SUMMARY
Epithelial cells of the thymus cortex express a unique proteasome particle involved in positive T cell selection. This thymoproteasome contains the recently discovered β5t subunit that has an uncharted activity, if any. We synthesized fluorescent epoxomicin probes that were used in a chemical proteomics approach, entailing activity-based profiling, affinity purification, and LC-MS identification, to demonstrate that the β5t subunit is catalytically active in the murine thymus. A panel of established protea-some inhibitors showed that the broad-spectrum inhibitor epoxomicin blocks the β5t activity and that the subunit-specific antagonists bortezomib and NC005 do not inhibit β5t. We show that β5t has a substrate preference distinct from β5/β5i that might explain how the thymoproteasome generates the MHC class I peptide repertoire needed for positive T cell selection.
INTRODUCTION
The ability to recognize nonself oligopeptides is a key feature of mammalian immunity. T cells that recognize antigenic oligopep-tides elicit a directed adaptive immune response aimed at the identification and eventual eradication of the invading pathogen that is the source of the nonself protein from which the antigenic oligopeptide is derived (Janeway and Bottomly, 1994; Medzhitov, 2007). T cell recognition is effected by binding of specific T cell receptors to the antigenic peptides that are complexed to either major hisocompatibility complex (MHC) class I or MHC class II molecules (Huseby et al., 2005; Takahama et al., 2008). MHC I molecules present oligopeptides derived from cytosolic and nuclear proteins to CD8+ cytotoxic T lymphocytes (CTL) and by this virtue report on the presence of virally encoded proteins (Kloetzel and Ossendorp, 2004). T cells specific for nonself peptides are produced by thymic selection. The generation in the thymus of nonself peptide-selective CTL proceeds in two discreet events (Nitta et al., 2008). Positive selection is mediated by cortical thymic epithelial cells. In this process, thymocytes expressing T cell receptors are confronted with tissues expressing MHC I molecules loaded with oligopeptides. Current understanding is that the MHC I/peptide antigen complexes produced by cortical thymic epithelial cells are low-affinity T cell receptor binders. Thymocytes passing through the thymic cortex that bind to MHC I molecules carrying a peptide load are selected from thymocytes expressing nonbinding receptors. In the ensuing negative selection step, mediated by medullary thymic epithelial cells, thymocytes from the positively selected pool that are responsive to MHC I molecules exposing self-peptides are eliminated.
Recently, Tanaka and co-workers made a major breakthrough toward understanding how positive selection proceeds (Murata et al., 2007). They found that epithelial cells at the thymic cortex express, next to the constitutive proteasome and the immunoproteasome, a third 20S proteasome particle which was dubbed the thymoproteasome. The 20S core particle of the proteasome is assembled from α and β subunits in a pattern of four, stacked, heptameric rings (α1-7, β1-7, β1-7, α1-7) generating a barrel-shaped structure that contains two copies of the catalytically active β subunits: β1 (post acidic), β2 (tryptic-like), β5 (chymotriptic-like) peptidase activities (Baumeister et al., 1998). The thymoproteasome contains the β1i and β2i subunits just like the immunoproteasome, with the important exception that the unique subunit β5t replaces the immunoproteasome-specific subunit, β5i.
The thymoproteasome is the most abundant proteasome species in cortical thymic epithelial cells (cTEC). Thymoprotea-some expression may have implications for the repertoire of oligopeptides presented by MHC I molecules on the surface of cTECs that might significantly differ from to the repertoire produced by medullary thymic epithelial cells. Closer inspection of the thymoproteasome 20S particle revealed that, in contrast to the constitutive and the immunoproteasome, it possessed little chymotryptic activity, a finding that seems to correlate with the hydrophilic nature of the putative substrate-binding site of β5t compared with β5/β5i (Murata et al., 2007). In theory, β5t can contribute in two ways to the generation of specific MHC I peptides used in positive T cell selection (Murata et al., 2008). It could act as an impassive, catalytically inactive bystander, in which case β1i/β2i produces the majority of MHC I peptides with a bias toward their substrate preferences. Alternatively, it could actively participate in protein degradation and assist in producing “nonself” peptides thanks to its intrinsic substrate preference, which then must be distinct from that of β5/β5i.
Activity-based probes are synthetic compounds bearing a reporter or affinity tag and an enzyme reactive group that can covalently bind to the active site of an enzyme (Cravatt et al., 2008). The tagged enzymatic activities can than be visualized by fluorescence or affinity purified, digested with trypsin, and identified by LC/MS analysis. We here demonstrate, by making use of activity-based proteasome probes (Verdoes et al., 2009), that β5t is in fact a catalytically active subunit and show that its preference toward established proteasome inhibitors differs substantially from those of β5/β5i.
RESULTS AND DISCUSSION
Activity-Based Profiling Reveals β5t Activity
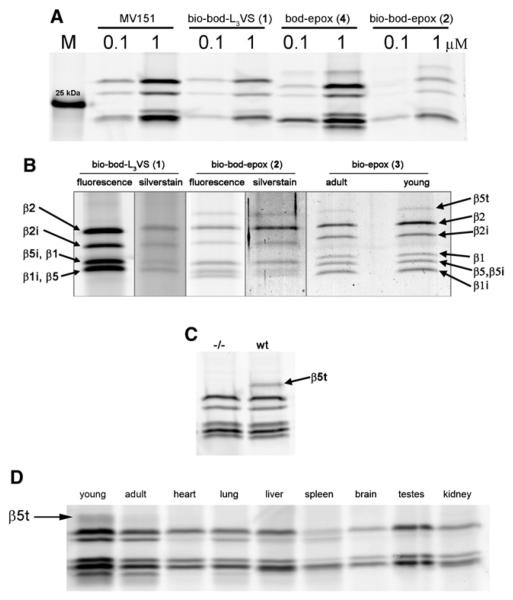
As the first experiment, we incubated whole tissue thymus homogenate from 3-week-old mice with the fluorescent broad-spectrum ABPs 1,(Verdoes et al., 2008) 2, 4, and MV151 (Verdoes et al., 2006) shown in Figure 1 (for the synthesis of probes 2 and 4, see Supplemental Experimental Procedures available online). Proteins were resolved by SDS-PAGE under reducing conditions and fluorescently labeled proteasome subunits were visualized by in-gel fluorescence scanning. In Figure 2A, MV151 shows the typical band pattern of staining that is similar to that of the EL4 cell line expressing the constitutive and the immunoproteasome (Kessler et al., 2001)(Figure S1) indicating that both particles are expressed in the thymus. Peptide vinyl sulphone 1, the biotinylated derivative of MV151, shows a similar pattern as MV151. Interestingly, the peptide epoxyketones 2 and 4 show two new bands that run below and above the constitutive and immunoproteasome subunits. Of these, the lower band corresponds to β1i.
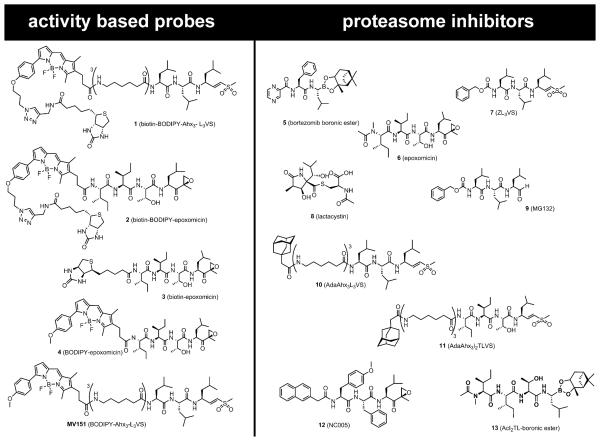
Figure 1. Activity-Based Probes and Proteasome Inhibitors Used in This Study.
In addition to the enzyme reactive group (warhead) and targeting sequence of the inhibitors, activity-based probes are equipped with a fluorophore for in-gel detection, a biotin tag for affinity purification or with both.
Figure 2. Activity-Based Profiling, Affinity Purification, and LC-MS Identification of Proteasome β Subunits in Murine Tissues Lysates.
(A) In-gel fluorescence detection of active proteasome β subunits in 3-week-old wild-type murine thymus homogenate after labeling with MV151, ABP 1, 2, and 4 (see also Figure S1). M indicates the molecular marker band of 25 kDa.
(B) In-gel fluorescence and silver stain detection of active proteasome β subunits in young and adult thymus after labeling with ABP 1, 2, 3, and affinity purification. Protein identification by LC-MS analysis of in-gel digested silver-stained bands (see Tables S1 and S2 for details).
(C) In-gel fluorescence detection with ABP 4 of β5t activity in wild-type and absence of activity in the (−/−) β5t knockdown thymus from 3-weeks-old mice.
(D) Activity-based proteasome profiling using ABP 4 shows β5t activity in murine thymus (young and adult) but not in heart, lung, liver, spleen, brain, testes, or kidney.
To ascertain whether the new, epoxyketone-sensitive protein, with a gel mobility corresponding to the predicted molecular weight of β5t (Murata et al., 2007), is indeed β5t and not a thymus-specific gene product unrelated to the thymoprotea-some, we performed a pull-down experiment by making use of the biotin moiety present in ABPs 1 and 2. Biotinylated proteins from thymus homogenate were captured by streptavidin-coated magnetic beads, resolved by SDS-PAGE and detected both by fluorescence and silver staining. Figure 2B shows the specific purification of several proteins that run in a pattern similar with that of Figure 2A. Protein ID, indicated by arrows in Figure 2B, was determined by on-bead (Table S2) and in-gel tryptic digestion followed by LC-MS/MS analysis. Oligopeptides corresponding to the expected constitutive proteasome (β1/β2/β5) and immunoproteasome (β1i/β2i/β5i) were captured by ABP 1 but no evidence for β5t was found. Peptides derived from β5t were found by affinity purification with ABP 2, indeed in the band running higher than the other active β subunits. However, the protein yield achieved by pull-down with ABP 1 and 2 was low, and we hypothesized that the short biotinylated epoxomicin ABP 3 might increase the pull-down efficiency (for the synthesis of probe 3, see Supplemental Experimental Procedures). ABP 3 performed as expected, showing bands of similar pattern as ABP 2, stronger signal in silver-stained gels and reliable LCMS identification of proteins (Table S1). Thymus from adult animals treated in the same fashion shows β5t activity as well, which suggests that the murine thymoproteasome remains active for at least 6 months. Next to the active proteasome β subunits, only four endogenously biotinylated background proteins were recovered with this method, a result that reflects the selectivity of ABPs 1, 2, 3, and 4 toward proteasomes. Thymus lysates of 2-week-old mice in which the β5t protein expression was genetically knocked down show normal activity of immuno- and constitutive proteasome compared with the wild-type, but complete absence of β5t activity (Figure 2C). To characterize the expression of β5t in murine tissues, we performed a tissue scan with ABP probe 4. Figure 2D shows that β5t activity is exclusively present in the young thymus and at lower activity in thymus of 6-month-old mice. Integration of the fluorescent signal from young thymus indicated that β5t contributes to some 4% of the total active signal in this full thymus lysate. Heart, lung, liver, spleen, brain, testes, and kidney do not show β5t activity. The presence of immunoproteasome bands in the heart, lung, liver, and spleen tissues is explained by the presence of lymphocytes in these organs.
LC-MS3 Analysis of the β5t Active-Site Peptide
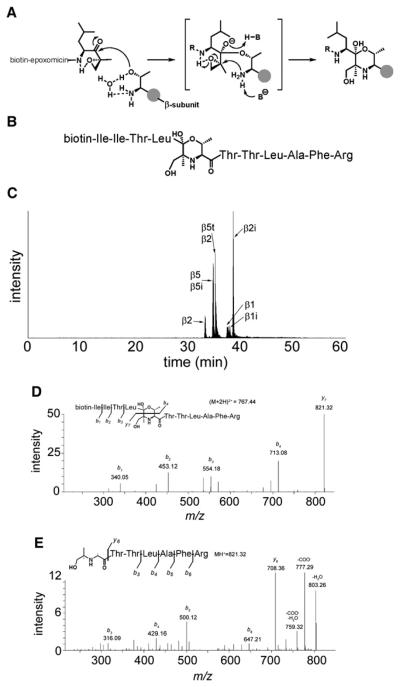
Isolation and analysis of the active-site peptide covalently bound to ABP probe 3 would be the ultimate proof for the β5t acitivity. Biotin-epoxomicin binds to the catalytic N-terminal threonine via an irreversible morpholino ring formation shown in Figure 3A. The β5t active-site peptide (Figure 3B) is generated after denaturation and tryptic digest of the thymoproteasome. Given that biotin-epoxomicin binds to all active β subunits, we expect to find six different active-site peptides because the tryptic peptides derived from β5 and β5i are identical (see Table S3). After LC-MS analysis, the active-site peptides were identified from the high resolution full MS scans by their exact mass and charge (Figure 3C). Further evidence was provided by the MS/MS (MS2) fragmentation that revealed the presence of the biotin-epoxomicin signature ions b1, b2, b3, and b4 from Figure 3D. In fact, the favored fragmentation of the morpholino ring due to push-pull radical stabilization of the ions (Carey and Sundberg, 2008) yields mainly two major ions b4 and y7, where y7 contains the peptide sequence of the β subunit active-site. By electrostatic trapping and further MS3 fragmentation of the y7 ion, the LAFR sequence of the β5t active-site peptide was identified (Figure 3E). Taken together, this data set demonstrates that β5t is, indeed, a reactive proteasome subunit.
Figure 3. Active-Site Peptide Identification and Determination of Proteasome β Subunits by Affinity Purification, Tryptic Digest, and LC-MS Analysis.
(A) Reaction mechanism of biotin-epoxomicin 3 with the catalytically active N-terminal Thr residue of active proteasome β subunits. The morpholino ring formation results in a covalent and irreversible binding.
(B) Schematic representation of the biotin-epoxomicin modified, N-terminal active-site tryptic peptide of β5t. Amino acid residues are represented in a three-letter code.
(C) LC-MS elution profile of the six unique biotinylated tryptic peptides derived from the active sites. Notice that β5 and β5i active-site peptides are identical (see also Table S3).
(D) LC-MS2 determination of the β5t active-site fragmentation pattern. The parent ion [m/z (M+2H)2+ = 767.44] was fragmented. The b1, b2, b3, and b4 ions are signature ions of the biotin-epoxomicin N-terminal part. The abundant y7 ion containing the β5t active-site peptide sequence was selected for further (MS3) fragmentation (see E).
(E) LC-MS3 determination of the y7 ion (MH+ = 821.32) revealing the β5t active-site peptide amino acid sequence.
Competitive Activity-Based Profiling Reveals β5t Substrate Specificity
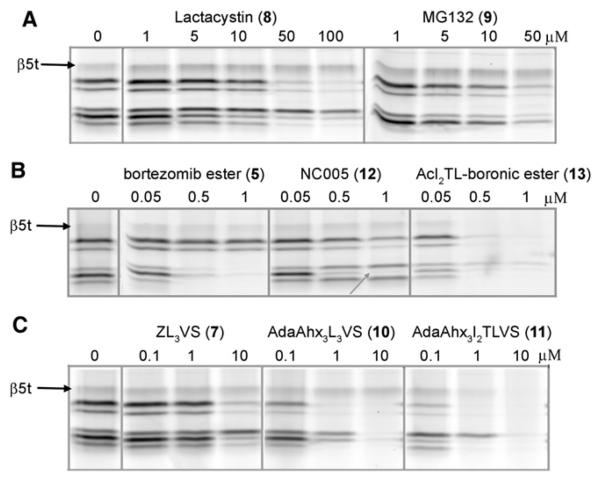
The finding that β5t reacts with epoxyketones 2, 3, and 4, but not with peptide vinyl sulphones 1 and MV151 gives a first indication of an altered substrate specificity compared to β5/β5i. With probe 4 in hand as readout, we set out to investigate the β5t substrate preference by competitive activity-based studies with established proteasome inhibitors of diverse chemical characteristics. Figure 4A shows the results of the most commonly used proteasome inhibitors lactacystin (Fenteany et al., 1995) and MG132 (Rock and Goldberg, 1999). Both require concentrations higher that 10 μM for broad-spectrum proteasome inhibition with marked affinity for β5 and β2 but do not inhibit the β5t activity. Figure 4B shows that the Bortezomib boronic ester 5 effectively blocks β1, β1i, β5, and β5i as previously described (Adams et al., 1998; Verdoes et al., 2007), while the subunit-specific inhibitor NC005 (Britton et al., 2009) selectively inhibits the β5/β5i subunits. Neither bortezomib nor NC005 appears to interact with the β5t. However, mixing of the potent boronic ester warhead with the AcI2TL peptide sequence inherent to epoxomicin as in compound 13 abolished the subunit preference of bortezomib and efficiently inhibited β5t. A similar effect is revealed in Figure 4C where the vinyl sulphone warhead (Bogyo et al., 1997) was equipped with the AdaAhx3 extended I2TL motif. Apparently, the presence of the hydrophilic threonine side chain at P2 in an inhibitor or ABP probe is favorable for affinity to the β5t subunit. From the results, some interesting trends pointing toward a substrate preference of β5t that is rather distinct to that of β5/β5i appear. Whereas β5t is sensitive toward the broad-spectrum proteasome epoxomicin 6, it is quite unreactive toward the β5/β5i-biased compounds. Bortezomib boronic ester 5 (which at the concentrations used disables β1/β1i/β5/β5i) and lactacystin 8 are unreactive toward β5t, as is the case with vinyl sulfone 7, peptide aldehyde 9, and epoxyke-tone 12.
Figure 4. Analysis of β5t Substrate Specificity in Juvenile Murine Thymus Lysates by Competitive Activity-Based Profiling with ABP 4.
(A) Lysates were exposed to increasing concentrations Lactacystin or MG132; residual β5t activity was stained with ABP 4 and visualized by in-gel fluorescence detection. The inhibitors are not reactive toward the β5t activity.
(B) Bortezomib efficiently inhibits β5, β5i, β1, and β1i activity but not β5t. NC005 specifically targets β5 (indicated by the gray arrow) but not β5t. The mixed inhibitor containing the boronic ester warhead equipped with the epoxomicin tail efficiently blocks β5t.
(C) In analogy to (B), installation of the epoxomicin IleIleThrLeu peptide targeting motif to the vinyl sulphone warhead affords a potent inhibitor of the β5t activity.
As a final research objective, we set out to establish whether β5t activity could be detected in fluorogenic substrate assays. In a first experiment, lysates of 3-week-old wild-type and β5t−/− murine thymi were preincubated with 1 μM 10, 6, 9, or the β5/β5i-specific inhibitor NC005 (12) prior to treatment with the reported β5-specific fluorogenic substrate, Ac-LLVY-amc (Kisselev and Goldberg, 2005). No differential signal could be detected in the wt versus the β5t−/− lysates (data not shown), indicating that this substrate does not react with β5t. We next synthesized two novel fluorogenic substrates, Ac-IITL-amc and Ac-SSTL-amc (Supplemental Experimental Procedures) that were expected to be cleaved by all catalytically active β subunits, including β5t. Preincubation of wild-type and β5t−/− lysates with 1 μM 10, 6, 9,or 12 followed by β5t read-out with Ac-IITL-amc or Ac-SSTL-amc again did not result in a discernible β5t-specific signal (data not shown). We next fractionated the lysates by 10%–40% sucrose gradient ultracentrifugation and used the enriched (thymo)proteasome fractions as determined by ABP profiling with 4 (Figure S2). The tryptic- and caspase-like activities were inhibited with 1 or 10 μM 6 or 7 and the β5t activity was assayed with Ac-IITL-amc or Ac-SSTL-amc (Figure S3). Again, no differences could be found between wild-type and β5t−/− (thymo)proteasomes.
In conclusion, we have demonstrated that β5t has an intrinsic reactivity resembling that of the other proteasome catalytic β5t subunits and have obtained evidence suggesting that this thymoproteasome-specific subunit plays an active role in positive T cell selection. Our discovery that β5t is an active species is based on activity-based probes, and we acknowledge the fact that the reactive groups differ in nature from the natural substrates (vinyl sulphone/epoxyketone as opposed to amide bonds). We note, however, that all known proteasome catalytic subunits are active toward these electrophilic traps and moreover that all nonactives are not. In fact, we argue that activity-based enzyme profiling is especially attractive for probing putative proteasome active sites, which are active only in the context of the 20S conglomerate as a whole and by activity-based profiling, can still be reported on individually. We indeed were unable to detect β5t activity by using fluorogenic substrates, due to the fact that the substrates we were able to obtain or design based on our data were either specific for β5/β5i (in which case no β5t activity is to be expected based on our inhibition studies) or are pan-reactive (in which case we could not discriminate between the different activities). More research is required to identify a fluorogenic substrate specific for β5t, and we argue this may be accomplished by screening (libraries of) designed inhibitors against thymoproteasomes in an activity-based profiling assay such as disclosed here. Altogether, our data, revealing that β5t is catalytically active toward inhibitors that contain a hydrophilic residue (Thr) in a hydrophobic stretch, point toward the involvement of β5t in the generation of a unique set of oligopeptides complexed to MHC I molecules for optimal positive T cell selection.
SIGNIFICANCE
We demonstrate for the first time to the best of our knowledge that thymoproteasome-specific β5t subunit is catalytically active. Interestingly, active β5t is also found in adult thymi but not in other murine organs. We provide the first insight into the nature of the substrate preference of β5t that is probably biased toward peptides with hydrophilic side chains. This body of evidence was made possible by the direct action of activity-based probes (ABP) with emphasis on the bifunctional ABPs that facilitate both readout and affinity purification. Moreover, the Bodipy-epoxomicin probe shows superior labeling and SDS-PAGE resolution for the active β subunits. The biotin-epoxomicin probe facilitated the LC-MS3 identification of the β5t active-site peptide that is the ultimate proof for the reactivity of this subunit. This work illustrates the versatility of fluores-cent activity-based probes and opens new avenues of detection as fluorescence microscopy or flow cytometry in living cells. Therefore, the probes presented here should be considered as a powerful addition to the toolbox for proteasome activity determination and quantification besides the classic fluorogenic substrate assays.
EXPERIMENTAL PROCEDURES
Animals and Tissues
Thymus and other organs were isolated from young (3 week old) or adult mice and kindly provided by Ine Tijdens, Chantal Pont, and Prof. Dr. Bob van de Water. Thymus from β5t knockout mice was kindly provided by Dr. Keiji Tanaka. Organ isolation was approved by the animal experimentation ethical committee of the Leiden University and Tokyo Metropolitan Institute of Medical Science.
Compounds
Design, synthesis, and mechanism of action of the activity-based probes 1–4, MV151, and the proteasome inhibitors 5–11, 13 is reviewed in Verdoes et al. (2009). NC005 is described in Britton et al. (2009). Lactacystin, MG132, and all other compounds of analytical grade were purchased from Sigma-Aldrich.
Activity-Based Profiling
Tissues were homogenized in 3 volumes of ice-cold lysis buffer (50 mM TrisHCl [pH 7.5], 250 mM sucrose, 5 mM MgCl2, 1 mM DTT, 2 mM ATP, 0.025% digitonin, 0.2% NP40; Kisselev and Goldberg, 2005) with a tissue homogenizer and further disrupted by 2 × 30 s sonication. Lysates were cleared by cold centrifugation at 13,000 × g, protein concentrations determined by Bradford assay and kept at −80°C until use. For comparative activity-based profiling, equal amounts of protein were incubated with ABPs for 1 hr at 37°C and resolved by 12.5% SDS-PAGE, and the wet gel slab was scanned on a Thyphoon scanner (GE Healthcare) with the TAMRA settings (λex = 530 nm, λem = 560 nm). Competitive activity-based profiling was done by first incubating thymus lysates with increasing concentrations of various proteasome inhibitors for 1 hr at 37°C, followed by 1 hr incubation with 0.5 μM ABP 4 for the in-gel detection of the residual proteasome activity. Images were acquired, processed, and quantified with Image Quant (GE Healthcare).
Affinity Purification
Approximately 1 or 2 mg of protein was incubated with 10 μM biotinylated ABPs 1, 2 or 3 for 1 hr at 37°C, denatured by boiling for 5 min with 1% SDS and precipitated with chloroform/ methanol (C/M, Wessel and Flügge, 1984). The protein pellet was rehydrated in 180 μl 8 M urea/100 mM NH4HCO3, reduced with 10 μl 90 mM DTT for 30 min at 37°C, alkylated with 15 μl 200 mM iodoacetamide at room temperature in the dark, cleared by centrifugation at 13,000 × g, and desalted by C/M. The pellet was dispersed in 25 μl PD buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl) with 2% SDS in a heated (37°C) sonic bath. Stepwise (3 × 25 μl, 4 × 100 μl, 1 × 500 μl) addition of PD buffer afforded a clear solution that was incubated with 50 μl MyOne T1 Streptavidin-grafted beads (Invitrogen) at room temperature with vigorous shaking for 2 hr. The beads were stringently washed with 2 × 300 μl PD buffer with 0.1% SDS, 2 × 300 μl PD buffer, 2 × 300 μl wash buffer I (4 M urea/50 mM NH4HCO3), 2 × 300 μl wash buffer II (50 mM Tris-HCl [pH 7.5], 10 mM NaCl), and 2 × 300 μl water. For in-gel analysis, two-thirds of the beads were eluted with 100 μl 1 × sample buffer containing 10 μM biotin by boiling for 5 min at 90°C and resolved by 12.5% SDS-PAGE. Proteins were visualized by fluorescence and silver stain, in-gel digested (Shevchenko et al., 2006), and desalted (Rappsilber et al., 2007). For on/bead digest, one-third of the beads were digested with 300 ng trypsin in 100 ml digest buffer (100 mM Tris-HCl [pH 7.8], 100 mM NaCl, 1 mM CaCl2, 2% ACN) overnight at 37°C. Peptides were collected and desalted on stage tips. The active-site peptides were eluted with 2 × 80 μl 10 μM biotin in 5% formic acid/25% ACN/70% H2O for 30 min at 37°C and desalted after ACN evaporation.
LC-MS Analysis
Trytic peptides were analyzed on a Surveyor nanoLC system (Thermo) hyphenated to a LTQ-Orbitrap mass spectrometer (Thermo). Gold and carbon coated emitters (OD/ID = 360/25 μm tip ID = 5 μm), trap column (OD/ID = 360/100 μm packed with 25 mm robust Poros10R2/ 15 mm BioSphere C18 5 μm 120 Å) and analytical columns (OD/ID = 360/75 μm packed with 20 cm BioSphere C18 5 μm 120 Å) were from Nanoseparations (Nieuwkoop, The Netherlands). The mobile phases (A: 0.1% FA/H2O, B: 0.1% FA/ACN) were made with ULC/MS grade solvents (Biosolve). The emitter tip was coupled end-to-end with the analytical column via a 15 mm long TFE Teflon tubing sleeve (OD/ID 0.3 × 1.58 mm, Supelco, USA) and installed in a stainless steel holder mounted in a nano-source base (Upchurch scientific, Idex, USA).
General mass spectrometric conditions were as follows: an electrospray voltage of 1.8 kV was applied to the emitter, no sheath and auxiliary gas flow, ion transfer tube temperature 150°C, capillary voltage 41 V, tube lens voltage 150 V. Internal mass calibration was performed with air-borne protonated polydimethylcyclosiloxane (m/z = 445.12002) and the plasticizer protonated dioctyl phthalate ions (m/z = 391.28429) as lock mass (Olsen et al., 2005).
For shotgun proteomics analysis, 10 μl of the samples was pressure loaded on the trap column with a 10 μl/min flow for 5 min followed by peptide separation with a gradient of 35 min 5%–30% B, 15 min 30%–60% B, 5 min A at a flow of 300 μl/min split to 250 nl/min by the LTQ divert valve. For each data-dependent cycle, one full MS scan (300–2000 m/z) acquired at high mass resolution (60,000 at 400 m/z, AGC target 1 × 106, maximum injection time 1000 ms) in the Orbitrap was followed by three MS/MS fragmentations in the LTQ linear ion trap (AGC target 5 × 103, maximum injection time 120 ms) from the three most abundant ions (Baek et al., 2008). MS2 settings were as follows: collision gas pressure 1.3 mT, normalized collision energy 35%, ion selection threshold of 500 counts, activation q = 0.25 and activation time of 30 ms. Fragmented precursor ions that were measured twice within 10 s were dynamically excluded for 60 s and ions with z < 2 or unassigned were not analyzed.
A parent ion list of the m/z ratios of the active-site peptides was compiled and used for LC-MS3 analysis in a data-dependent protocol. The parent ion was electrostatically isolated in the ion trap of the LTQ and fragmented by MS2, and the most intense peak was isolated and further fragmented in MS3 to reveal the amino acid sequence of the active-site peptide. Data from MS2 and MS3 were validated manually.
Supplementary Material
ACKNOWLEDGMENTS
Financial support from the Netherlands Organisation for Scientific Research (NWO, to H.S.O.), the Netherlands Genomics Initiative (NGI, to B.I.F., M.V. and H.S.O.) and the National Cancer Institute (NCI grant 5R01CA124634 to AFK) is gratefully acknowledged. Chi-Lin (Ethan) Kuo, Eric van Gilzel, Ine Tijdens, Chantal Pont, and Bob van de Water are acknowledged for the technical assistance, and Gerrit Lodder for the scientific discussion.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Material includes Supplemental Experimental Procedures, three figures, and three tables and can be found with this article online at doi:10.1016/j.chembiol.2010.05.027.
REFERENCES
- Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton M, Lucas MM, Downey SL, Screen M, Pletnev AA, Verdoes M, Tokhunts R, Amir O, Goddard A, Pelphrey P, et al. Selective inhibitor of proteasome's caspase-like sites sensitize cells to the specific inhibitor of the chymotrypsin-like sites. Chem. Biol. 2009;16:1278–1289. doi: 10.1016/j.chembiol.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey FA, Sundberg RJ. In Advanced Organic Chemistry Part A: Structure and Mechanism. Springer Science; Charlottesville, VA: 2008. Radical intermediates; pp. 311–318. [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr., Bottomly K. Signals and signs for lymphocyte responses. Cell. 1994;76:275–285. doi: 10.1016/0092-8674(94)90335-2. [DOI] [PubMed] [Google Scholar]
- Kessler BM, Tortorella D, Altun M, Kisselev AF, Fiebiger E, Hekking BG, Ploegh HL, Overkleeft HS. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic beta-subunits. Chem. Biol. 2001;8:913–929. doi: 10.1016/s1074-5521(01)00069-2. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- Murata S, Takahama Y, Tanaka K. Thymoproteasome: probable rolein generating positively selecting peptides. Curr. Opin. Immunol. 2008;20:192–196. doi: 10.1016/j.coi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Nitta T, Murata S, Ueno T, Tanaka K, Takahama Y. Thymic microenvironments for T-cell repertoire formation. Adv. Immunol. 2008;99:59–94. doi: 10.1016/S0065-2776(08)00603-2. [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC classI-presented peptides. Annu. Rev. Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Takahama Y, Tanaka K, Murata S. Modest cortex and promiscuous medulla for thymic repertoire formation. Trends Immunol. 2008;29:251–255. doi: 10.1016/j.it.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, Van der Linden WA, Van den Nieuwendijk A, Hofmann T, Berkers CR, van Leeuwen FWB, et al. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem. Biol. 2006;13:1217–1226. doi: 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, Hillaert U, Willems LI, van der Linden WA, Sae-Heng M, Filippov DV, Kisseley AF, van der Marel GA, Overkleeft HS. Azido-BODIPY acid reveals quantitative StaudingerBertozzi ligation in two-step activity-based proteasome profiling. ChemBioChem. 2008;9:1735–1738. doi: 10.1002/cbic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, van der Linden WA, Renou D, van den Nieuwendijk A, van der Marel GA, Overkleeft HS. Mixing of peptides and electrophilic traps gives rise to potent, broad-spectrum proteasome inhibitors. Org. Biomol. Chem. 2007;5:1416–1426. doi: 10.1039/b702268a. [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, van der Marel GA, Overkleeft HS. Chemical tools to study the proteasome. Eur. J. Org. Chem. 2009:3301–3313. [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.