Abstract
The artificial inhibition of expression of genes in Saccharomyces cerevisiae is not a widespread, useful phenomenon. The external guide sequence (EGS) technology, which is well-proven in bacteria and mammalian cells in tissue culture and in mice, can also be utilized in yeast. The TOP2 and SRG1 genes can be inhibited by ∼30% with EGSs in vivo. Results in vitro also show convenient cleavage of the relevant transcripts by RNase P and appropriate EGSs. The feasible constructs shown to date have an EGS covalently linked to M1 RNA, the RNA subunit of RNase P from Escherichia coli. Greater efficiency in cleavage of transcripts can be fashioned using more than one EGS targeted to different sites in a transcript and stronger promoters controlling the EGS constructs.
Keywords: RNase P, target RNA, TOP2 gene, SRG1 gene
INTRODUCTION
RNase P, which is responsible for generation of the matured 5′ end of tRNAs in prokaryotes and eukaryotes, consists of one RNA and up to 10 protein subunits in eukaryotes (Altman and Kirsebom 1999). The RNA components of archaeal and eukaryotic RNase P are inefficient catalysts in vitro in the absence of their respective protein subunits, unlike that of the RNA subunit of bacterial RNase P, and show some activity at high salt concentrations (Kikovska et al. 2007). Studies of substrate recognition by M1 RNA, the RNA subunit of Escherichia coli and RNase P have led to the development of a general strategy of gene targeting in which M1 RNA and RNase P can be used as tools to cleave any specific RNA sequence (Li et al. 1992; Yuan et al. 1992; Guerrier-Takada and Altman 2000) as mentioned above. The target RNA is cleaved by RNase P and remains untranslated.
In several recent studies, the external guide sequence (EGS)-based cleavage by RNase P (Forster and Altman 1990) has been used successfully to inhibit gene expression in bacterial, animal, and plant cells (Guerrier-Takada et al. 1997; Plehn-Dujowich and Altman 1998; Raj et al. 2001; Dreyfus et al. 2004; Rangarajan et al. 2004; Pei et al. 2008), while its utility in yeast remains unexplored. Therefore, we undertook this study to test the viability of this approach for disrupting expression of host genes in Saccharomyces cerevisiae.
The utility of S. cerevisiae as an organism for elucidating genetic and metabolic pathways cannot be overestimated. Gene knockdowns have been employed with considerable efficiency in this regard. Artificial methods of inactivating gene expression have not been so reliable in yeast. In this report two yeast genes were chosen as the targets: the TOP2 gene, which is essential for cell viability, encodes the topoisomerase II (1428 amino acids) for catalyzing of the interconversion between topological states of DNA by breaking and rejoining DNA strands (Goto and Wang 1984) and the SRG1 gene (SER3 regulatory gene 1, a noncoding regulatory RNA) expresses a 553-nt non-protein-coding RNA to repress expression of its neighboring gene SER3 via a transcription interference mechanism (Martens et al. 2004). We now show that TOP2 and SRG1 RNAs can be cleaved in vitro by a construct that includes M1 RNA, the RNA subunit of E. coli RNase P, covalently linked to an EGS. The expression of the two genes has been inactivated by ∼30% compared to normal expression as measured by Northern blots, but the effect on cell growth is more dramatic during the relevant time period as is shown below.
MATERIALS AND METHODS
Materials
Competent E. coli DH5α, S. cerevisiae strain INVSc1 (his−, leu−, trp−, ura−), yeast expression vector pYES2.0 and S.c. EasyComp Transformation Kit were from Invitrogen. Yeast expression vector pCu426CUP1 was a gift from Dr. Dennis J. Thiele (Duke University). The composition of yeast media and general manipulations were as described in S.c. EasyComp Transformation Kit handbook. Yeast extract-peptone-dextrose (YPD) and synthetic complete (SC) media lacking uracil were used for yeast growth and plasmid maintenance. Glucose was used as the carbon source unless otherwise indicated. DNA restriction and other enzymes reacting with DNA or RNA were obtained from New England Biolabs unless described otherwise. QIAquick gel extraction kit and QIAprep spin miniprep kit were purchased from Qiagen. Sephadex G-50 Quick Spin columns and Ampli-Taq DNA polymerase were purchased from Roche Applied Science. Radiochemicals were purchased from Perkin-Elmer. Hybridization buffer Rapid-hyb was from GE Healthcare UK Limited. Oligonucleotides were synthesized by the Keck Facility at Yale University.
Construction of plasmids
DNA fragments that encode TOP2 and SRG1 were isolated by PCR amplification using S. cerevisiae genome DNA as template and oligonucleotide sets: TOP2-F (5′-CGGGGTACCACGACAATTTTAACTTTCTGTTTCAGTTAAAG-3′)/TOP2-R (5′-CCGGAATTCAGTGGAAAATATATTACAAAGCTTAGCACC-3′), SRG1-F (5′-CGGGGTACCTAATGCCTTTGTTTGGCCAAGC-3′)/SRG1-R (5′-CGCGGATCCACAGCGCCTGGGGAG-3′) as primers, respectively. The amplified fragments were purified and digested by KpnI/EcoRI or KpnI/BamHI and inserted into the same sites of the pBluscript II SK (+) vector, respectively. The new constructs were named pBSKTOP2 and pBSKSRG1, which contained the yeast TOP2 or SRG1 parts of the gene under the control of the T7 promoter. EGSs against TOP2 and SRG1 mRNA were designed based on the rEGS assay and partial RNase T1 assay according to previous reports (Lundblad et al. 2008). To create the plasmids producing EGS RNAs, PCR amplification was carried out using pUC19TyrT DNA containing the gene for precursor tRNATyr (pTyr) from E. coli as a template, and the following oligonucleotide sets as primers: T12-F (5′-AAACTGCAGATGGAGCAGACTCTAAATC-3′)/T12-R (5′-TCCGGTGACCTGGTGCACGGCGAAGGATTCGAACC-3′), T28-F (5′-AAACTGCAGGTCGAGCAGACTCTAAATC-3′)/T28-R (5′-TCCGGTGACCTGGTGAGTTTAGAAGGATTCGAACC-3′), S15-F (5′-AAACTGCAGCACAAGCAGACTCTAAATC-3′)/S15-R (5′-TCCGGTGACCTGGTGGCCAAGGAAGGATTCGAACC-3′).
The PCR products were subcloned as PstI/BstEII fragments into the appropriate sites in the pUCT7/AEFRNAHHT7T (Xiao et al. 2008) under the control of T7 promoters, respectively, generating the 3/4EGS plasmids, pUCT7/T12, pUCT7/T28, or pUCT7/S15. The definition of 3/4EGS is given in Guerrier-Takada and Altman (2000). The word “M1 3/4EGS” merely means the 3/4EGS that is attached to the 3′ end of M1 RNA. The same PCR fragments were also individually subcloned into pUCT7/M1AEFRNAHHT7T (Xiao et al. 2008) to construct M1 3/4EGS plasmids pUCT7/M1T12, pUCT7/M1T28, or pUCT7/M1S15.
The constructs (pUCT7/T12, pUCT7/T28, pUCT7/M1T12, pUCT7/M1T28) were further digested with PvuII/BamHI and individually inserted into pYES2.0 vector (Invitrogen) under the control of GAL1 promoter to create galactose-induced yeast expressional plasmids T12, T28, M1T12, and M1T28. In the same manner, pUCT7/S15 and pUCT7/M1S15 were digested by BamHI/SalI and subcloned into the expression vector pCu426CUP1 under the control of the CUP1 promoter, respectively, to create Cu2+-induced 3/4EGS and M1 3/4EGS plasmids (S15 and M1S15) for tests in vivo.
Transcription of substrates in vitro
Plasmid DNA of pBSKTOP2 and pBSKSRG1 were digested by EcoR1 or BamH1 for transcription in vitro of RNA substrates by T7 RNA polymerase. The constructs pUCT7/T12, pUCT7/T28, pUCT7/M1T12, pUCT7/M1T28, pUCT7/S15, and pUCT7/M1S15 that had been linearized by treatment with BstEII were used as temples for transcription in vitro by T7 RNA polymerase to produce 3/4EGS or M1 3/4EGS RNAs. pTyr and yeast precursor tRNASer (pSupS1) were prepared as described (Krupp et al. 1986; Ko and Altman 2007). The transcription reactions were also carried out as described previously (Yuan et al. 1992), and transcripts were purified by using Sephadex G-50 Quick Spin columns (Roche Applied Science). For 5′ end labeling, the transcribed RNA was first dephosphorylated with 0.01 units of calf intestinal phosphatase/pmol RNA, and then 5′ end-labeled by T4 polynucleotide kinase in the presence of [γ-32P] ATP. pTyr, pSupS1, and target RNA used for cleavage assays in vitro were internally labeled by transcription with [α-32P] GTP. The labeled RNAs were purified on 5% polyacrylamide/7 M urea gel at room temperature.
RNase P cleavage assay in vitro
EGS activity assays were performed according to a previous report (Lundblad et al. 2008). The EGS in 10-, 50-, or 100-fold molar excess was mixed with 10 nM of internally labeled target RNAs in PA100 buffer (50 mM Tris-Cl, pH 7.5, 100 mM MgCl2, 100 mM NH4Cl). Subsequently, 10 nM of M1 RNA or crude yeast RNase P (a gift of D. Wesolowski, Yale University) was added to the mixture and further incubated for 30 min at 37°C. For reactions with M1 3/4EGS RNA, no additional M1 RNA was added. Reactions in 10 μL volume were performed at 37°C for 30 min, and terminated by adding 10 μL of 10 M urea/10% phenol solution. The products were separated on 5% polyacrylamide gels that contained 7 M urea and were then exposed to BioMax film (Kodak).
Northern blot analysis
Preparation and transformation of competent S. cerevisiae cells were performed as described in the S.c. EasyComp Transformation Kit manual. After transformation with the galactose inducible plasmid DNA (M1T12 or M1T28) or Cu2+ inducible plasmid DNA (S15 or M1S15), single-clone transformants were cultured in SC liquid medium for two days, and then diluted to OD560 = 0.5 by galactose inducible medium or Cu2+ inducible (with or without 1 mM Cu2+) SC liquid medium. After growing for one more day, yeast pellets were obtained by centrifugation and subjected to total RNA extraction by mRNA Midi Kit (Oligotex) according to the commercial handbook. Total RNAs were separated in 1% agarose gel (15 μg per sample) and transferred onto a positively charged nylon membrane (Roche Diagnostics). Cross-linking was employed using the Stratalinker (Stratagene) and the hybridization reaction was performed in Rapid-hyb buffer as described in the commercial manual. The PCR products of TOP2 and SRG1 gene by primer sets TOP2-F/TOP2-R and SRG1-F/SRG1-R were labeled by Prime-a-Gene Labeling System (Promega) and used as probes. The probe for detection of EGSs was amplified by PCR from the conserved internal region of pTyr that was part of the mature tRNA sequence. Actin gene RNA was used as an internal control and amplified by primer set Act-F: 5′-ACGTCGCCTTGGACTTCGAACA-3′/Act-R: 5′-TGGAAGATGGAGCCAAAGCGGT-3′. After hybridization, the resulting signal was detected on the membrane using a phosphorimager (Fuji) and quantitated by image analysis software (Fuji ImageGauge).
RESULTS
Selection of target genes
TOP2 was chosen as the target for EGS-based cleavage because it is essential for yeast viability (Goto and Wang 1984). Currently, most EGS or other RNA-based techniques target coding mRNA, and reports involved in noncoding RNA, aside from miRNA, only focused on experiments in vitro (Yang and Altman 2007). Therefore, SRG1 was chosen as a non-mRNA target.
Mapping of EGS sites in vitro and preparation of EGS RNAs
EGS sites against TOP2 and SRG1 mRNA were designed by using the general methods as previously reported (Guerrier-Takada and Altman 2000). An accessible region near the start codon was identified as an ideal target for EGS binding by employing computer-aided secondary structure predictions (Zuker et al. 1999). More importantly, rapid rEGS assay (Lundblad et al. 2008) also yielded several cleavage sites in TOP2 and SRG1 mRNAs. Therefore, site 12 and site 28, which are located 12 and 28 nucleotides (nt) upstream of TOP2 start codon, respectively, as well as site 15, which is located 15 nt downstream from the start transcription point of SRG1 gene were chosen for EGS design. At each site, 3/4EGSs were then designed and synthesized by PCR amplification as described in Materials and Methods.
Cleavage of TOP2 and SRG1 RNAs in vitro
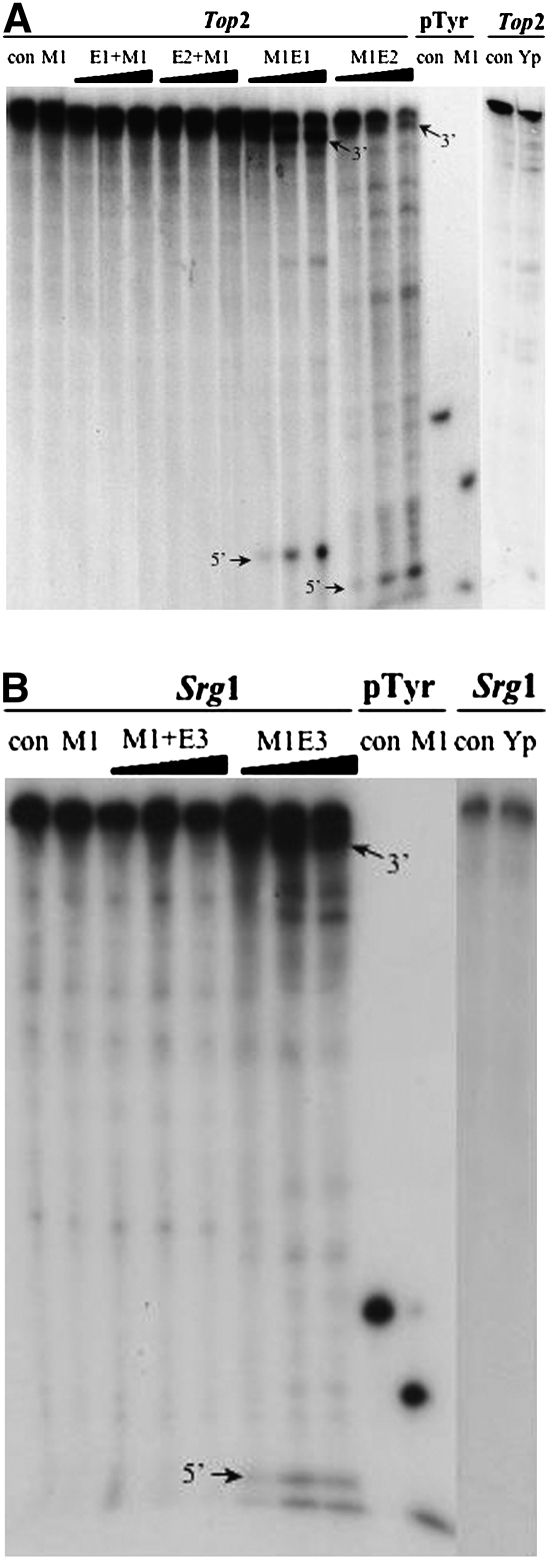
Internally labeled TOP2 and SRG1 RNA substrates were employed for RNase P cleavage assays in vitro under conditions described previously (Lundblad et al. 2008). As shown in Figure 1A, cleavage of fragments was detected when a labeled TOP2 RNA was mixed with the recombinant M1 3/4EGS12 (M1E1) or M1 3/4EGS28 (M1E2), alone. While the reaction of M1+3/4EGS12 (M1+ indicates a noncovalent addition of M1 RNA) and an EGS, for example, (M1+E1) or M1+3/4EGS28 (M1+E2) with target RNAs did not produce the expected cleavage fragments. As a control for activity of M1 RNA, internally labeled pTyr was also incubated with M1 RNA under identical conditions and yielded the predicted fragments. TOP2 RNA was also subject to cleavage in vitro by crude yeast RNase P in the presence of 3/4EGS, but no products were detected (data not shown).
FIGURE 1.
Cleavage in vitro of the RNA substrates by 3/4EGSs and M1 3/4EGSs. (A) Cleavage in vitro of the TOP2 RNA by M1 3/4EGSs. (B) Cleavage in vitro of the SRG1 RNA by M1 3/4EGSs. EGSs in 10-, 50-, or 100-fold molar excess were mixed with 10 nM of internally labeled target RNAs in PA100 buffer (50 mM Tris-Cl, pH 7.5, 100 mM MgCl2, 100 mM NH4Cl). Internally labeled pTyr was used as a positive control to check for activity of M1 RNA or M1 3/4EGS RNA. con: negative control, without enzyme or EGS; M1: E. coli M1 RNA; E1: 3/4EGS12; E2: 3/4EGS28; E3: 3/4EGS15; +M1: 3/4EGS plus extra 0.1 pmol M1 RNA; M1E: recombinant M1 3/4EGS RNA; Yp: crude yeast RNase P. When cleaved by Yp, the concentration of MgCl2 was reduced to 10 mM in the incubation buffer.
The cleavage in vitro of labeled SRG1 RNA is shown in Figure 1B. Cleavage of the target by M1 3/4EGS15 alone was most efficient and yielded the expected products (3′ fragment/5′ fragment) as well as four minor, unexpected cleaved fragments. This result indicated that there are two other cleavage sites in SRG1 mRNA for cleavage by M1¾EGS. The amount of all cleavage products increased with M1 3/4EGS15 concentration. In contrast, no cleavage fragments were obtained when SRG1 RNA and 3/4EGS15 in the presence of M1 RNA were incubated together. Cleavage of the same substrates by crude yeast RNase P was not detected in the presence of ¾EGSs (data not shown). These results showed that M1 3/4EGSs were efficient in cleavage of the targets, but 3/4EGSs are completely inactive. The cleavage efficiency of the crude yeast RNase P on target RNAs is not observable in these experiments compared to that on pSupS1 substrate (>95% for pSupS1 in 30 min at 37°C; data not shown).
M1 3/4EGS inhibits the growth of certain S. cerevisiae transformants
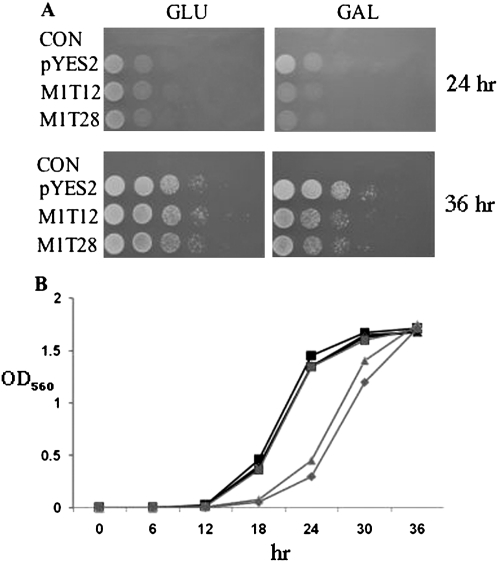
M1T12 and M1T28 plasmids, which produced M1 3/4EGS RNAs targeting TOP2 mRNA under the galactose inducible GAL1 promoter, were transformed into S. cerevisiae cells individually to check the effect of M1 3/4EGSs in vivo. The transformants grew to saturation in SC liquid medium, and then were serially diluted and spotted on SC plates that contained galactose for 24 and 36 h after incubation at 30°C. In Figure 2A, the cells containing M1T12 or M1T28 plasmid grew as well as the control cells on plates that contained glucose as the sole carbon source. When transferred to plates that contained galactose as the sole carbon source, M1T12 and M1T28 transformants grew relatively poorly in comparison to the control. The growth of cells that expressed TOP2 M1 3/4EGS in vivo is thus clearly inhibited when the EGSs are expressed.
FIGURE 2.
Growth inhibition of S. cerevisiae cells by TOP2 M1 3/4EGS RNA. (A) Phenotype investigation of M1 3/4EGSs transformants on plates. Cells were transformed with empty vector pYES2 or plasmids (M1T12 and M1T28) containing the galactose-inducible GAL1 promoter. After growth to saturation in SC liquid medium, cells were serially diluted (1/10, 1/100, 1/1000, and 1/10,000), and spotted onto SC plates containing 2% glucose (to repress expression of M1 3/4EGS) or galactose (to induce transcription). The results are shown after 24 and 36 h of incubation at 30°C. At least three independent experiments were carried out and a representative result is shown. (B) Growth profile of M1 3/4EGSs transformants in liquid medium. Cells with the indicated plasmids were incubated in SC liquid medium at 30°C to saturation, and then diluted the saturated culture to OD560 = 0.1 with SC medium containing 2% glucose or galactose. The OD560 values of each culture were measured every 6 h over 2 d. Values presented here represent means of three independent incubations of the same preparation. M1T12 and M1T28 transformants in galactose medium (triangles and pluses).
M1T12 and M1T28 transformants as well as the control strain were cultured in liquid medium at 30°C to saturation, and then individually diluted to OD560 = 0.1 with SC medium that contained either 2% glucose or galactose. The two EGS-plasmid transformants grew as fast as the control cells in glucose-containing medium (Fig. 2B). In medium that contained galactose as the sole carbon source, the growth rate of control cells was the same as in the glucose medium, but M1T12 and M1T28 transformants grew significantly more slowly. At 18 and 24 h, the OD560 value of these cultures was one-fourth the value of the control strain. At 34 h, late log phase, their OD560 values were similar.
Expression of M1 3/4EGSs in vivo leads to the reduction of TOP2 and SRG1 RNAs
To confirm that the growth inhibition of S. cerevisiae transformants was due to the reduction of the TOP2 mRNA in vivo, M1T12 and M1T28 strains were further cultured in glucose or galactose containing medium (Materials and Methods). Total mRNA was extracted and subjected to Northern blot analysis. The expression of 3/4EGS, M1 3/4EGS, and TOP2 mRNAs were normalized to a control actin mRNA.
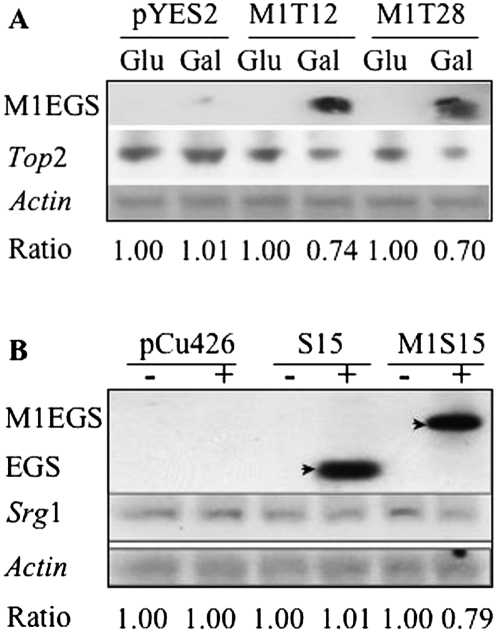
Bands that represent M1 3/4EGS RNAs are visible in the lanes fourth and sixth from the left (Fig. 3A), in which RNA samples were from galactose-induced M1T12 and M1T28 transformants. The level of TOP2 mRNA in lanes 4 and 6 was also reduced. The ratio for cells carrying empty plasmid and grown in noninducible medium was chosen as 1 (using actin mRNA as a control) in order to calculate the relative TOP2 mRNA level in other samples. The reductions of mRNA in the fourth and sixth lanes from the left were 26% and 30%, respectively.
FIGURE 3.
Expression of M1 3/4EGS in vivo leads to the reduction of TOP2 and SRG1 RNA. (A) Measurement of the steady-state levels of M1 3/4EGS and TOP2 RNAs in vivo. Cells transformed with empty vector pYES2.0, M1T12, or M1T28, grew in SC liquid medium for 2 d, and then diluted to OD560 = 0.5 by SC medium containing 2% glucose (Glu) or galactose (Gal) for one more day. Cells were then collected for total RNA extraction and Northern blot analysis. The ratio for cells carrying pYES2 plasmid and grown in glucose-containing medium was chosen as 1 to calculate the relative TOP2 mRNA level in other samples. (B) The steady-state levels of 3/4EGS15 and M1 3/4EGS15 RNAs in vivo. Cells transformed with empty vector pCu426CUP1 and Cu2+-inducible plasmids, S15 or M1S15, grew on SC liquid medium for 2 d, and were then diluted to OD560 = 0.5 with SC medium in the presence (+) or absence (−) of 1 mM Cu2+ for one more day. After that, cells were collected for total RNA extraction and Northern blot analysis. The ratio for cells carrying pCu426CUP1 plasmid and grown in the absence of Cu2+ medium was chosen as 1 with actin mRNA as a control to calculate the relative SRG1 RNA level in other samples.
The expression of SRG1 RNA in the strains that were transformed with Cu2+-inducible plasmid DNA (pCu426CUP1, S15, or M1S15) was also checked. The strains were cultured with or without 1 mM Cu2+ in Sc liquid medium for 1 d. Total mRNA was extracted and Northern blot analysis was performed on the RNA. Hybridization signals were detected in the lanes fourth and sixth from the left (Fig. 3B), and indicated that the expected sizes of 3/4EGS and M1 3/4EGS RNA were transcribed by Cu2+ induction in cells containing S15 or M1S15 plasmids, respectively. The full-length M1 3/4EGS is readily observable in the sixth lane from the left. In the fourth lane the 3/4EGSS15 can be seen. Normalized to actin mRNA control, SRG1 RNA level in lane 6 was found to be reduced to 79%. SRG1 RNA in cells that contained 3/4EGSS15 was not reduced within experimental error in expression.
The results show that M1 3/4EGS RNA was capable of reducing target mRNA in vivo but 3/4EGS RNA could not. These data are consistent with RNase P mediated cleavage in vitro of TOP2 and SRG1 RNAs, in which M1 3/4EGSs are efficient in mediating cleavage of the target RNA but 3/4EGSs were inactive in this respect.
DISCUSSION
Although the utility of EGS-directed cleavage by RNase P had been demonstrated in bacterial, animal, and plant cells (Guerrier-Takada et al. 1995; Plehn-Dujowich and Altman 1998; Raj et al. 2001; Dreyfus et al. 2004; Rangarajan et al. 2004; Pei et al. 2008), similar possibilities for inhibition of gene expression in yeast cells remained unclear. We show here that M1 3/4EGS RNA alone is efficient in cleavage of yeast target RNA in vitro, and was capable of reducing the target mRNA in vivo by up to 30% as measured by Northern blots. The cleavage by E. coli M1 RNA or crude yeast RNase P with ¾EGS is unable to cleave both targets in vitro and the enzyme cannot do this in vivo. Since the cleavage in vitro of target mRNA by crude yeast RNase P was not detected in the presence of 3/4EGSs (or in vivo), yeast RNase P must have a different mode of specificity for recognition of substrates compared to the M1 3/4EGS. This manner of specificity is also different from that observed in mammalian cells (Guerrier-Takada and Altman 2000).
The data presented here also show that the efficiency of EGS-directed cleavage by RNase P is somewhat limited, because the level of target RNAs TOP2 or SRG1 in vivo was only reduced to 70% and 79% compared to normal levels, respectively, in contrast to much larger effects of EGSs in E. coli and human tissue culture cells. The methodology we used can certainly be developed further by identification of several sites in the same RNA that can be cleaved by different EGSs, construction of multiple EGS expression plasmids, and the use of a more powerful inducible promoter or higher-copy vector.
ACKNOWLEDGMENTS
We thank members of our laboratory for helpful discussions and the members of Shirleen Roeder's laboratory, especially Dr. C. Yellman (Yale University) for advice. X.C. and J.-H.K. were supported in part by funds from the Armed Forces Institute of Pathology. We declare no conflict of interest.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2538711.
REFERENCES
- Altman S, Kirsebom L 1999. Ribonuclease P. In The RNA world, 2nd ed (ed. Gesteland RF et al. ), pp. 351–380 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Dreyfus DH, Matczuk A, Fuleihan R 2004. An RNA external guide sequence ribozyme targeting human interleukin-4 receptor alpha mRNA. Int Immunopharmacol 4: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Forster AC, Altman S 1990. External guide sequences for an RNA enzyme. Science 249: 783–786 [DOI] [PubMed] [Google Scholar]
- Goto T, Wang JC 1984. Yeast DNA topoisomerase II is encoded by a single-copy essential gene. Cell 36: 1073–1080 [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Altman S 2000. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol 313: 442–456 [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Li Y, Altman S 1995. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc Natl Acad Sci 92: 11115–11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Salavati R, Altman S 1997. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc Natl Acad Sci 94: 8468–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikovska E, Svärd SG, Kirsebom LA 2007. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci 104: 2062–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Altman S 2007. OLE RNA, an RNA motif that is highly conserved in several extremophilic bacteria, is a substrate for and can be regulated by RNase P RNA. Proc Natl Acad Sci 104: 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp G, Cherayil B, Frendewey D, Nishikawa S, Söll D 1986. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J 5: 1697–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guerrier-Takada C, Altman S 1992. Targeted cleavage of mRNA in vitro by RNase P from Escherichia coli. Proc Natl Acad Sci 89: 3185–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad EW, Xiao G, Ko JH, Altman S 2008. Rapid selection of accessible and cleavable sites in RNA by Escherichia coli RNase P and random external guide sequences. Proc Natl Acad Sci 105: 2354–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Pei DS, Sun YH, Long Y, Zhu ZY 2008. Inhibition of no tail (ntl) gene expression in zebrafish by external guide sequence (EGS) technique. Mol Biol Rep 35: 139–143 [DOI] [PubMed] [Google Scholar]
- Plehn-Dujowich D, Altman S 1998. Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc Natl Acad Sci 95: 7327–7332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj ML, Pulukkunat DK, Reckard JF 3rd, Thomas G, Gopalan V 2001. Cleavage of bipartite substrates by rice and maize ribonuclease P. Application to degradation of target mRNAs in plants. Plant Physiol 125: 1187–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan S, Raj ML, Hernandez JM, Grotewold E, Gopalan V 2004. RNase P as a tool for disruption of gene expression in maize cells. Biochem J 380: 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Lundblad EW, Izadjoo M, Altman S 2008. Inhibition of expression in Escherichia coli of a virulence regulator MglB of Francisella tularensis. PLoS ONE 3: e3719–e3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Altman S 2007. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA 13: 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Hwang ES, Altman S 1992. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci 89: 8006–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH 1999. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA Biochemistry and Biotechnology (ed. Barciszewski J, Clark BFC), NATO ASI Series. Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]