E, Killian JA, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta. 2004;1666:275–88. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Mesmin B, Drin G, Levi S, Rawet M, Cassel D, Bigay J, Antonny B. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 2007;46:1779–90. doi: 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- 32.Sackmann E. Physical basis of self-organization and function of membranes: physics of vesicles. In: Lipowsky R, Sackmann E, editors. Biological Membranes Architecture and Function. Vol. 1. Elsevier Science B.V, Garching; Germany: 1995. pp. 213–304. [Google Scholar]
- 33.Nicot AS, Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–9. doi: 10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai H. Molecular mechanisms of α-tocopherol transfer protein (α-TTP)-dependent α-tocopherol transfer in hepatocytes. FASEB J. 2006;20:LB44. [Google Scholar]
- 35.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5:835–41. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 36.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106:13541–5. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroeder F, Barenholz Y, Gratton E, Thompson TE. A fluorescence study of dehydroergosterol in phosphatidylcholine bilayer vesicles. Biochemistry. 1987;26:2441–8. doi: 10.1021/bi00383a007. [DOI] [PubMed] [Google Scholar]
- 38.Rouser G, Siakotos AN, Fleischer S. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1966;1:85–6. doi: 10.1007/BF02668129. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, Guo L, Ding JL, Ho B, Feng SS, Popplewell J, Swann M, Wohland T. Interaction of an artificial antimicrobial peptide with lipid membranes. Biochim Biophys Acta. 2009;1788:333–44. doi: 10.1016/j.bbamem.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Terry CJ, Popplewell JF, Swann MJ, Freeman NJ, Fernig DG. Characterisation of membrane mimetics on a dual polarisation interferometer. Biosens Bioelectron. 2006;22:627–32. doi: 10.1016/j.bios.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Cross GH, Reeves AA, Brand S, Popplewell JF, Peel LL, Swann MJ, Freeman NJ. A new quantitative optical biosensor for protein characterisation. Biosens Bioelectron. 2003;19:383–90. doi: 10.1016/s0956-5663(03)00203-3. [DOI] [PubMed] [Google Scholar]
- 42.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 43.Kucerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophys J. 2008;95:2356–67. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asmis R. Physical partitioning is the main mechanism of alpha-tocopherol and cholesterol transfer between lipoproteins and P388D1 macrophage-like cells. Eur J Biochem. 1997;250:600–7. doi: 10.1111/j.1432-1033.1997.0600a.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.