Abstract
Pseudohypoparathyoridism type Ib (PHP-Ib) typically defines the presence of end-organ resistance to parathyroid hormone in the absence of Albright's hereditary osteodystrophy. Patients affected by this disorder present with imprinting defects in the complex GNAS locus. Microdeletions within STX16 or GNAS have been identified in familial cases with PHP-Ib, but the molecular cause of the GNAS imprinting defects in sporadic PHP-Ib cases remains poorly defined. We now report a case with sporadic PHP-Ib for whom a SNPlex analysis revealed loss of the maternal GNAS allele. Further analysis of the entire genome with a 100K SNP chip identified a paternal uniparental isodisomy affecting the entire chromosome 20 without evidence for another chromosomal abnormality. Our findings explain the observed GNAS methylation changes and the patient's hormone resistance, and furthermore suggest that chromosome 20 harbors, besides GNAS, no additional imprinted region that contributes to the clinical and laboratory phenotype.
Keywords: Pseudohypoparathyroidism, uniparental disomy, GNAS, imprinting, parathyroid hormone, stimulatory G protein
INTRODUCTION
The transmission of both homologous chromosomes, or segments thereof, from a single parent to its offspring is described as uniparental disomy (UPD). UPDs are known to be responsible for a limited number of human diseases that often involve developmental defects [1, 2], and some recent studies have also implicated acquired, somatic UPDs in tumorigenesis [3, 4]. The clinical phenotypes associated with UPDs are caused by homozygosity of recessive disease-causing mutations or due to aberrant expression of an imprinted gene. In the latter case, the loss or duplication of one parental allele results in the deficiency or overexpression of the imprinted gene product(s), respectively.
GNAS is a complex imprinted locus on chromosome 20q13.3 that encodes the α-subunit of the stimulatory G protein (Gsα) and several additional imprinted transcripts (reviewed in [5, 6]). Derived exclusively from the paternal allele are XLαs, A/B, and antisense transcripts, and from the maternal allele is NESP55. Consistent with this imprinted expression pattern, the promoters of these imprinted transcripts are methylated on the silenced allele. In contrast, the Gsα promoter is not methylated, and accordingly, Gsα expression is biallelic in most tissues. However, it shows predominantly maternal expression in certain tissues, including the renal proximal tubules, thyroid, and pituitary.
Defects in GNAS imprinting are found in patients with PHP-Ib [7, 8], a disorder of hormone resistance characterized by PTH- and, sometimes, TSH-resistance (reviewed in [9, 10]). Unlike in patients with PHP-Ia, who carry loss-of-function mutations in Gsα-coding GNAS exons, PHPIb patients typically lack the features of AHO, although some PHP-Ib cases with GNAS imprinting defects have recently been shown to have AHO features [11-14]. Nearly all patients with autosomal dominant PHP-Ib show a loss of methylation at GNAS exon A/B, which is combined, in some patients, with loss of methylation at exon XL and the promoter of the antisense transcript, and with gain of methylation at exon NESP55 [7, 8]. Deletions involving the NESP55 differentially methylated region or deletions in the gene encoding syntaxin-16 (STX16), which is located ~200 kb centromeric of GNAS, are responsible for the disease in patients with autosomal dominant PHP-Ib [15-17]
Most cases of PHP-Ib are sporadic and many such cases also demonstrate broad GNAS methylation defects [9, 10]. However, in contrast to the autosomal dominant form of PHP-Ib, no specific gene mutation has been identified as a cause of the broad GNAS methylation defects associated with sporadic PHP-Ib. In fact, only two patients with this relatively frequent form of PHP-Ib have been defined at the molecular level thus far, both revealing a duplicated long arm of the paternal chromosome 20 (patUPD20q), i.e. the region which comprises the GNAS locus [18, 19]. We now describe another patient with sporadic case of PHP-Ib who has paternal UPD affecting the entire chromosome 20.
MATERIALS AND METHODS
The study was approved by the Massachusetts General Hospital Institutional Review Board, and written informed consent was obtained from the patient and her parents. Genomic DNA was extracted from blood leukocytes, as described [20]. Multiplex analysis of all 23 SNPs was custom-designed and performed at Harvard-Partners Center for Genetics and Genomics (www.hpcgg.org). Microsatellites were analyzed as previously described [20]. Genotyping with 100K SNP chip analysis was performed as previously described [21].
RESULTS
Patient description and clinical course
The clinical and laboratory findings of the patient have been described previously [22, 23]. Briefly, she presented with a gradually worsening limp, leg pain, and “knock knees” at the age of 3 years. No AHO features were evident, but initial lab data revealed hypocalcemia, hyperphosphatemia, and elevated serum PTH, which was consistent with the diagnosis of PHPIb. Treatment with calcium carbonate and increasing doses of calcitriol has been initiated, which reduced the bone pain, normalized serum calcium, and suppressed serum PTH. However, subsequent non-compliance to the treatment led to the recurrence of bone pain and radiological evidence of bilateral slipped capital femoral epiphysis.
She underwent bilateral percutaneous in-situ fixation of her slipped capital femoral epiphyses with two minor subsequent revisions and is maintained on medical management. Her regimen now consists of both Rocaltrol and calcium supplements. Her serum calcium levels have ranged from 10-11 mg/dl with phosphorous levels ranging from 4.4-5.7 mg/dl and PTH levels in the normal or somewhat above the normal range. Her urinary calcium to creatinine ratios have been normal with the most recent 5 years after diagnosis. Her 25-vitamin D level currently is 26 ng/ml with a 1,25 dihydroxy vitamin D of 38 pg/ml (Table 1). She has had no further bone pains or fractures, and has shown both clinical and biochemical improvement. She continues to grow just above the 97th percentile for height and her weight has stabilized at the 95th percentile for her age. She remains prepubertal and has demonstrated radiographical improvement in the appearance of her skeletal X-rays on follow-up studies.
Table 1.
Follow-up Laboratory Course and Growth of Patient
| Pre-Treatment (months) | Post Treatment (months) | |||||
|---|---|---|---|---|---|---|
| 0 | 25 | 34 | 46 | 60 | 68 | |
| Chronological Age | 3y3m | 5y3m | 6y2m | 7y1m | 8y3m | 8y11m |
| Calcium (mg/dl) (normal: 8.4-10.2) | 7.2 | 10.2 | 10 | 10.9 | 11 | 10.6 |
| Phosphate (mg/dl) (normal: 2.5-5.6) | 8.0 | 5.7 | 5.4 | 5.5 | - | 4.4 |
| Albumin (gm/dl) (normal: 3.0-5 0) | 4.3 | - | - | 4.5 | 4.6 | 4.6 |
| Alkaline Phosphatase (IU/L) (normal: 100-320) | 643 | 258 | 170 | 120 | 147 | 121 |
| Creatinine (mg/dL) (normal: 0.3-0 8) | 0.3 | 0.4 | - | 0.7 | 0.8 | 0.6 |
| 25(OH)D3 (ng/mL) (normal: 9-52) | 20 | 30 | 15 | 25 | 19 | 26 (8y6m) |
| 1,25(OH)D3 (pg/mL) (normal: 15-90) | 50 | 15 | 36 | - | - | 38 (8y6m) |
| PTH (pg/mL) (normal: 10-65) | 3685 | 136 | 141 | 29 | 76 | 165 |
| Urine Ca/Urine Cr ratio (mg/mg) | - | 0.0157 | 0.039 | 0.201 | 0.099 | 0.147 |
| TSH (uU/mL) (prepubertal normal: 0.6-5.5) | - | - | - | 1.75 | - | - |
| fT4 | 1 | - | - | 1.14 | - | - |
| Height (cm) (%ile) | 107 (>97%ile) | 125.2 (>97%ile) | 129.4 (>97%ile) | 135.4 (>97%ile) | 142.3 (>97%ile) | 144.3 (>97%ile) |
| Weight (kg) (%ile) | 26.2 (>97%ile) | 38.1 (>97%ile) | 40.7 (>97%ile) | 36.2 (>97%ile) | 42.4 (>97%ile) | 41.6 (95th%ile) |
| BMI (kg/m2) | 22.9 | 24.3 | 24.3 | 19.7 | 20.9 | 20 |
| Bone Age | - | - | 10y | - | - | 10y |
| Calcium Carbonate | - | 625mg TID | 625mg TID | 625mg TID | 625mg TID | 625mg TID |
| Calcitriol (daily) | 0.1 mcg | 0.8 mcg | 1.0 mcg | 1.0 mcg | 1.0 mcg | 0.5 mcg |
Molecular findings
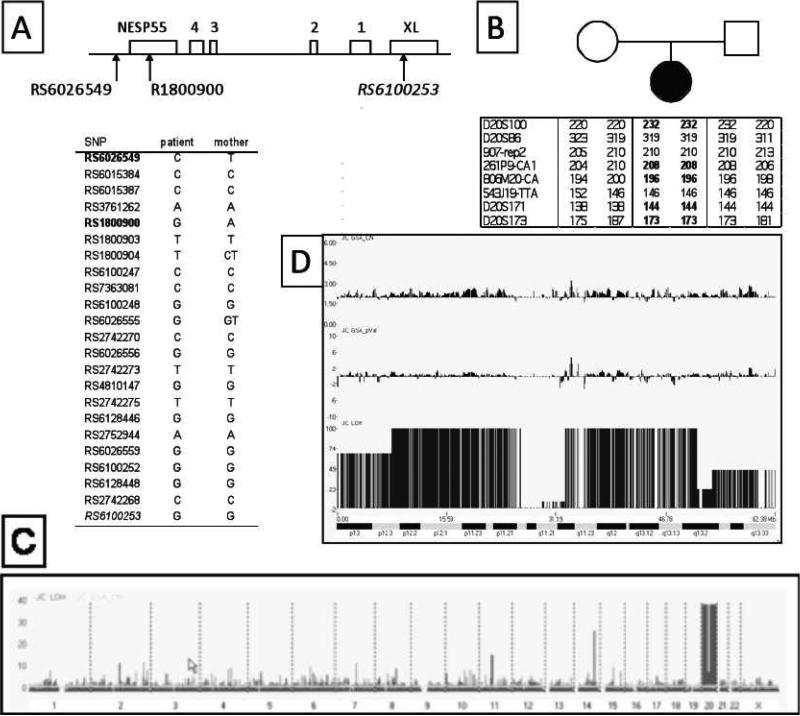
The frequent 3-kb deletion within STX16 was previously ruled out in this patient [22], and analysis of GNAS methylation showed that the patient has loss of exon A/B methylation, combined with a loss of methylation at exon XL and the promoter of the antisense transcript and a gain of methylation at exon NESP55 [23]. To search for the genetic defect responsible for the hormone resistance and this methylation pattern, we examined genomic DNA from the patient and her mother. Analysis of SNPs between GNAS exon A/B and exon NESP55 and of microsatellites within the chromosomal region comprising GNAS revealed loss of heterozygosity and allelic discordance between the patient and her mother (Figure 1A,B). 100K SNP chip analysis of DNA from the patient and her parents showed that the loss of heterozygosity affects the entire chromosome 20, but it also indicated no changes in copy number (Figure 1C,D). These findings thus indicated that the patient has a paternal uniparental isodisomy of the entire chromosome 20.
Figure 1. Identification of a paternal UPD of entire chromosome 20 in the index patient.
A. Analysis of SNPs within the upstream region of GNAS. Genomic DNA from the patient and her mother were genotyped for 23 SNPs clustered within the centromeric portion of GNAS that includes exons NESP55, XL, and exons 1-4 of the antisense transcript, revealing two discordant SNPs (bold) located upstream or within NESP55.
B. Analysis of polymorphisms in the chromosomal region comprising GNAS. The patient and her parents were also genotyped for microsatellites within the telomeric end of chromosome 20q, demonstrating a loss of the maternal allele for five of eight informative markers (bold).
C. 100K SNP Chip analysis of leukocyte genomic DNA. Genome-wide analysis with LOH probability (black) and the copy number (CN; grey) were plotted together.
D. Analysis of chromosome 20. The probability of LOH (JC LOH) is significantly elevated, while the copy number (JC GSA_CN) and the p-value (JC GSA_pVal) for the same chromosome are not different from other chromosomes. The gap represents the centromeric region. The cytobands of chromosome 20 are displayed below. Analysis was performed concomitantly on DNA samples from the patient and her parents, but the results shown here were obtained with DNA from the patient. The parental DNA did not reveal any abnormalities (data not shown).
DISCUSSION
The identified paternal UPD20 in our patient is consistent with the observed methylation defects at the GNAS locus and hormone resistance, and re-emphasizes the important role of GNAS methylation in silencing Gsα expression in the proximal renal tubules and thus the development of PTH-resistant hypocalcemia and hyperphosphatemia. One previously reported sporadic case of PHP-Ib was shown to have paternal UPD involving chromosome 20 [18]. In addition, paternal UPD20 causing PHP-Ib has also been identified in a family in which three additional members were coincidentally affected by PHP-Ia [19]. In these two UPD cases, the long arm of chromosome 20 was duplicated, whereas the UPD in our patient involves the entire chromosome. Based on the similarity of the clinical and laboratory findings in the two patients with patUPD20q and the patient with patUPD20 described herein, it appears unlikely that duplication of the paternal short arm of chromosome 20 results in significant clinical phenotypes. Nonetheless, it remains conceivable that aberrant gene expression from as-yet-undefined imprinted loci and/or unmasked recessive traits located on this chromosomal region contributed to our patient's severe bone defects, which are predicted to be a consequence of secondary hyperparathyroidism associated with renal PTH-resistance [24], yet were not reported for both patients with patUPD20q [18, 19].
Paternal UPD involving chromosome 20q and, particularly, the GNAS locus, appears to be a rare cause of PHP-Ib. Nevertheless, we suggest that this genetic abnormality should be considered as a possible molecular cause in sporadic PHP-Ib cases, particularly since it has important implications for genetic counseling. In fact, four patients with PHP-Ib due to partial paternal uniparental disomy of chromosome 20 were recently described [25], thus confirming our point that patients with broad GNAS methylation defects should be evaluated regarding this genetic defect.
ACKNOWLEDGEMENTS
The authors would like to thank Michael P. Goldman, BS, for his help in reviewing the patient's clinical and laboratory findings.
Funding support: This work was funded by research grants from National Institutes of Diabetes and Digestive and Kidney Diseases (R01 DK073911 to MB and R37 DK46718 to HJ)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None
REFERENCES
- 1.Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. BioEssays. 2000;22:452–459. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 2.Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, Pollack MS, O'Brien WE, Beaudet AL. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–26. [PMC free article] [PubMed] [Google Scholar]
- 3.Bacolod MD, Schemmann GS, Giardina SF, Paty P, Notterman DA, Barany F. Emerging paradigms in cancer genetics: some important findings from high-density single nucleotide polymorphism array studies. Cancer Res. 2009;69:723–7. doi: 10.1158/0008-5472.CAN-08-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuna M, Knuutila S, Mills GB. Uniparental disomy in cancer. Trends Mol Med. 2009;15:120–8. doi: 10.1016/j.molmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Plagge A, Kelsey G, Germain-Lee EL. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J Endocrinol. 2008;196:193–214. doi: 10.1677/JOE-07-0544. [DOI] [PubMed] [Google Scholar]
- 6.Peters J, Williamson CM. Control of imprinting at the Gnas cluster. Adv Exp Med Biol. 2008;626:16–26. doi: 10.1007/978-0-387-77576-0_2. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Litman D, Rosenberg M, Yu S, Biesecker L, Weinstein L. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J. Clin. Invest. 2000;106:1167–1174. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastepe M, Pincus JE, Sugimoto T, Tojo K, Kanatani M, Azuma Y, Kruse K, Rosenbloom AL, Koshiyama H, Jüppner H. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum. Mol. Genet. 2001;10:1231–1241. doi: 10.1093/hmg/10.12.1231. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein LS, Chen M, Xie T, Liu J. Genetic diseases associated with heterotrimeric G proteins. Trends Pharmacol Sci. 2006;27:260–6. doi: 10.1016/j.tips.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Bastepe M. The GNAS locus and pseudohypoparathyroidism. Adv Exp Med Biol. 2008;626:27–40. doi: 10.1007/978-0-387-77576-0_3. [DOI] [PubMed] [Google Scholar]
- 11.de Nanclares GP, Fernandez-Rebollo E, Santin I, Garcia-Cuartero B, Gaztambide S, Menendez E, Morales MJ, Pombo M, Bilbao JR, Barros F, Zazo N, Ahrens W, Jüppner H, Hiort O, Castano L, Bastepe M. Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab. 2007;92:2370–3. doi: 10.1210/jc.2006-2287. [DOI] [PubMed] [Google Scholar]
- 12.Mariot V, Maupetit-Mehouas S, Sinding C, Kottler ML, Linglart A. A maternal epimutation of GNAS leads to Albright osteodystrophy and parathyroid hormone resistance. J Clin Endocrinol Metab. 2008;93:661–5. doi: 10.1210/jc.2007-0927. [DOI] [PubMed] [Google Scholar]
- 13.Unluturk U, Harmanci A, Babaoglu M, Yasar U, Varli K, Bastepe M, Bayraktar M. Molecular diagnosis and clinical characterization of pseudohypoparathyroidism type-Ib in a patient with mild Albright's hereditary osteodystrophy-like features, epileptic seizures, and defective renal handling of uric acid. Am J Med Sci. 2008;336:84–90. doi: 10.1097/MAJ.0b013e31815b218f. [DOI] [PubMed] [Google Scholar]
- 14.Mantovani G, de Sanctis L, Barbieri AM, Elli FM, Bollati V, Vaira V, Labarile P, Bondioni S, Peverelli E, Lania AG, Beck-Peccoz P, Spada A. Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab. 2010;95:651–8. doi: 10.1210/jc.2009-0176. [DOI] [PubMed] [Google Scholar]
- 15.Bastepe M, Fröhlich LF, Hendy GN, Indridason OS, Josse RG, Koshiyama H, Körkkö J, Nakamoto JM, Rosenbloom AL, Slyper AH, Sugimoto T, Tsatsoulis A, Crawford JD, Jüppner H. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 2003;112:1255–63. doi: 10.1172/JCI19159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastepe M, Fröhlich LF, Linglart A, Abu-zahra HS, Tojo K, Ward LM, Jüppner H. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type-Ib. Nat Genet. 2005;37:25–37. doi: 10.1038/ng1487. [DOI] [PubMed] [Google Scholar]
- 17.Chillambhi S, Turan S, Hwang DY, Chen HC, Juppner H, Bastepe M. Deletion of the Noncoding GNAS Antisense Transcript Causes Pseudohypoparathyroidism Type Ib and Biparental Defects of GNAS Methylation in cis. J Clin Endocrinol Metab. 2010;95:3993–4002. doi: 10.1210/jc.2009-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastepe M, Lane AH, Jüppner H. Paternal uniparental isodisomy of chromosome 20q (patUPD20q) - and the resulting changes in GNAS1 methylation - as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68:1283–1289. doi: 10.1086/320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecumberri B, Fernandez-Rebollo E, Sentchordi L, Saavedra P, Bernal-Chico A, Pallardo LF, Jimenez Bustos JM, Castano L, De Santiago M, Hiort O, Perez de Nanclares G, Bastepe M. Coexistence of two different pseudohypoparathyroidism subtypes (Ia and Ib) in the same kindred with independent Gs{alpha} coding mutations and GNAS imprinting defects. J Med Genet. 2009 doi: 10.1136/jmg.2009.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jüppner H, Schipani E, Bastepe M, Cole DEC, Lawson ML, Mannstadt M, Hendy GN, Plotkin H, Koshiyama H, Koh T, Crawford JD, Olsen BR, Vikkula M. The gene responsible for pseudohypoparathyroidism type Ib is paternally imprinted and maps in four unrelated kindreds to chromosome 20q13.3. Proc. Natl. Acad. Sci. USA. 1998;95:11798–11803. doi: 10.1073/pnas.95.20.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altug-Teber O, Dufke A, Poths S, Mau-Holzmann UA, Bastepe M, Colleaux L, Cormier-Daire V, Eggermann T, Gillessen-Kaesbach G, Bonin M, Riess O. A rapid microarray based whole genome analysis for detection of uniparental disomy. Hum Mutat. 2005;26:153–9. doi: 10.1002/humu.20198. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal C, Seigle R, Agarwal S, Bilezikian JP, Hyman JE, Oberfield SE. Pseudohypoparathyroidism: a rare cause of bilateral slipped capital femoral epiphysis. J Pediatr. 2006;149:406–8. doi: 10.1016/j.jpeds.2006.04.057. [DOI] [PubMed] [Google Scholar]
- 23.Linglart A, Bastepe M, Jüppner H. Similar clinical and laboratory findings in patients with symptomatic autosomal dominant and sporadic pseudohypoparathyroidism type Ib despite different epigenetic changes at the GNAS locus. Clin Endocrinol (Oxf) 2007;67:822–31. doi: 10.1111/j.1365-2265.2007.02969.x. [DOI] [PubMed] [Google Scholar]
- 24.Farfel Z. Pseudohypohyperparathyroidism-pseudohypoparathyroidism type Ib. J. Bone Miner. Res. 1999;14:1016. doi: 10.1359/jbmr.1999.14.6.1016. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Rebollo E, Lecumberri B, Garin I, Arroyo J, Bernal-Chico A, Goni F, Orduna R, Castano L, Perez de Nanclares G. New mechanisms involved in paternal 20q disomy associated with pseudohypoparathyroidism. Eur J Endocrinol. doi: 10.1530/EJE-10-0435. In press. [DOI] [PubMed] [Google Scholar]