Abstract
Esa1, an essential MYST histone acetyltransferase found in the yeast piccolo NuA4 complex (picNuA4), is responsible for genome-wide histone H4 and histone H2A acetylation. picNuA4 uniquely catalyzes the rapid tetra-acetylation of nucleosomal H4, though the molecular determinants driving picNuA4 efficiency and specificity have not been defined. Here, we show through rapid substrate-trapping experiments that picNuA4 utilizes a non-processive mechanism, where picNuA4 dissociates from substrate after each acetylation event. Quantitative mass spectral analyses indicate that picNuA4 randomly acetylates free and nucleosomal H4, with a small preference for lysines 5, 8, and 12 over 16. Using a series of 24 histone mutants of H4 and H2A, we investigated the parameters affecting catalytic efficiency. Most strikingly, removal of lysine residues did not substantially affect the ability of picNuA4 to acetylate remaining sites, and insertion of an additional lysine into the H4 tail led to rapid quintuple-acetylation. Conversion of the native H2A tail to an H4-like sequence resulted in enhanced multi-site acetylation. Collectively, the results suggest picNuA4’s site selectivity is dictated by accessibility on the nucleosome surface, the relative proximity from the histone fold domain, and a preference for intervening glycine residues with a minimal (n + 2) spacing between lysines. Functionally distinct from other HAT families, the proposed model for picNuA4 represents a unique mechanism of substrate recognition and multisite acetylation.
Eukaryotic DNA is packaged in the nucleoprotein structure chromatin, the smallest component of which is the nucleosome core particle (NCP). Each core particle consists of 147 bp of DNA wrapped 1.7 times around an octamer of the four canonical histone proteins H4, H3, H2A and H2B (1, 2). Histones can be post-translationally modified on their N-terminal tail domains as well as their globular core domains. Numerous histone modifications have been identified, including methylation, phosphorylation, ubiquitination, sumoylation and acetylation (3). These modifications not only affect structural and intra-molecular interactions (i.e. octamer destabilization, tail-DNA association), but also serve as molecular signals whereby other proteins dynamically associate with chromatin. Mounting evidence suggests that this network of modifications coordinate gene expression via an epigenetic code, in which demarcations are read by enzyme complexes, resulting in altered phenotypes through differential gene activation or repression (4).
Histone acetyltransferases (HATs) are a class of enzymes that catalyze the transfer of the acetyl group from acetyl coenzyme-A (AcCoA) to the ε-amino group of lysines in histones. HAT-containing complexes are categorized into five distinct families according to the sequence homology of their catalytic domains and their shared substrate specificity. To date, the Gcn5-related N-acetyltransferase (GNAT) and MYST (MOZ, Ybf2/Sas3, Sas2 and Tip60) families have been the most well-characterized. Esa1 (KAT5), a MYST Tip60 homolog, remains the only essential acetyltransferase found in S. cerevisiae, where it is required for cell viability (5, 6), ribosomal gene transcription (7), DNA double strand break repair (8) and cell cycle progression (9). Esa1 can be co-purified in two evolutionarily conserved complexes, 1) a 1.3 MDa nucleosome acetyltransferase of histone H4 (NuA4) complex that contains 13 total subunits, and 2) a small, heterotrimeric complex dubbed piccolo NuA4 (picNuA4) that contains Esa1, Epl1 and Yng2 (8, 10). Recent studies have implicated NuA4 in locus-specific recruitment and transcriptional activation, resulting in targeted H4 and H2A acetylation both in vitro and in vivo (7, (11), while picNuA4 catalyzes non-targeted global H4 acetylation in yeast (12, 13).
The molecular determinants dictating substrate specificity for HAT enzyme complexes are still poorly understood. Much of the current information on substrate specificity and recognition originate from crystallographic studies describing Gcn5 catalytic domains bound with H3 tail peptide substrates (14–17). These findings suggested that the tail peptide sequence alone is sufficient for efficient recognition and catalysis by GNAT HAT active sites (18, 19), a supposition supported by the crystal structure of the MYST Esa1 catalytic domain (20). Consistent with this model, Utley et al. has shown that H4 peptides containing S1 phosphorylation can oblate efficient peptide acetylation by NuA4 (21). But recently, Berndsen et al. showed that histone tail peptides alone are surprisingly poor substrates for the MYST picNuA4 complex and that the histone fold domain of H4 is required for efficient nucleosomal recognition (22). Thus, picNuA4 appears to bind substrates in a bipartite manner in which both the histone tails and the histone fold domain are required for efficient acetylation.
Unlike Gcn5 acetyltransferases, which preferentially acetylate H3K14 10–100 times more efficiently than other candidate lysines (23), the picNuA4 complex rapidly tetra-acetylates the four sites on the H4 tail at comparable levels of efficiency (22). Here, we have investigated the molecular mechanisms that allow for the efficient tetra-acetylation of H4 by picNuA4. Our results indicate that picNuA4 utilizes a non-processive mechanism of acetylation for nucleosomal substrates, in which picNuA4 dissociates from substrate after each acetylation event. Rapid tetra-acetylation does not require sequential order of acetylation, as picNuA4 randomly acetylates H4 tail lysines (K5, K8, K12 and K16) within the nucleosome. Kinetic analyses of histone mutants suggest a specificity model where site preference is dictated by accessibility on the surface of the nucleosome, the relative proximity from the histone fold domain, and a preference for glycine residues with a minimal (n + 2) spacing between lysines.
EXPERIMENTAL PROCEDURES
Materials
Acetyl-Coenzyme A (AcCoA), dithiothreitol (DTT), Tris-Cl, benzamidine, P81 cellulose discs and other reagents were purchased from either Sigma-Aldrich or Fisher unless otherwise noted. SYPRO Ruby protein stain was purchased from Invitrogen, [3H]-Acetyl-CoA was obtained from Morevek, and Quikchange site-directed mutagenesis kits were purchased from Stratagene.
Enzyme expression and purification
The enzyme expression and purification conditions for both yeast and human picNuA4 complexes closely mimic those published (13). Briefly, a truncated yeast picNuA4 complex (containing Esa1, Yng2 (residues 1–218) and His-tagged Epl1 (residues 51–380)) or human picNuA4 complex (containing Tip60b, Ing3 (residues 1–300) and His-tagged Epc1 (residues 1–400)) were expressed using a polycistronic expression system developed in the laboratory of Dr. Song Tan (24). The complex was expressed in BL21(DE3) PlysS cells, grown in 2x YT media until OD600 = 0.4 – 0.8, and induced at 37 °C for 4 hours with 0.4 mM isopropyl-β-D-thigalactopyranoside (IPTG). Harvested cells were resuspended in 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM benzamidine, 1 mM β;-mercaptoethanol and 10 % (v/v) glycerol, sonicated on ice, and centrifuged for 20 min at 60,000 × g. Cell supernatant was loaded on a nickel-saturated 5 mL HiTrap HP chelating column (GE Healthsciences) and washed free of contaminants. picNuA4 was eluted using a linear gradient of 0 – 200 mM imidazole. Fractions containing the complex, as determined by SDS-PAGE, were pooled and dialyzed overnight against lysis buffer to remove imidazole. Concentrated fractions were further purified by size-exclusion chromatography on a XK-26 column packed with Sephacryl 200 (EMD Biosciences). Fractions were analyzed by SDS-PAGE to verify the integrity of the picNuA4 complex, concentrated, and stored in aliquots at −20 °C. Enzyme concentrations were determined by Bradford and verified with activity assay. Yeast picNuA4 complex was used in all reactions unless otherwise noted.
Purification of histones and NCPs
Recombinant Xenopus laevis core histones (H4, H3, H2A and H2B) were purified as referenced in (22). All point mutant and insertion/deletion histone constructs were made with QuikChange mutagenesis kits according to manufacturer instructions and were verified by sequencing. Protein concentrations were derived using published extinction coefficients (25). Purified core histones and mutants were reconstituted into nucleosome core particles (NCPs) using similar methods as published (25).
Triton-acidic acid-urea (TAU) gel electrophoresis
Gel matrices consisting of 7 M urea, 1 % (v/v) Triton X-100, and 5 % (v/v) acetic acid were made as outlined (26). All TAU gels were loaded with 1.3 μg substrate per lane, run at 10 mA for 22–70 hours and stained with Invitrogen SYPRO Ruby stain. Unless indicated otherwise, time course experiments were conducted using catalytic amounts of picNuA4 (0.15 – 1 μM), saturating amounts of substrate (NCP, histone, or mutant) and AcCoA, 50 mM Tris (pH 7.5), 1 mM DTT, and were quenched using 2 × TAU sample buffer. Limiting AcCoA experiments contained 1 μM picNuA4, 10 μM substrate (or 2 μM NCP) and increasing amounts of AcCoA in 50 mM Tris (pH 7.5) and 1 mM DTT. Reactions were quenched with liquid nitrogen after 5 minutes, dried in a speedvac, and resuspended in TAU sample buffer.
TAU Densitometry
A processing efficiency equation (1) was utilized to determine the efficiency of histone and nucleosomal substrates. SYPRO-Ruby stained TAU gels were normalized across all lanes and quantified by densitometry
| (1) |
where t1/2 describes the time (in seconds) required for half of the initial substrate (S0) to be converted to the fully-acetylated state (SAc) by picNuA4. Gels were imaged on an Epi Chemi II Darkroom platform and analyzed using Lab Works Software (UVP).
Radioactive filter binding assay
Reactions were conducted as outlined by Berndsen et al. (27). All reactions were conducted in 50 mM Tris (pH 7.5) and 1 mM DTT at 25 °C, with catalytic amounts of picNuA4 (0.1 – 1.0 μM), saturating amounts of AcCoA (50 – 200 μM) and saturating amounts of substrate (2 – 200 μM, depending on substrate). Saturation and time course experimental data were fit to the Michaelis-Menten (2) or integrated Michaelis-Menten (3) equations using Kaleidagraph software (Synergy Software). Kinetic parameters kcat, Km and kcat/Km were determined as outlined in (28).
| (2) |
| (3) |
Rapid quench flow experiments
A Hi-Tech RQF-63 apparatus was used to conduct substrate trapping experiments. To prepare for substrate trapping experiments, pre-steady state time points were quenched with 2 % (v/v) TFA, dried on phosphocellulose disks, and quantified by liquid scintillation (29). The product formed and rates were analyzed using Kaleidagraph software, to determine the flux value used for subsequent trapping experiments (Supplemental Figure 1). Substrate trapping reactions contained 1 μM picNuA4, 4 μM nucleosome substrate or 8 μM H4, and saturating amounts of AcCoA (40 μM), and were conducted in 50 mM Tris (pH 7.5) at 25 °C for 350 ms, a value slightly lower than the flux value (385ms). This ensured mono-acetylation took place prior to exposure to trapping agent (Supplemental Figure 1). The quench reservoir was filled with either 50 mM Tris, pH 7.5 (Mock Quench), 2 % (v/v) TFA (Chemical Quench), or 200 μM globular domain of H4 (gH4, residues 20–102) competitive inhibitor (Trap) in 50 mM Tris, pH 7.5. Reactions were manually stopped after 3 sec using 100 μL 2 % (v/v) TFA and concentrated with a speedvac. Reactions were run on 15 % SDS-PAGE gel, to separate H4 from other reaction constituents, after which H4 was excised from the gel and analyzed by liquid chromatography coupled to tandem MS (LC-MS/MS) using an Eksigent nanoLC coupled to Thermo LTQ linear ion trap.
Mass Spectrometry
MS/MS-based sequencing was used to determine both the site and relative ratio of picNuA4 enzymatic acetylation, essentially as described (30, 31). Briefly, picNuA4 acetylation reactions were quenched, chemically acetylated to generate fully acetylated peptides, and analyzed by LC-MS/MS. Acetylation sites were identified and quantified using peptide fragmentation within the MS/MS spectra (Supplemental Figure 2). Note that chemical acetylation added deuterated acetyl groups (+ 45 Da) that are easily distinguished from the enzymatic acetylation (+ 42Da, Supplemental Figure 2).
picNuA4 acetylation reactions were conducted in 50 mM Tris (pH 7.5), 1 mM DTT, with enzyme and substrate concentrations as indicated, quenched using 2 % (v/v) TFA, and run on 15 % SDS-PAGE gels. Gels were stained with Coomassie blue or SYPRO Ruby before histone bands were extracted from the gel. Gel bands were then chemically acetylated using deuterated acetic anhydride (Acros, 99.5% atom purity) and deuterated acetic acid (Acros, 98.5% atom purity) for 6 hours at 25 °C (31) followed by in-gel trypsin digestion at 37 °C for 18 hours (30). Extracted peptides were purified using OMIX C18 tips (Varian), dried in a speedvac, and resuspended in 0.1 % (v/v) formic acid for analysis by LC-MS/MS. LC-MS/MS conditions were as previously outlined (30) and acetylation sites were identified and quantified as described previously (31).
LC-MS/MS data was processed using Thermo Bioworks software. First, the histone peptides (Supplemental Figure 2A) were identified using Thermo Bioworks software by comparing the experimental MS/MS spectra to the theoretical human protein database (NCBI, non-redundant, downloaded February 2007), with enzymatic and chemical acetylation (+ 42 Da and + 45 Da, respectively), methionine oxidation (+16 Da), and cysteine carboxyamidomethylation (+57 Da) as variable modifications. Search parameters contained a mass accuracy tolerance of ≤ 2.0 amu for peptides and ≤ 1.0 amu for fragment ions. Next, the amount of picNuA4-mediated acetylation at each lysine residue was quantified by averaging the MS/MS spectra for each acetylation state and integrating the fragment ions peak areas corresponding to enzymatic or chemical acetylation species (Supplemental Figure 2C). Whenever possible, multiple independent peaks were integrated for each lysine. The percent picNuA4-mediated acetylation was obtained by dividing the intensity of the enzymatic acetylation by the total acetylated species for each lysine residue (31). All values, unless otherwise noted, indicate the average level of enzymatic, picNuA4-mediated acetylation on each site. Supplemental Figure 2D shows that site acetylation trends for independent experimental replicates correlate very well, with standard deviations ranging from 4 to 8 % for each site. Similar errors were observed for all MS experiments in this study, with representative data sets shown.
RESULTS
Nucleosomal structure restricts picNuA4 acetylation of histones
Previous work has shown that the NuA4 complex preferentially acetylates histones H4 and H2A on oligonucleosomes (10), while chromatin immunoprecipitation studies have linked Esa1 activity to the acetylation state of H4, H2A and H2B in vivo (32). Recombinant Esa1 alone is able to efficiently acetylate individual free H4, H3 and H2A histones in vitro (5, 9). It remains unclear, however, how the structure of the nucleosome substrate dictates site selectivity.
To investigate the impact of substrate structure on site selection by picNuA4, we characterized the kinetics of individual histone and nucleosome core particle acetylation using triton-acetic acid-urea (TAU) gel electrophoresis, a method that separates proteins based on mass and charge. Thus, the picNuA4-catalyzed acetylation of each histone can be resolved as a function of time, allowing the relative efficiency of each acetylation event to be measured. Substrate processing was quantified by TAU densitometry, in which we utilized an efficiency constant, t1/2, to quantify the time required for half of the substrate to be converted to fully acetylated product. To determine the identity of each modified histone band, intermediate acetyl-species were excised and analyzed by MS.
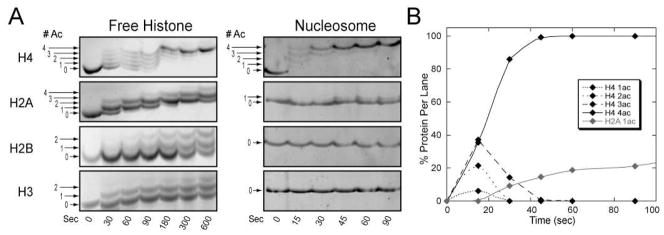
Figure 1A depicts time course reactions for both free histone and reconstituted nucleosomal substrates under saturating substrate conditions. Our analyses indicate that free H4 is tetra-acetylated in a highly efficient manner, and that intermediate acetyl-species (mono-, di- and tri-acetylated) are formed prior to the occurrence of tetra-acetylated forms (Figure 1A). TAU densitometry and LC-MS/MS confirmed that all four sites on free H4 (K5, K8, K12 and K16) are completely acetylated by 600 s, exhibiting a processing efficiency of t1/2 = 72.8 ± 12.2 s (Figure 1A and data not shown). picNuA4 can also tetra-acetylate free H2A (K5, K9, K13, K15), though the pattern of acetylation indicates that only 2 sites are processed efficiently (Figure 1A and data not shown). Thus, free H2A exhibits a significantly higher t1/2 (> 600 s) and is tetra-acetylated at least 8-fold slower than free H4. In contrast to H4 and H2A, the picNuA4 complex can only diacetylate free H3 (K9, K14) and H2B (K16), as seen in Figure 1A and data not shown. TAU acetylation patterns are consistent with only 1 preferred acetylation site for free H3 and H2B substrates, and neither substrate underwent complete mono- or di-acetylation under these conditions (Figure 1A). Collectively, free H3 and H2B exhibit very inefficient processing, with a lower limit of t1/2 > 1500 s for both substrates.
Figure 1. Structure of nucleosome core particle restricts picNuA4 histone acetylation.
A) Free histone and nucleosome core particle (NCP) acetylation by picNuA4 was resolved on SYPRO Ruby stained TAU gels, in which increasing levels of acetylation (0 – 4 ac) are formed as a function of time. B) Graph depicts densitometry analysis of each acetylated species from the nucleosome TAU gel (Figure 1A) plotted as total percent acetylated species per lane. Reaction conditions: 200 nM picNuA4, 200 μM AcCoA, 20 μM histone; 30 nM picNuA4, 200 μm AcCoA, 2 μM nucleosome, both in 50 mM Tris (pH 7.5) and 1 mM DTT. TAU gel time course studies are representative of at least 3 independent experiments.
The above TAU analyses were validated on a more physiological substrate, the nucleosome core particle. Using a salt (NaCl) dialysis method, mononucleosomes were reconstituted from the four core histones and palindromic 146-bp fragment from the human α-satellite DNA (25). Because free histones are purified from recombinant bacterial expression system, reconstituted mononucleosomes are an ideal homogenous substrate that lack post-translation modifications. Nucleosomal H4 exhibits similar acetylation kinetics compared to free H4 (Figure 1A), as both substrates are rapidly tetra-acetylated and display mono, di, and tri intermediate states that exist at near-equivalent levels under early time points. These results indicate that each acetylation event occurs with similar levels of efficiency, and are in good agreement with previous kinetic findings which show that the efficiency of initial acetylation (kcat/Km) and total average efficiency (kcat/Kavg) are comparable for both H4 and nucleosome substrates (22). TAU densitometry indicates that nucleosomal H4 (t1/2 = 17.2 ± 2.7) is tetra-acetylated much more efficiently than free H4 (4.2 ± 1.0 fold faster, Figure 1A), consistent with previously published kcat/kavg values for H4 and nucleosome substrates (22). Our MS analyses confirm that picNuA4 site selectivity for nucleosomal H4 is identical to free H4 in solution (K5, K8, K12 and K16, data not shown). In contrast to H4, nucleosomal H2A is restricted to mono-acetylation at K5 within the nucleosome (Figure 1A, data not shown), and displays slower processing efficiency (t1/2 > 90 s). Densitometry analyses indicate that nucleosomal H4 is multiply acetylated prior to significant acetylation of H2A (Figure 1). Surprisingly, we find that neither H3 nor H2B are acetylated in the nucleosome. Collectively, these findings are consistent with a model in which the structure of the core particle restricts access of picNuA4 to H3, H2B and all sites on H2A, except for K5.
picNuA4 utilizes a dissociative mechanism of acetylation
TAU gel analyses indicate that tetra-acetylation of nucleosomal H4 proceeds in a highly efficient manner (Figure 1A), in which the acetylated states (mono-, di-, tri- and tetra-) appear at near-equivalent rates of formation (Figure 1). To explain the efficiency of H4 tetra-acetylation by picNuA4, a processive-like and/or cooperative mechanism of acetylation has been proposed (21, 22, 33). With a processive model, the enzyme remains bound to the histone substrate for multiple rounds of acetylation prior to dissociation. In contrast, a dissociative mechanism involves independent binding events in which the enzyme dissociates from the substrate after each round of catalysis.
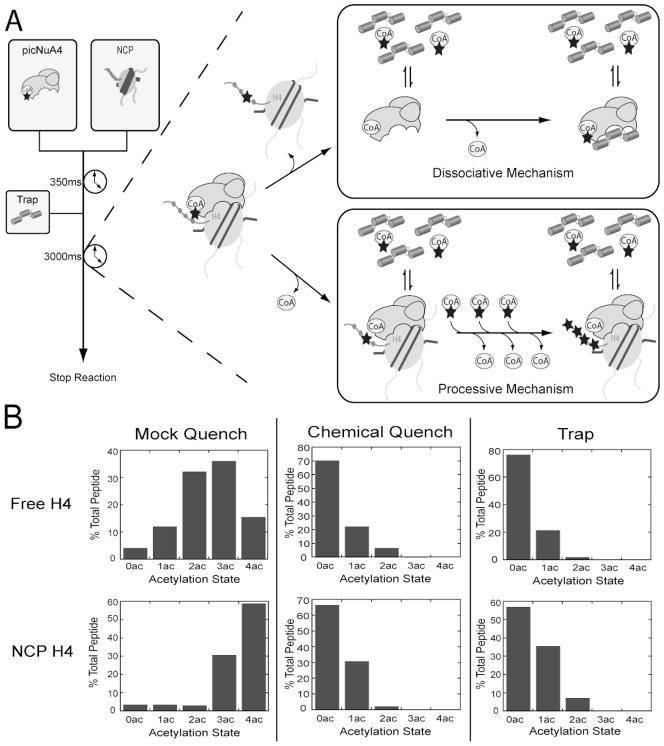
To determine the mechanism of H4 acetylation by picNuA4, we utilized substrate trapping methodology, an effective approach used to study DNA polymerase processing (34). Reactions are conducted in the presence of substrate mimics, which “trap” free enzyme and prevent re-association of the enzyme to substrate (34–36). Here, we have adapted this method to measure the total number of acetylation cycles that take place after a single binding event, by preventing dissociated picNuA4 from binding new or previously acetylated substrate. A rapid quench-flow apparatus was used to rapidly introduce pre-equilibrated picNuA4 and AcCoA with H4 or nucleosome substrates (Figure 2A). Reactions were carried out for 350 ms, a time point experimentally determined to allow the formation of not more than 1 acetylation event (Supplemental Figure 1), after which a trapping agent was introduced (Figure 2A). We have utilized a tailless H4 construct for substrate trapping (H4 residues 20–102, hereafter gH4), which has been shown previously to competitively inhibit picNuA4 with an inhibition constant of 1.5 ± 0.6 μM (22). After addition of the trapping agent, reactions were allowed to continue for 3 seconds and were then manually quenched by 2 % (v/v) TFA.
Figure 2. Substrate trapping indicates picNuA4 dissociates after each acetylation event.
Substrate trapping methodology allows for the determination of the total number of acetylations taken place per binding event. A) Rapid quench flow was used to rapidly mix pre-equilibrated picNuA4 and AcCoA with H4 or nucleosomal substrate. Reactions were carried out for 350 ms and quenched with buffer (50 mM Tris, pH 7.5), 2 % (v/v) TFA, or competitive inhibitor trap (200 μM gH4, ~100x excess Ki). Reaction conditions: 1 μM picNuA4, 40 μM AcCoA, 8 μM H4 (or 4 μM NCP), 50mM Tris (pH 7.5) and 1 mM DTT. B) Graphs depict percent of total peptide per acetylation state as determined from MS analyses of rapid quench flow samples. MS results were analyzed with Thermo Bioworks and filtered with peptide probability < 0.01, Xcorr vs. charge +1 ≥ 2.0, +2 ≥ 2.7, +3 ≥ 3.5 and with ΔCn ≥ 0.2. Data sets are representative of n = 2 for both H4 and NCP experiments. Substrate trapping MS experimental replicates differed no more than 5% (H4) and 7% (NCP) per acetylation state.
To confirm that picNuA4 is able to tetra-acetylate free or nucleosomal H4, we omitted the gH4 trap and manually quenched reactions after 3 seconds. A quantitative MS method was employed (discussed below) to confirm that picNuA4 tetra-acetylates both H4 and nucleosomal H4 in the absence of gH4 and under the conditions of the experiment (Mock Quench, Figure 2B). To determine the number of acetylations formed when a chemical denaturant was used, gH4 was replaced with 2 % (v/v) TFA. These reactions yielded predominantly unacetylated and mono-acetylated H4, as expected (Chemical Quench, Figure 2B). Substrate trapping reactions conducted in the presence of gH4 resulted in similar mono-acetylated species as that observed in the chemical quench reactions for both H4 and nucleosomal H4 (Trap, Figure 2B). These findings suggest that picNuA4 dissociates from both H4 and nucleosomal H4 after the formation of mono-acetylated product. To ensure substrate acetylation states were a product of picNuA4 activity, we conducted reactions in which enzyme or AcCoA were omitted (Supplemental Figure 3). As expected, both control reactions did not contain acetylated species. Taken together, these results are consistent with a non-processive mechanism for picNuA4 acetylation of free or nucleosomal H4. The rapid quenching results are in good agreement with TAU observations (Figure 1A), as the clear formation of mono-, di-, and tri-acetyl-species are consistent with a dissociative mechanism of acetylation.
picNuA4 randomly acetylates free and nucleosomal H4
NuA4 complexes can acetylate K5, K8, K12, and K16 of H4 both in vivo and in vitro (10, 21, 32, 33, 37, 38); however, the order of acetylation and the effect of the nucleosome structure on site specificity for picNuA4 have not been elucidated. The TAU progress curves presented in Figure 1 suggest that picNuA4 is able to rapidly tetra-acetylate all four sites on free H4 and nucleosomal H4 with similar efficiently as the first acetylation event (Figure 1A, see also (22)). It is possible that the rapid tetra-acetylation of H4 requires a sequential order of acetylation that might implicate a cooperative model.
To investigate the order of acetylation by picNuA4 on free H4 and nucleosomes, we have adapted a previously published MS methodology that allows for the direct quantification of lysine acetylation on histone tails (31). Aliquots from acetylation reactions were quenched, chemically acetylated using deuterated acetic anhydride, and in-gel digested. MS/MS fragmentation of isotopically-labeled peptides yield unique fragment ions that reveal both identity and quantification of acetylation at each lysine position. Here, to capture various acetylation states, different limiting amounts of AcCoA were added to acetylation reactions, resolved by TAU electrophoresis, and analyzed by LC-MS/MS (Figure 3A). Our results indicate that picNuA4 can individually acetylate all four sites on free H4 under limiting AcCoA conditions, consistent with random first site acetylation (see mono-acetylated states, Figure 3B). The four sites on free H4 are not kinetically equivalent, however, as K5, K8 and K12 were preferred over K16 by at least 3-fold (Figure 3B, Supplementary Figure 2D). Similarly, we find that the first site of acetylation for nucleosomal H4 was also random (Figure 3C), and displayed similar site preference compared to free H4. Interestingly, we note that site preference for K5/8/12 over K16 is also reflected in the di- and tri-acetylated species of the nucleosome (Figure 3C).
Figure 3. picNuA4 randomly acetylates free and nucleosomal H4.
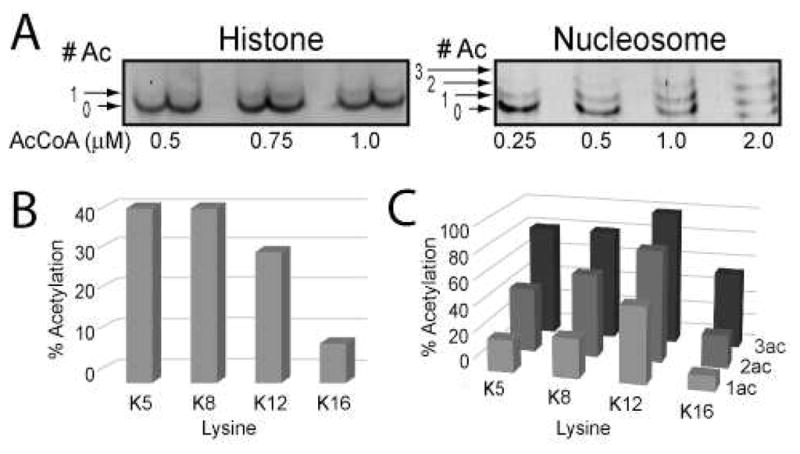
A) picNuA4 acetylation reactions, in which limiting amounts of AcCoA were added to reactions and visualized by SYPRO Ruby stained TAU electrophoresis. Acetylation bands were excised, deuterated with acetic anhydride and analyzed by LC-MS/MS. Graphs depict percent picNuA4-acetylated peptide in free H4 (B), and H4 within the NCPs (C), plotted by lysine position and acetylation state. Reaction conditions: 1 μM picNuA4, 10 μM H4 or 2 μM NCP in 50 mM Tris (pH 7.5) and 1 mM DTT. Data sets are representative of n = 3 for H4 and n = 1 for NCP experiments. MS experimental replicates differed no more than 6% per acetylation site for H4 (see Supplemental Figure 2D).
To investigate whether free H4 tetra-acetylation proceeds through a fully random mechanism, reactions were performed using saturating amounts of AcCoA and the levels of acetylation at each site were determined during the time-course for complete acetylation. Among the di- and tri-acetylated species, all four sites (K5, K8, K12 and K16) were represented in the MS analyses, though acetylation levels at K16 were ~ 2.5-fold lower than other sites (Supplemental Figure 4). The combined results (Figure 3B-C, and Supplemental Figure 4) are consistent with fully random tetra-acetylation of both free and nucleosomal H4 with a slight bias against K16.
Multiple lysine residues are dispensable for efficient acetylation
Having established that picNuA4 randomly acetylates all four sites on the H4 tail (Figure 3, Supplemental Figure 4), we sought to investigate the importance of individual lysines on recognition and catalysis. A series of arginine and alanine substitutions were generated at all four lysines on the H4 tail and analyzed for acetylation efficiency by measuring the steady-state kcat/Km values. A comparison of the kcat/Km values indicates that acetylation efficiency at the first site is not substantially affected by the loss of other lysines, as the most severe substitution (K5R) yielded a modest 3.5-fold decrease compared to native H4 (Figure 4). The severity of individual tail substitutions correlates well with MS trends (Figure 3B, Supplemental Figure 4), as the more N-terminal K5, K8, and K12 tail mutants exhibit slightly lower kcat/Km values compared to K16. All mutants exhibited a decrease in both kcat/Km and kcat values, with no significant effect on Km values. Collectively, these findings indicate that the charge of the histone tail does not contribute substantially to recognition, as K→R mutants are consistently within 2-fold of K→A mutants (Figure 4). MS analyses demonstrated that site preference is not significantly impacted by lysine mutation, as positions K5, K8, and K12 were consistently preferred over K16 acetylation (Supplemental Table 1). To test if multiple lysine mutations would result in lowered affinity, we characterized several double arginine and alanine tail mutants, all of which exhibit modest 4-fold decreases in kcat/Km values (Supplemental Figure 5). These data suggest that neutralization of charge on the H4 tail does not substantially affect the ability of picNuA4 to catalyze rapid multiple acetylations of the H4 tail, and are consistent with the primary H4 binding interaction residing proximal to the histone tail domain (22). picNuA4 prefers a minimal lysine spacing of (n + 2). Previously, we have shown that the deletion of the H4 histone fold domain results in a 1000-fold decrease in acetylation efficiency on the remaining tails (22). The mutational studies described above are consistent with the H4 histone fold domain being the high-affinity binding site for picNuA4, as substitution of lysine residues within the tail resulted in no more than a 4-fold loss of acetylation efficiency at the remaining lysine residues (Figure 4, Supplemental Figure 5). Next, the effects of tail length, spacing between lysines, and primary sequence on acetylation efficiency were determined. We hypothesized that the conserved (n + 2 or 3) register between lysine residues in the H4 tail sequence was an important molecular determinant. A small library of H4 and H2A mutants was constructed in which intervening residues were altered, and these mutants were characterized by TAU gel electrophoresis (Figure 5A and Supplemental Figure 6A). Histone mutants that displayed significantly altered acetylation kinetics (t1/2 values) compared to WT were reconstituted in nucleosomes for validation (Figure 5).
Figure 4. Multiple individual tail lysine residues are dispensable for efficient acetylation.
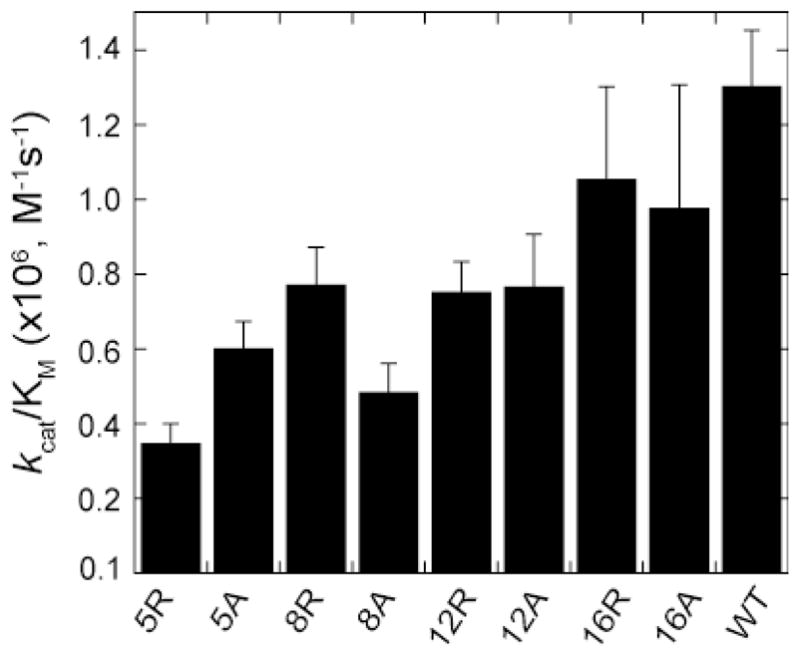
Independent saturation curves were conducted using filter binding assays on tail lysine mutants. Data was fit to the Michaelis-Menten equation in Kaleidagraph to determine the specificity constant, kcat/Km. All reactions contained 50 nM picNuA4, 20 μM histone substrate, 100 μM AcCoA, 50 mM Tris (pH 7.5) and 1 mM DTT. Each data point represents an average of 3 or more assays with error bars representing one standard deviation.
Figure 5. picNuA4 prefers tail lysines with a minimal (n + 2) spacing.
Histone mutants with altered lysine spacing show defects or gain of function in picNuA4 processing, as seen by SYPRO Ruby-stained TAU gels (A) and filter binding assays (B, C). A) TAU gels of free histone and nucleosomal substrates of H4 and H2A. Reaction conditions: 150 nM picNuA4, 200 μM AcCoA, 20 μM histone or 30 nM picNuA4, 200 μM AcCoA, 2 μM nucleosome in 50 mM Tris (pH 7.5) and 1 mM DTT. Data is representative of at least 3 independent histone experiments and 2 independent experiments for nucleosomal substrates. B) picNuA4 time course studies utilized filter binding assays to investigate the amount of product formed versus time for free WT H4 (●), H4313-14 (□), and H4GGAKins (◇). C) Graph depicts time courses for free WT H2A (●) and H2AGGins (◇) spacing mutant. A representative graph is shown from one of three independent experiments for all time course reactions. Data points were fit to the integrated form of the Michaelis-Menten equation. Reactions contained 100 nM picNuA4, 20 μM histone substrate, 200 μM AcCoA, 50 mM Tris (pH 7.5) and 1 mM DTT.
TAU analyses revealed that picNuA4 efficiently acetylates lysine residues that fulfill a minimal (n + 2) spacing requirement within the tail (Figure 5). To examine the shortest spacing register required for efficient acetylation, we deleted two glycine residues from the H4 tail (H4313-14), altering the lysine spacing to (n + 1). This mutant exhibited significant processing defects both free and in the nucleosome, with t1/2 values ~2-fold slower than free H4 and ~4.8-fold slower than nucleosomal H4 (Supplemental Figure 6B). TAU acetylation patterns indicate that a significant amount of the mutant remains stalled at the tri-acetylated state under these conditions (Figure 5A). Time courses of free H4313-14 acetylation confirm only 3 sites are acetylated efficiently (Figure 5B), and MS analyses indicate that 94 % of the histone contained only K5, K8 and K12 acetylation (data not shown). To test the effect of inserting an additional spacer residue, two mutants were prepared in which a glycine or glutamine were inserted at position 13 of the native tail sequence (H4G13ins, H4Q13ins). Densitometry confirmed that mutant t1/2 values were within error of free WT H4, indicating that a (n + 4) register does not impair processing significantly (Supplemental Figure 6B). Taken collectively, our findings suggest that picNuA4 prefers tail lysines with a minimal (n + 2) spacing, though longer registers are accommodated by picNuA4.
The H4 tail contains numerous evolutionarily conserved glycine residues that exist between lysines. To investigate the importance of glycine spacers between substrate lysines, we mutated glycine 13 to an alanine, glutamine or arginine residue and measured the t1/2 values of tetra-acetylation (Supplemental Figure 6). H4G13A exhibited a modest decrease in efficiency (t1/2 = 152.7 ± 14.9 s) compared to WT H4 (t1/2 = 110.6 ± 9.5 s), while H4G13Q and H4G13R were tetra-acetylated at similar rates (t1/2 = 192 ± 11.3 s, and 180 ± 12.4 s, respectively) and were 1.6–1.7-fold slower than with WT H4. Next, we substituted both intervening glycine residues (G13 and G14) to alanine and determined the t1/2 of tetra-acetylation. The resulting H4G13,14A mutant exhibited ~4-fold decrease in processing efficiency (t1/2 = 468.3 ± 53.1 s) with substantial stalling at the tri-acetylation state. Collectively, these results demonstrate a preference for intervening glycine residues proximal to tail lysines, and suggest that glycine’s small size and conformational flexibility contribute to efficient multi-site acetylation of the H4 tail.
Our findings suggest that picNuA4 could accommodate altered tail lengths as long as a minimal (n + 2) register is maintained. To test this, a deletion mutant was made in which residues 6–8 were deleted (H436-8) and thus contained only 3 acetylatable sites with (n + 3) spacing. TAU analysis confirmed that this mutant was efficiently acetylated on 3 sites and exhibits very efficient processing (t1/2 = 43.3 ± 3.8) compared to WT H4 (Supplemental Figure 6). To investigate whether picNuA4 can accommodate longer tails, we constructed a H4 tail mutant in which the sequence GGAK was inserted at position 13 of H4, lengthening the tail by 4 amino acids (H4GGAKins). This mutant contained an additional lysine residue, and maintained a (n + 3) register with small aliphatic spacers. TAU analysis clearly showed that the H4GGAKins mutant undergoes robust acetylation both free and in the nucleosome, in which 5 lysines are acetylated with similar t1/2 values compared to WT H4 (Figure 5A, Supplemental Figure 6B). Consistent with this, reaction time courses confirmed 5 sites are efficiently acetylated for the free H4GGAKins mutant (Figure 5B).
To validate these molecular requirements on a non-H4 substrate, we constructed an H2A mutant in which two glycine residues were inserted at position 14 of the native tail sequence, providing a (n + 3) register between K13 and K15 (Figure 5A). TAU analysis demonstrated that free H2AGGins mutant displays robust product formation compared to free WT H2A (Figure 5A), and exhibits a similar t1/2 (83.3 ± 14.7 s) compared to WT H4 (t1/2 = 100.9 ±7.8 s, Supplemental Figure 6B). This gain-of-function was confirmed by time course analyses, which revealed that 4 sites are acetylated on H2AGGins compared to only 2 in WT H2A (Figure 5C). Interestingly, when reconstituted into the nucleosome, this gain-of-function is not recapitulated, as the H2AGGins mutant displays similar acetylation kinetics compared to WT (Figure 5A), and MS analyses confirmed only K5 acetylation was observed.
DISCUSSION
Knowledge of HAT substrate specificity has largely come from crystal structures of catalytic domains in complex with small peptide substrates (14–20). These studies have provided evidence that active-site residues participate in multiple interactions with peptide substrates, leading some to postulate that primary sequence plays the major role in substrate recognition (14–16). Such conclusions may have limited interpretive value and may require validation with more complex chromatin substrates. Indeed, other evidence suggests that HAT complexes show stark differences in substrate specificity compared to recombinant HAT catalytic domains. For example, Gcn5p is able to poly-acetylate free H3 but only mono-acetylates H3 within the nucleosome (39). When in the context of the SAGA complex, however, Gcn5 shows enhanced H2B nucleosomal acetylation (40, 41). Thus altered specificity appears to be dictated in part by subunit-substrate interactions, as well as the structure of the nucleosome.
Here, we investigated the mechanism of rapid H4 tetra-acetylation and the molecular determinants of site-selectivity by the MYST family HAT complex picNuA4. The specificity of picNuA4 is controlled in part by the inherent nucleosome structure, as a variety of free histones are multiply acetylated, but site accessibility is restricted to H4 tetra-acetylation and H2A mono-acetylation within the nucleosome (Figure 1A). The unique specificity of picNuA4 for nucleosomal H4 requires the solvent-accessible histone fold domain of H4 (22), which likely serves as the primary binding interaction on the nucleosome. Our findings further support this model (Figure 4, Supplemental Table 1, Supplemental Figure 5), and the TAU results suggest that H4 undergoes multiple rounds of acetylation prior to H2A, though H2A mono-acetylation is processed much less efficiently (Figure 1A). These findings suggest that picNuA4 might bind at the H4 histone fold domain and acetylate H2A K5. In this model, most sites on H2A are sterically occluded by the nucleosome structure, and only the most N-terminal site can be accessed by the picNuA4 active site. Consistent with this, the nucleosomal H2AGGins mutant did not demonstrate enhanced acetylation kinetics compared to WT H2A (Figure 5A). Collectively, these findings indicate that interaction with the globular region of the NCP restricts substrate access to those lysines proximally located on the same face of the nucleosome.
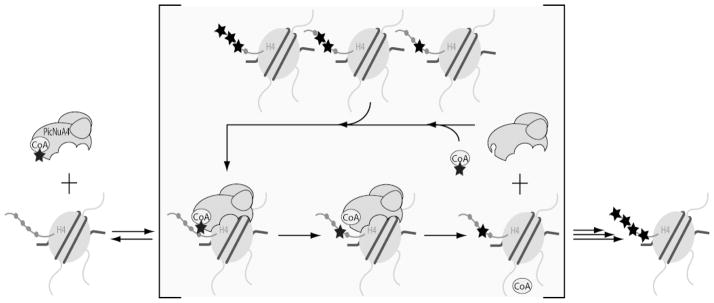
The efficient tetra-acetylation of nucleosomal substrates has been proposed to involve a processive or cooperative mechanism of acetylation (21, 22, 33). Here, we present detailed kinetic analyses that strongly suggest picNuA4 does not utilize a processive mechanism of acetylation, but rather dissociates from nucleosomes after each acetylation event (Figure 6). The ability of the substrate trap (gH4) to essentially stop the acetylation of nucleosomes (Figure 2B) as well as the evidence of intermediate acetyl-species formed during acetylation reactions (Figures 1–3, 5A) supports a dissociative mechanism of multiple acetylations (Figure 6).
Figure 6. Proposed dissociative model of nucleosome acetylation by picNuA4.
Diagram describes several kinetic steps required for recognition and processing of nucleosomal substrates by picNuA4. After binding AcCoA, picNuA4 binds the H4 histone fold domain while the active site effectively scans the histone tail for lysine residues surrounded by small, uncharged aliphatic residues. Preferred sites of acetylation maintain a minimal (n + 2) register between lysines. Upon catalysis, picNuA4 releases both the acetylated product and CoA before the next cycle of catalysis.
Utley et al. showed that NuA4 is able to acetylate H4 tail peptide sequences regardless of the number of pre-acetylated lysines (21), leading the authors to suggest that acetylation may have a cooperative effect on subsequent reactions. Our findings do not support an obvious cooperative component to multi-site acetylation. The H4 K→A mutants, which mimic the loss of charge similarly to acetylated lysine, show no increase in Km values compared to WT H4 (Figure 4, Supplemental Figure 5). Additionally, analysis of several H4 K→Q mutants did not reveal increased Km values (data not shown). Most importantly, the fact that picNuA4 displays the ability to randomly acetylate all four lysines on H4 (Figure 3, Supplemental Figures 2, 7) argues against a cooperative model where a sequential order of acetylation would be predicted. We propose that the efficiency with which picNuA4 tetra-acetylates nucleosomal H4 is due to its high affinity for nucleosomes (Km value < 190 nM) (22), and because the primary binding site (H4 histone fold domain) lies proximal to the sites of acetylation.
Using TAU and quantitative MS/MS analyses (Figure 3, Supplemental Figures 4, 7), we demonstrated that picNuA4 does not exhibit a preferred order of acetylation, and can mono-acetylate K5, K8, K12 and K16 on free and nucleosomal H4. Random first site acetylation may provide an important physiological function, as it would allow the enzyme to catalyze multiple cycles regardless of pre-existing tail modifications. The H4 tail is the site of numerous modifications (3, 42–45), some of which could act antagonistically to rapid sequential tetra-acetylation. Indeed, H4 S1 phosphorylation has been shown to decrease efficient peptide acetylation by NuA4 (21). Our findings suggest that alterations in lysine modification status have minimal effect on subsequent acetylation events, allowing for rapid acetylation to occur during stimulated chromatin remodeling (Figure 4, Supplemental Figure 5). Bird et al. have shown that NuA4 activity is essential for DNA double-strand break repair (8, 46, 47), while Downs et al. reported that NuA4 localization at sites of DNA repair results in rapid H4 acetylation leading to critical changes in chromatin structure (47). Interestingly, the Tip60-containing human picNuA4 complex is also able to individually acetylate all four sites on free H4 (Supplemental Figure 7), suggesting a conserved function for this essential HAT family.
Though picNuA4 randomly acetylates all four sites on H4, it exhibits a significant preference for K5, K8, and K12 over K16 on free and nucleosomal substrates (Figure 3B, Supplemental Figures 2D). Our findings are in good agreement with previously published in vitro studies using recombinant Esa1, where free enzyme showed a marked preference for K5, K8, and K12 over K16 on free H4 (5, 9). This specificity appears to be a hallmark of many MYST HATs, as HBO1 prefers K5, K8 and K12 over K16 in vitro (48), as does the human picNuA4 complex containing Tip60 (Supplemental Figure 7). Within the nucleosome, picNuA4 shows a subtle preference for K12 acetylation (Figure 3C), consistent with a recent study linking Esa1 activity to K12 acetylation in vivo (49). Moreover, Vogelauer et al. have shown that esa1ts (temperature sensitive) cells display decreased K5ac, K8ac and K12ac acetylation at non-permissive temperatures, while K16ac is largely unaffected (38). Collectively, these studies underscore the connection between Esa1 activity and global K5, K8, and K12 acetylation in vivo. That K16 is the poorest site is not functionally surprising, as Esa1 is not responsible for bulk K16 acetylation in S. cerevisiae, where it is regulated by the opposing activities of Sas2 and the HDAC Sir2 (50–52). The crystal structure of the nucleosome shows that residues 16–25 on H4 can interact with the H2A-H2B dimer interface (2), possibly interfering with access to K16.
Our findings strongly support a bipartite model of substrate recognition for picNuA4, as all point mutations analyzed here exhibited no more than a 4-fold loss in acetylation efficiency (Figure 4–5, Supplemental Figure 5–6). Because the primary site of picNuA4-histone interaction occurs within the histone fold domain, alterations to the primary sequence that slightly increase or decrease the conformational entropy of the tail do not dramatically affect the overall binding and acetylation efficiency. Analyses of 24 different histone variants suggest picNuA4 prefers glycine residues between acetylation sites (Figure 5, Supplemental Figure 6). This conclusion is supported by the x-ray structure of the Esa1 catalytic domain (20), which suggests that bulky intervening amino acids are poorly accommodated in the active site. Together, the results suggest that glycine’s small size and conformational flexibility contribute to efficient multi-site acetylation of the H4 tail. Our results indicate that the more N-terminal sites on the H4 tail are preferred (Figure 4), and suggest a minimal distance from the histone fold domain might be important for acetylation efficiency. Consistent with this, MS/MS analyses of the nucleosomal H4GGAKins mutant shows that the most N-terminal sites are acetylated 4-fold faster compared to K16 (data not shown). Because the primary site of picNuA4-histone interaction occurs within the histone fold domain, it is unclear whether the parameters that influence chromatin acetylation efficiency can be extrapolated to substrate specificity for non-histone substrates (53).
Although our findings suggest that many pre-existing modifications on the H4 tail would not significantly impact acetylation efficiency, it is not known how modifications to other histones might affect the kinetics of nucleosome acetylation by picNuA4. The picNuA4 and NuA4 complexes harbor a PHD finger on Yng2 that binds H3K4me3 (53, 54). Such modifications clearly affect HAT complex targeting and localization in vivo (55–57). Indeed, Li et al. reported that swapping the PHD finger of native Yng2 (H3K4me3) with Rco1 (H3K36me) resulted in increased affinity for nucleosomal substrates and altered targeting of the NuA4 complex to H3K36me-enriched regions (58). Additional work will be necessary to investigate how motif recognition by chromatin binding modules may alter the mechanisms of acetylation for HAT complexes.
Supplementary Material
Acknowledgments
This work was supported by NIH funds (Grant GM059785). The authors would like to acknowledge the UW-Madison Human Proteomics Program, funded by the Wisconsin Partnership Fund for a Healthy Future, for support in obtaining mass spectrometry data, particularly Matthew Lawrence. We thank the Song Tan lab for helpful discussions and reagents during the early stage of this project, as well as the Karolin Luger lab for providing the α-satellite DNA plasmid. Lastly, we thank present and past members of the John Denu laboratory, particularly Chris Berndsen and Casey Hallows, for helpful discussions.
This work was supported by funds from NIH GM059785
Abbreviations
- NuA4
nucleosome acetyltransferase of histone H4
- HFD
histone fold domain
- picNuA4
piccolo nucleosome acetyltranferase of histone H4
- NCP
nucleosome core particle
- HAT
histone acetyltransferase
- AcCoA
acetyl-Coenzyme A
- Esa1
essential Sas2-related acetyltransferase 1
- Epl1
enhancer of polycomb-like 1
- Yng2
yeast inhibitor of growth 2
- DTT
dithiothreitol
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TAU
Triton-acetic acid-urea
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- TFA
trifluoroacetic acid
- gH4
globular domain of histone H4
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional experimental details and supplemental figures are available free of charge via the Internet at http://pubs.acs.org
References
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 8.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 9.Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Nourani A, Utley RT, Allard S, Cote J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selleck W, Fortin I, Sermwittayawong D, Cote J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 15.Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell. 2003;12:461–473. doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 16.Clements A, Marmorstein R. Insights into structure and function of GCN5/PCAF and yEsa 1 histone acetyltransferase domains. Methods Enzymol. 2003;371:545–564. doi: 10.1016/S0076-6879(03)71041-6. [DOI] [PubMed] [Google Scholar]
- 17.Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poux AN, Cebrat M, Kim CM, Cole PA, Marmorstein R. Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor. Proc Natl Acad Sci U S A. 2002;99:14065–14070. doi: 10.1073/pnas.222373899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poux AN, Marmorstein R. Molecular basis for Gcn5/PCAF histone acetyltransferase selectivity for histone and nonhistone substrates. Biochemistry. 2003;42:14366–14374. doi: 10.1021/bi035632n. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 21.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berndsen CE, Selleck W, McBryant SJ, Hansen JC, Tan S, Denu JM. Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry. 2007;46:2091–2099. doi: 10.1021/bi602366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 24.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 26.Lennox RW, Cohen LH. Analysis of histone subtypes and their modified forms by polyacrylamide gel electrophoresis. Methods Enzymol. 1989;170:532–549. doi: 10.1016/0076-6879(89)70063-x. [DOI] [PubMed] [Google Scholar]
- 27.Berndsen CE, Denu JM. Assays for mechanistic investigations of protein/histone acetyltransferases. Methods. 2005;36:321–331. doi: 10.1016/j.ymeth.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyler RC, Bitto E, Berndsen CE, Bingman CA, Singh S, Lee MS, Wesenberg GE, Denu JM, Phillips GN, Jr, Markley JL. Structure of Arabidopsis thaliana At1g77540 protein, a minimal acetyltransferase from the COG2388 family. Biochemistry. 2006;45:14325–14336. doi: 10.1021/bi0612059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15:948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith CM. Quantification of acetylation at proximal lysine residues using isotopic labeling and tandem mass spectrometry. Methods. 2005;36:395–403. doi: 10.1016/j.ymeth.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 33.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryant FR, Johnson KA, Benkovic SJ. Elementary steps in the DNA polymerase I reaction pathway. Biochemistry. 1983;22:3537–3546. doi: 10.1021/bi00284a001. [DOI] [PubMed] [Google Scholar]
- 35.Patwardhan P, Miller WT. Processive phosphorylation: mechanism and biological importance. Cell Signal. 2007;19:2218–2226. doi: 10.1016/j.cellsig.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchetot C, Chagnon M, Dube N, Halle M, Tremblay ML. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005;35:44–53. doi: 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Ohba R, Steger DJ, Brownell JE, Mizzen CA, Cook RG, Cote J, Workman JL, Allis CD. A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol Cell Biol. 1999;19:2061–2068. doi: 10.1128/mcb.19.3.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 39.Tse C, Georgieva EI, Ruiz-Garcia AB, Sendra R, Hansen JC. Gcn5p, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J Biol Chem. 1998;273:32388–32392. doi: 10.1074/jbc.273.49.32388. [DOI] [PubMed] [Google Scholar]
- 40.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Garcia AB, Sendra R, Pamblanco M, Tordera V. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 1997;403:186–190. doi: 10.1016/s0014-5793(97)00049-5. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 43.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 45.Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, Dorsey JA, Peterson CL, Berger SL, Allis CD. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 46.Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 47.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF, Jr, Fukusato T, Smith MM. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436:108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang CS, Pillus L. Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae. Genetics. 2009;183:149–160. doi: 10.1534/genetics.109.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 52.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 53.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 56.Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.