Abstract
The opportunistic pathogen Candida albicans undergoes a parasexual mating cycle in which cells must switch from the conventional “white” form to the alternative “opaque” form to become mating competent. Pheromones secreted by opaque cells induce the formation of polarized mating projections and result in cell–cell conjugation. In contrast, white cells are unable to undergo mating, but can still respond to pheromone by expression of adhesion genes that promote biofilm formation. In this study, we have analyzed the dual ability of pheromones to activate mating by opaque cells and biofilm formation by white cells. We first show that there is considerable plasticity in interactions between the α pheromone and its receptor, Ste2, by analysis of analogs of the α pheromone. Significantly, substituted forms of α pheromone can induce a response in opaque cells and this is sufficient to drive same-sex a-a cell fusion and homothallic mating. In addition, pheromone analogs were able to induce adhesion and biofilm formation in white cells of C. albicans. Because of the observed plasticity in pheromone signaling, we subsequently tested putative pheromones from multiple Candida species and identified nonnative ligands that can induce self-mating and biofilm responses in C. albicans. Our findings demonstrate that environmental signals can initiate C. albicans parasexual reproduction and biofilm formation, and highlight the role of the pheromone-signaling apparatus in mediating these functions.
Keywords: microbiota, pathogenesis, phenotypic switch
Many human pathogens have predominantly clonal lifestyles yet still retain the ability to undergo sexual or parasexual reproduction (1). In the case of the fungal pathogen Candida albicans, this species was long thought to be an obligate asexual organism, but a mating cycle has been established that shows fundamental differences from that of the related hemiascomycete, Saccharomyces cerevisiae (for reviews, see refs. 2–4). C. albicans mating occurs between a and α diploids and is dependent on a unique phenotypic switch; cells must convert from the more common white form to the alternative opaque form to undergo efficient cell conjugation (5–8). The switch between states is a stochastic, heritable event that occurs approximately once every 10,000 cell generations. White and opaque cells exhibit differential expression of more than 400 genes, with opaque cells showing higher expression of several genes that are directly implicated in mating (9, 10).
C. albicans mating is driven by sex-specific pheromones that are expressed by the two cell types; α cells secrete α pheromone and respond to a pheromone via the Ste3 receptor; a cells produce a pheromone and respond to α pheromone via the Ste2 receptor (11–13). Pheromone binding activates a conserved MAPK cascade, leading to the formation of mating projections and subsequent cell–cell conjugation (14, 15). Although only opaque cells exhibit the mating response to pheromone, white cells can also respond by undergoing increased cohesion and biofilm formation (16). Recent studies have shown that both opaque and white cells use the same conserved MAPK cascade for pheromone signaling, but whereas signaling activates the Cph1/Ste12 transcription factor in opaque cells, the Tec1 transcription factor is activated in white cells (17, 18). Curiously, pheromone signaling by white a cells also requires a specific region in the Ste2 receptor (intracellular region 1) that is not necessary for signal transduction in opaque cells (19).
The α pheromone secreted by C. albicans cells is a 13-amino acid peptide that is produced by proteolytic processing of the MFα gene product (11, 13). In both S. cerevisiae and C. albicans, mature α pheromone is targeted by the Bar1 protease. This aspartyl protease is secreted by a cells and inactivates the α pheromone, thereby enabling nonmating cells to overcome pheromone-induced cell-cycle arrest (20–22). Bar1 activity may also sharpen the gradient of α pheromone, promoting accurate partner recognition (23). Recent studies in C. albicans have established that Bar1 also regulates a novel program of homothallic mating in this yeast. It is now apparent that opaque a cells secrete both a and α pheromones; inhibition of self-mating requires that Bar1 degrade α pheromone to prevent autocrine activation of the mating response (24). In the absence of Bar1, self-fertilization of opaque a cells can occur, leading to the formation of same-sex tetraploid a-a cells. Same-sex mating has also been described in another prominent human fungal pathogen, Cryptococcus neoformans (25–28). It therefore appears that flexibility in sexual programs can have significant advantages for microbial pathogens (1).
In this work, we show that the C. albicans pheromone/receptor interaction exhibits considerable plasticity, and that multiple pheromone analogs are able to activate the signaling machinery. We also establish that pheromone signaling alone is sufficient to induce same-sex mating between opaque cells and the productive formation of tetraploid cells. White cells of C. albicans respond to pheromone analogs by undergoing increased cell–cell cohesion and biofilm formation. Moreover, pheromones from related Candida pathogens can induce both same-sex mating and biofilm formation in opaque and white cells of C. albicans, respectively. We therefore discuss the potential roles for interspecies signaling in driving adaptive events in C. albicans biology.
Results
Analysis of Pheromone Analogs on C. albicans Mating.
C. albicans α pheromone is a 13-amino acid peptide (GFRLTNFGYFEPG) that is sensed by its cognate receptor, Ste2, on MTLa cells (11, 13). To determine the specificity of the α pheromone for mating signaling, we analyzed the activity of a series of di-alanine–substituted pheromone analogs (Fig. 1A) for their ability to induce a mating response in opaque cells of C. albicans. As shown in Fig. 1B, each of the 12 di-alanine–substituted analogs were still able to induce a morphological response in opaque a cells, as evidenced by the production of wrinkled colonies because of filament-like mating projections (Fig. 1C and Fig. S1) (24). Each of these responses was strictly dependent on the Ste2 receptor for the α pheromone, as addition of synthetic peptides to Δste2 cells resulted in no change in phenotype (Fig. 1B).
Fig. 1.
Substituted pheromone peptides retain activity in C. albicans. (A) Sequences of di-alanine derivatives of the 13 amino acid C. albicans α pheromone. (B) Substituted pheromones induce a morphological change (wrinkled colonies) because of induction of mating. Colony images of opaque C. albicans a cells exposed to 25 μg of the indicated pheromone daily for 3 d. (Holes in colonies indicate where pheromone was added.) (C) Cells exposed to pheromones form filament-like mating projections. Images of cells taken from the colonies shown in B. (Scale bar, 16 μm.) (D) Quantification of mating gene expression induced by pheromone analogs using a pFIG1-GFP reporter strain (DSY700). Opaque a cells were incubated with 10 μg/mL of peptide in Spider medium for 24 h, then analyzed for GFP fluorescence by flow cytometry. Values are mean fluorescence of the entire cell population (arbitrary units). (E) Sequences of α pheromone analogs of different lengths. (F) Quantification of mating-gene expression induced by pheromone analogs A to G. Cells treated with C. albicans native α pheromone (Ca), cells treated with S. cerevisiae α pheromone (Sc). Error bars represent SEM from a minimum of three replicate experiments. *P < 0.05 vs. DMSO control.
Although all 12 di-alanine analogs induced the mating pathway, there were clear differences in the efficiency of the response by opaque a cells (Fig. 1B). To quantify the effectiveness of pheromone analogs at inducing mating, a GFP reporter under the control of the FIG1 promoter was used (24) (Fig. 1D). Addition of the C. albicans α pheromone (10 μg/mL) to opaque a cells induced FIG1 expression, whereas addition of the S. cerevisiae α pheromone (WHWLQLKPGQPMY) did not. Each of the di-alanine–substituted pheromone analogs also induced significant expression of the FIG1 reporter over the DMSO control, although peptides 10 and 11, with substitutions at amino acids 10 to 12, were the weakest inducers of the mating response (Fig. 1D). Notably, peptide 2 induced stronger FIG1 expression than the native C. albicans pheromone. A similar phenomenon was previously described in S. cerevisiae, where several single alanine-substituted analogs showed increased bioactivity by more than threefold (29).
To also investigate the effect of peptide length on pheromone signaling, we used a series of peptide analogs ranging from 10 to 17 amino acids (Fig. 1E). Removal of three amino acids from the N terminus of α pheromone (peptide A) abolished FIG1 expression, but removal of one or two amino acids from the N terminus (peptides B and C) reduced FIG1 expression but still retained significant activity (Fig. 1F). Furthermore, the addition of two amino acids to the N terminus (peptide E) only reduced FIG1 expression slightly. These findings support the observations with alanine-substituted pheromones that manipulation of the N terminus of the peptide has only a modest effect on mating signaling. In contrast, manipulation of the C terminus, either by deletion of one amino acid (peptide D) or by addition of two amino acids (peptides F and G) resulted in a marked reduction in mating signaling (Fig. 1F). These results indicate that there is considerable plasticity in the pheromone–receptor interaction in C. albicans and that the organism can respond to a variety of pheromone analogs.
Pheromone Signaling Drives Homothallic a-a Mating.
One intriguing feature of C. albicans mating is the ability of cells to undergo primary homothallism via same-sex mating. Opaque a cells lacking the Bar1 protease secrete α pheromone, and this initiates an autocrine feedback loop resulting in same-sex a-a mating (24, 30). To test if pheromone is sufficient to drive the homothallic program of mating, we cocultured two auxotrophic a strains in the presence of varying concentrations of synthetic pheromone, and potential a-a mating products were detected by plating onto selective media. In the absence of synthetic pheromone, no mating products were obtained. However, in the presence of the C. albicans α pheromone, mating products were detected after 5 d culture on Spider medium (Fig. 2). Mating products were analyzed by cell cytometry to confirm that they were tetraploid and by PCR to confirm their MTLa genotype (Fig. 2 A and B). These results establish the ability of α pheromone to induce homothallic mating of opaque a cells. The demonstration of wild-type a cells mating in the absence of α partners confirms that activation of pheromone signaling is sufficient to drive self-fertilization in C. albicans.
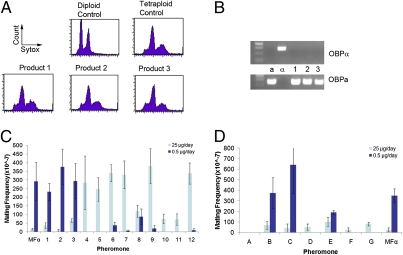
Fig. 2.
Addition of α pheromone or analogs induces same-sex mating of C. albicans a cells. (A) Addition of pheromone to C. albicans opaque a cells results in cell–cell conjugation and progeny that have tetraploid DNA content. Flow cytometry plots showing DNA content of cells fixed and stained with Sytox Green. Three representative mating product strains are shown. (B) PCR confirming the mating type of a-a matings using primers directed at OBPa (Lower) and OBPα (Upper). Control a and α strains together with three pheromone-induced a-a mating products. (C) Frequency of mating induced by di-alanine–substituted pheromones. Opaque a cells from strains RBY1118 (-his) and RBY1179 (-arg) were coincubated with either 25 μg (light bars) or 0.5 μg (dark bars) of pheromone daily for 5 d on Spider medium. Mating products were detected by plating cells on selective medium (-his, -arg), as described (24). (D) Frequency of mating induced by pheromones of different lengths. Error bars represent SEM from a minimum of three experiments.
Comparison of mating efficiencies in the presence of varying concentrations of native pheromone and pheromone analogs was revealing. First, all 12 di-alanine–substituted analogs were still able to induce homothallic mating, albeit to varying degrees (Fig. 2C). Again, peptides 10 and 11 containing substitutions near the C terminus of the peptide were the least effective at inducing conjugation, consistent with low induction of the mating response (Fig. 1D). Peptides that were of different lengths at the C terminus (peptides D, F, and G) also tended to be weak inducers of same-sex mating (Fig. 2D). Curiously, high concentrations (25 μg/d) of the native pheromone and peptides 1, 2, and 3, resulted in fewer mating events than lower concentrations (0.5 μg/d) of these same peptides. Similar results were seen with peptides missing amino acids at the N terminus, as peptides B and C showed more frequent mating at lower pheromone concentrations than at higher concentrations. Significantly, these peptides were also the strongest inducers of the mating program based on FIG1 expression, and indicates that overstimulation of the pheromone-signaling pathway depresses overall mating frequencies. This finding is consistent with previous studies, where both C. albicans and S. cerevisiae mutants lacking the Bar1 protease showed a decreased frequency of a-α conjugation, presumably because of excessive stimulation of pheromone signaling (22, 31).
We note that each of the pheromone analogs is capable of inducing polarized mating projections, activating mating gene expression, and even productive self-mating events. The only peptide that was not active was peptide A, which is lacking the first three residues at the N terminus. Substitutions at position 11 of the α pheromone (in peptides 10 and 11) also had a very significant affect on pheromone activity, and reduced mating signaling to a larger extent than that seen with the corresponding substitution in S. cerevisiae (29). Overall, these data indicate that an important region for bioactivity exists near position 11, but the first three amino acids in particular do not contain critical amino acid contacts for signaling through C. albicans Ste2.
Pheromone Analogs Increase Cell Adhesion by White Cells of C. albicans.
Opaque cells are the mating-competent form of C. albicans, and yet recent experiments demonstrate that a productive pheromone-signaling pathway also operates in white cells. Work by Daniels et al. showed that white cells exhibit increased adhesion in the presence of pheromone, promoting attachment to each other and to synthetic surfaces and resulting in biofilm formation (16). Interestingly, the white response is dependent on a specific domain in Ste2, intracellular region 1, which is dispensable for pheromone signaling in opaque cells (19).
We tested the ability of white cells to form biofilms in response to pheromone analogs by measuring adherence to plastic (17–19). As shown in Fig. 3, white a cells did not adhere to plastic in the absence of pheromone, whereas addition of the C. albicans α pheromone resulted in efficient formation of a cohesive film. Comparison of the di-alanine peptide analogs revealed that all of the analogs were capable of increasing biofilm response by white cells. Similar to the native α pheromone, peptide analogs 1, 2, and 3 induced the strongest adherence to plastic and did so even at low concentrations (Fig. 3 and Fig. S2). The majority of the remaining analogs induced strong biofilm formation when used at higher concentrations (10 μg/mL) (Fig. 3 A and B) but not at lower concentrations (0.2 μg/mL) (Fig. S2). In contrast, peptides 4, 10, and 11 induced only weak adherence compared with the native pheromone. Overall, the results with white cells show strong parallels to those obtained with opaque cells, including the fact that peptides 10 and 11 are the weakest inducers of a cellular response.
Fig. 3.
Pheromone analogs induce adhesion in C. albicans white cells. (A) Images of white cells adhering to plastic. C. albicans white P37005 a cells were grown in Lee's medium for 24 h in the presence of di-alanine pheromone analogs (10 μg). Nonadherent cells were removed by washing and pictures of the wells taken. (B) Quantification of adherent white cells shown in A. (C) Images of adherent white cells after exposure to 10 μg of pheromones A-G for 24 h. (D) Quantification of adherent white cells shown in C. Error bars represent SEM from a minimum of three experiments.
Analysis of pheromone analogs of different lengths also revealed that white and opaque cells exhibit similar responses. For example, removal of three amino acids from the N-terminal region of the α pheromone (peptide A) abolished induction of adherence to plastic, whereas other manipulations at the N terminus of the pheromone did not negatively affect white cell signaling (see peptides B, C, and E) (Fig. 3 and Fig. S2). Analogs that were shorter or longer at the C terminus were also able to induce weak adherence to plastic (peptides D, F, and G) (Fig. 3 and Fig. S2). These results indicate that pheromone analogs that induce signaling in opaque cells are also able to do so in white cells. Multiple peptides are therefore able to activate the pheromone-signaling cascade in C. albicans, either inducing the mating response in opaque cells or biofilm formation in white cells.
Interspecies Pheromone Signaling Can Induce Biofilm Formation and Homothallic Mating in C. albicans.
C. albicans coexists with a number of yeast and bacterial species as part of the human microbiota. The discovery that alternative pheromone analogs can successfully signal to C. albicans a cells suggested that pheromones from different Candida species might induce responses in C. albicans. With the availability of genome sequences for multiple Candida species and conservation of pheromone-processing proteins (32), we identified potential α pheromones for several hemiascomycete species (Fig. 4A; for details on pheromone identification, see Fig. S3).
Fig. 4.
Pheromones from other Candida species induce same-sex mating and adherence in C. albicans. (A) Phylogenetic tree showing relationship between Candida species and their predicted α pheromones (not to scale). Two potential α pheromones are shown for C. dubliniensis and C. tropicalis. (B) Pheromones from multiple other Candida species induce a morphological change in C. albicans opaque a cells. Images of colonies incubated with 25 μg of pheromone daily for 3 d on Spider medium. (C) Quantification of FIG1 gene expression induced by 10 μg/mL pheromone for 24 h in Spider medium (arbitrary units). (D) Pheromone analogs induce C. albicans a-a conjugation. Quantification of mating products after 5 d culture on Spider medium in the presence of 25 or 0.5 μg pheromone per day. (E) Alternative pheromones induce adherence of C. albicans white cells. Images of adherent P37005 white cells grown in 12-well plastic dishes in Lee's medium and exposed to 10 μg of pheromone for 24 h. (F) Quantification of adherent white cells shown in E. Error bars represent SEM from a minimum of three experiments. *P < 0.05 vs. DMSO control.
The response of C. albicans opaque a cells to each synthetic pheromone was first monitored by examination of colony morphology (Fig. 4B) and by pFIG1-GFP expression (Fig. 4C). Notably, even highly divergent pheromones from species such as Candida parapsilosis and Lodderomyces elongisporus were able to induce a mating response in C. albicans, as evidenced by both morphological changes and FIG1 expression. These responses were specific, as α pheromones from more divergent species, including Candida lusitaniae, Kluyveromyces lactis, and S. cerevisiae, did not induce mating (Fig. 4C). Furthermore, none of the peptides were able to induce a mating response in Δste2 cells. Although the responses to Candida dubliniensis and C. parapsilosis pheromones was relatively strong, Candida tropicalis and L. elongisporus peptides were less efficient at inducing FIG1 expression (Fig. 4C). Significantly, the alternative pheromones were also successful at inducing same-sex mating of C. albicans cells. Thus, C. dubliniensis and C. parapsilosis pheromones, and to a lesser extent C. tropicalis and L. elongisporus pheromones, could induce fusion of opaque a cells and formation of stable a-a tetraploid cells (Fig. 4D). These results support our conclusion that activation of the mating program alone is sufficient to drive homothallic mating of C. albicans cells.
We also examined the potential for interspecies signaling to activate biofilm formation by white cells of C. albicans. As expected, pheromones from multiple Candida species were able to induce adhesion of C. albicans white a cells to plastic (Fig. 4 E and F). Again, the efficiency of biofilm formation varied depending on the pheromone, but showed parallels to the response in opaque cells. Thus, pheromone-induction of biofilm formation decreased in the order C. dubliniensis > C. parapsilosis > L. elongisporus > C. tropicalis. Biofilm formation was not induced by S. cerevisiae pheromone or from any other species that was tested. These results demonstrate that a divergent set of pheromone sequences from different Candida species are able to signal to C. albicans and induce a biological response in both white and opaque cells.
Discussion
Plasticity of the Candida Pheromone–Receptor Interaction.
In this work, we investigated the specificity of the ligand–receptor interaction that regulates the pheromone-sensing pathway of C. albicans. We demonstrate that there is considerable plasticity in the interaction between the Ste2 receptor and its pheromone ligand. Thus, substitution of any of the 13 amino acids that make up the cognate pheromone can occur without a complete loss of signaling activity. Substitutions at the C terminus of the pheromone sequence had the most negative effect on signaling through Ste2. In contrast, di-alanine substitutions at the N terminus had only modest effects on bioactivity, and in some cases even resulted in increased signaling through Ste2. There was also considerable flexibility in signaling via analogs of different lengths. Subtraction of one or two amino acids from the N terminus, or addition of two amino acid residues to the N terminus, did not greatly affect pheromone performance. Modifications to the C terminus were tolerated less favorably, as the addition or subtraction of amino acids had a significant effect on bioactivity, although functionality was still maintained.
Studies on S. cerevisiae Ste2 have shown that this receptor can also tolerate substitutions in its ligand. In this case, single alanine substitutions revealed that no side chain is absolutely required for activity (29, 33). In general, alanine analogs resulted in one order of magnitude variation in bioactivity, and included analogs with both increased and decreased activity. A genetic screen for agonists and antagonists of Ste2 also demonstrated flexibility in ligand recognition, although the vast majority of agonists differed, at most, at only 3 of the 13 amino acid residues in the native α pheromone (34). Notably, pheromone bioactivity did not correlate with binding affinity of the ligand for the receptor, and this may represent a requirement for ligand-induced receptor isomerization for signaling activity (33).
Pheromone Signaling Drives Same-Sex Homothallic Mating in C. albicans.
In contrast to S. cerevisiae, we demonstrate that the addition of α pheromone to C. albicans cells not only activates pheromone signaling but results in productive same-sex fusion of opaque cells. Self-fertilization required Ste2, confirming that activation of the canonical pheromone signaling pathway was required for mating. These results therefore establish that pheromone signaling is sufficient to drive the program of homothallic self-mating in C. albicans (24, 30). Same-sex fertilization due to pheromone alone has, to our knowledge, not been observed in other hemiascomycete yeast. In contrast, in the filamentous ascomycete Ustilago maydis, addition of mating pheromone was shown to induce conjugation tube formation and fusion of cells from the same mating type (35). In addition, pheromones have been implicated in homothallic mating in other ascomycete species. For example, in Aspergillus nidulans, activation of pheromone signaling is central to self-fertility and expression of pheromones and receptors in a single cell type drives this process (36, 37). Programming of pheromone signaling pathways therefore plays a key role in determining whether mating is homothallic or heterothallic in fungi. In the case of C. albicans, both modes of reproduction are possible in a cells. Heterothallic a-α mating is favored in the presence of α cells, but homothallic mating can occur in Δbar1 cells (24) or, as we now show, in wild-type cells in the presence of pheromones from other Candida species.
Biofilm Formation due to Pheromone Signaling in C. albicans White Cells.
Recent studies have revealed that C. albicans white cells, although infertile, can respond to pheromone by expressing genes involved in biofilm formation (16–18). In contrast to pheromone signaling in opaque cells that targets the Ste12/Cph1 transcription factor, signaling in white cells results in Tec1 activity and up-regulation of adhesion genes (17). A particular region of the Ste2 receptor, intracellular domain 1 (located between transmembrane domains 1 and 2) was implicated in the pheromone response of white cells but not opaque cells, which may be indicative of unique interactions between Ste2 and the downstream signaling machinery in white cells (19). In this work, we show that pheromone analogs that induce mating in opaque cells also stimulate biofilm formation in white cells. In general, a strong correlation was observed between analog activity in white and opaque cells, indicating ligand-Ste2 interactions are similar in the two cell types. One exception was peptide 4, which contained alanine residues at positions 4 and 5; this peptide produced virtually no biofilm in white cells, yet was still relatively active in inducing mating in opaque cells. Overall, these results demonstrate that white and opaque cells recognize pheromone analogs with a similar specificity, although some minor differences may exist between pheromone signaling in the two cell types.
Cross-Species Signaling Drives C. albicans Self-Mating and Biofilm Formation.
The most revealing aspect of the current work is the demonstration that the pheromone-signaling machinery in C. albicans can respond to signals other than the cognate ligand. C. albicans is a commensal species of warm-blooded hosts, where it is exposed to a variety of stimuli from the host and other microbes. The ability of C. albicans to adapt allows it to thrive in multiple niches in the human body and is a major reason for the success of this species as an opportunistic pathogen. The plasticity inherent in the ligand–Ste2 interaction led us to test C. albicans responses to pheromones from species with which it coexists. We show that pheromones from several related species, including C. dubliniensis, C. parapsilosis, C. tropicalis, and L. elongisporus, induce mating and same-sex conjugation in C. albicans opaque cells and biofilm formation in C. albicans white cells. Multiple Candida species (e.g., C. albicans, C. parapsilosis, and C. tropicalis) have been shown to coinhabit the same niches (38–41), while L. elongisporus was recently identified from bloodstream infections (42), confirming that these species can encounter one another in nature.
Early studies in Saccharomyces established that pheromones from different yeast species can act interspecially. For example, Hisatomi et al. (43) showed that synthetic pheromones from S. cerevisiae, Saccharomyces kluyveri, and Saccharomyces exiguus could all act upon one another. These peptides were highly homologous, with S. kluyveri and S. exiguus differing at only four or five amino acid residues, respectively, with the S. cerevisiae pheromone. In contrast, two of the peptides active on C. albicans (C. parapsilosis and L. elongisporus pheromones) show much greater divergence, differing at 8 of the 13 amino acid residues in the C. albicans sequence (Fig. 4). Moreover, whereas interspecies signaling events were futile in Saccharomyces, we emphasize that pheromone signaling results in productive homothallic mating events in C. albicans, as well as increased biofilm formation in white cells.
We propose that these studies extend the conditions under which sex and sex-related processes act in Candida and other fungal species. Our discovery that alternative pheromones can activate the program of homothallic mating indicates that mating could be more frequent than previously anticipated. Thus, even in the absence of mating partners (and in a BAR1+ strain background) activation of the mating program can occur because of environmental telesensing. Ligand signaling can drive C. albicans same-sex mating, generating tetraploid cells that are competent to undergo the parasexual program of chromosome reduction, and resulting in diploid and aneuploid forms of the species (44, 45). Given the complex niche that C. albicans inhabits, we speculate that pheromone-like signals from the host or other microbiota could also activate the pheromone-sensing machinery and drive homothallism. Ongoing studies into the human microbiome will therefore be potentially revealing in identifying novel ligands that could stimulate C. albicans mating pathways.
Taken together, our findings show that diverse signals activate C. albicans pheromone signaling, and that the downstream consequences of signaling include both homothallism and biofilm formation. These results further the conditions under which the sexual machinery is active, and we speculate that interspecies signaling events will similarly direct mating in other fungi. It will therefore be important to examine all aspects of C. albicans’ mating cycle in the context of its complex environment in the mammalian host. Based on our findings, it will be particularly exciting to examine other constituents of the host microbiota to determine what ligands are recognized by C. albicans and can activate (or inhibit) its sexual biology.
Materials and Methods
Media and Reagents.
Media was prepared as described previously (46–48). Peptides were synthesized by Genemed Synthesis. Peptides were resuspended at a concentration of 10 mg/mL in 10% DMSO and used at the indicated concentrations.
Pheromone Response Assays.
Overnight cultures of opaque cells from strain RBY1118 (a/a) were grown in SCD medium at room temperature. Cells were washed with water and resuspended in Spider medium (48) at a concentration of 2 × 109 cells/mL. Twenty microliters of this culture was added to Spider plates and incubated at room temperature. Next, 25 μg of pheromone was added daily for 3 d; colonies were imaged using a Zeiss Stemi 2000-C stereoscope with an Infinity 1 camera. Cells were taken from colonies and examined using a Zeiss Axioplan 2 microscope equipped with a Hamamatsu-ORCA camera. For quantification of the response to pheromone treatment, a GFP reporter under the control of the FIG1 promoter was used (24). Opaque cells of DSY700 (pFIG1-GFP) were grown overnight at room temperature in SCD medium, cells washed, and resuspended in Spider medium. Then, 6 × 107 cells were added to 3 mL of fresh Spider medium, 10 μg/mL pheromone added, and cells grown for 24 h at room temperature. Cells were added to SCD medium, sonicated and fluorescence measured in the FL1 channel on a Becton Dickinson FACSCalibur.
Quantitative Mating Assays.
Overnight cultures of opaque strains with different auxotrophic markers (RBY1118 and RBY1179) (22) were grown in SCD medium at room temperature. Cells were washed with water and resuspended in Spider medium at a concentration of 1 × 109 cells/mL. Ten microliters of each culture was mixed and plated on a nitrocellulose filter on Spider plates. Pheromone was added daily at the indicated concentration for 4 d. On day 5, cells were removed from the filter, resuspended in water, and sonicated to disperse clumps. Cells were then plated on selective media and analyzed for the formation of mating products, as previously described (24).
Biofilm Assays.
Adherence assays were performed essentially as described (16, 18) with the following modifications. Overnight cultures of white cells from strain P37005 were grown in YPD at room temperature. Cells were washed with PBS (phosphate-buffered saline) pH 7.4 and resuspended in Lee's medium to a concentration of 5 × 107 cells/mL. One milliliter of cells was then added to 12-well culture dishes (Costar, Corning Inc.) to which the indicated amount of pheromone was added. Plates were left at room temperature overnight, wells were gently washed with PBS, and plates scanned using a HP Scanjet G3010. Adherent cells were also scraped off the well and quantified at OD600 using a spectrophotometer.
Supplementary Material
Acknowledgments
Work in the laboratory of K.A. and R.J.B. was supported by National Institutes of Health Grants R21AI081560, AI081560, and AI081704 (to R.J.B.) and F31DE019752 (to K.A.). R.J.B. also holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017234108/-/DCSupplemental.
References
- 1.Heitman J. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe. 2010;8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alby K, Bennett RJ. Sexual reproduction in the Candida clade: Cryptic cycles, diverse mechanisms, and alternative functions. Cell Mol Life Sci. 2010;67:3275–3285. doi: 10.1007/s00018-010-0421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 4.Soll DR. Why does Candida albicans switch? FEM Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- 5.Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 6.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 7.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 8.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 9.Lan CY, et al. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci USA. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: A case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 11.Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol. 2002;46:1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- 13.Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot Cell. 2003;2:1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett RJ, Miller MG, Chua PR, Maxon ME, Johnson AD. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol Microbiol. 2005;55:1046–1059. doi: 10.1111/j.1365-2958.2005.04466.x. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart SR, Daniels KJ, Zhao R, Wessels D, Soll DR. Cell biology of mating in Candida albicans. Eukaryot Cell. 2003;2:49–61. doi: 10.1128/EC.2.1.49-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahni N, et al. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: Insights into the evolution of new signal transduction pathways. PLoS Biol. 2010;8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi S, et al. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell. 2008;19:957–970. doi: 10.1091/mbc.E07-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi S, et al. A Candida albicans-specific region of the alpha-pheromone receptor plays a selective role in the white cell pheromone response. Mol Microbiol. 2009;71:925–947. doi: 10.1111/j.1365-2958.2008.06575.x. [DOI] [PubMed] [Google Scholar]
- 20.Hicks JB, Herskowitz I. Evidence for a new diffusible element of mating pheromones in yeast. Nature. 1976;260:246–248. doi: 10.1038/260246a0. [DOI] [PubMed] [Google Scholar]
- 21.Manney TR. Expression of the BAR1 gene in Saccharomyces cerevisiae: Induction by the alpha mating pheromone of an activity associated with a secreted protein. J Bacteriol. 1983;155:291–301. doi: 10.1128/jb.155.1.291-301.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer D, Côte P, Whiteway M, Bennett RJ. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryot Cell. 2007;6:907–918. doi: 10.1128/EC.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkai N, Rose MD, Wingreen NS. Protease helps yeast find mating partners. Nature. 1998;396:422–423. doi: 10.1038/24760. [DOI] [PubMed] [Google Scholar]
- 24.Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryot Cell. 2008;7:1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser JA, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 27.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 28.Lin X, et al. Diploids in the Cryptococcus neoformans serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abel MG, Zhang YL, Lu HF, Naider F, Becker JM. Structure-function analysis of the Saccharomyces cerevisiae tridecapeptide pheromone using alanine-scanned analogs. J Pept Res. 1998;52:95–106. doi: 10.1111/j.1399-3011.1998.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 30.Heitman J. Microbial genetics: Love the one you're with. Nature. 2009;460:807–808. doi: 10.1038/460807a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giot L, DeMattei C, Konopka JB. Combining mutations in the incoming and outgoing pheromone signal pathways causes a synergistic mating defect in Saccharomyces cerevisiae. Yeast. 1999;15:765–780. doi: 10.1002/(SICI)1097-0061(19990630)15:9<765::AID-YEA418>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naider F, Becker JM. The alpha-factor mating pheromone of Saccharomyces cerevisiae: A model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Manfredi JP, et al. Yeast alpha mating factor structure-activity relationship derived from genetically selected peptide agonists and antagonists of Ste2p. Mol Cell Biol. 1996;16:4700–4709. doi: 10.1128/mcb.16.9.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spellig T, Bölker M, Lottspeich F, Frank RW, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paoletti M, et al. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Seo JA, Han KH, Yu JH. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol Microbiol. 2004;53:1611–1623. doi: 10.1111/j.1365-2958.2004.04232.x. [DOI] [PubMed] [Google Scholar]
- 38.Benjamin DK, Jr., et al. Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network Neonatal candidiasis: Epidemiology, risk factors, and clinical judgment. Pediatrics. 2010;126:e865–e873. doi: 10.1542/peds.2009-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins M, et al. Oral Candida carriage of patients attending a dental clinic in Braga, Portugal. Rev Iberoam Micol. 2010;27:119–124. doi: 10.1016/j.riam.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Melton JJ, et al. Recovery of Candida dubliniensis and other Candida species from the oral cavity of subjects with periodontitis who had well-controlled and poorly controlled type 2 diabetes: A pilot study. Spec Care Dentist. 2010;30:230–234. doi: 10.1111/j.1754-4505.2010.00159.x. [DOI] [PubMed] [Google Scholar]
- 42.Lockhart SR, Messer SA, Pfaller MA, Diekema DJ. Lodderomyces elongisporus masquerading as Candida parapsilosis as a cause of bloodstream infections. J Clin Microbiol. 2008;46:374–376. doi: 10.1128/JCM.01790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hisatomi T, Yanagishima N, Sakurai A, Kobayashi H. Interspecific actions of alpha mating pheromones on the a mating-type cells of three Saccharomyces yeasts. Curr Genet. 1988;13:25–27. doi: 10.1007/BF00365752. [DOI] [PubMed] [Google Scholar]
- 44.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forche A, et al. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bedell GW, Soll DR. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: Evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun. 1979;26:348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- 48.Liu H, Köhler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.