Abstract
IL-6 gene expression is controlled by a promoter region containing multiple regulatory elements such as NF-κB, NF-IL6, CRE, GRE, and TRE. In this study, we demonstrated that TRE, found within the IL-6 promoter, is embedded in a functional antioxidant response element (ARE) matching an entire ARE consensus sequence. Further, point mutations of the ARE consensus sequence in the IL-6 promoter construct selectively eliminate ARE but not TRE activity. Nrf2 is a redox-sensitive transcription factor which provides cytoprotection against electrophilic and oxidative stress and is the most potent activator of ARE-dependent transcription. Using Nrf2 knock-out mice we demonstrate that Nrf2 is a potent activator of IL-6 gene transcription in vivo. Moreover, we show evidence that Nrf2 is the transcription factor that activates IL6 expression in a cholestatic hepatitis mouse model. Our findings suggest a possible role of IL-6 in oxidative stress defense and also give indication about an important function for Nrf2 in the regulation of hematopoietic and inflammatory processes.
Keywords: Antioxidant, Gene Expression, Interleukin, Liver Injury, Oxidative Stress, Transcription Promoter, Antioxidant Response Element, Nrf2, Nuclear Factor Erythroid 2-related Factor 2
Introduction
Because of its diverse biological function and simultaneous description in different studies, IL-6 was initially assigned several names. It was identified as a T-cell-derived B-cell differentiation factor, as it induced activated B-cells into antibody-producing cells: interferon-β2 (26 kDa protein), a hybridoma/plasmacytoma growth factor and a hepatocyte-stimulating factor. The name IL-6 was proposed when the cDNA nucleotide sequences for these proteins had been determined, and the molecules were found to be identical (1). In addition, IL-6 plays a key role in inflammation, being the main inducer of fibrinogen, serum amyloid A protein, the acute phase response and is one of the most important mediators of fever. In muscle and fatty tissues, IL-6 stimulates energy mobilization.
The IL-6 promoter is rapidly activated by cytokines, including IL-1 and TNF-α, as well as by phorbol esters and cyclic AMP. The promoter-region of the IL-6 gene contains multiple regulatory elements such as nuclear factor-κB (NF-κB), nuclear factor-IL6 (NF-IL6) (also referred to as C/EBPβ), cAMP response element (CRE), TPA (12-O-tetradecanoylphorbol-13-acetate) responsive element (TRE; also referred to as the AP-1 binding site), and the glucocorticoid response element (GRE) (2).
Structurally related to TRE is the antioxidant responsive element (ARE,2 also referred to as the electrophile responsive element (EpRE)) (3, 4). Some TREs are found to be embedded within an ARE, such as in the promoter region of human NAD(P)H:quinine oxidoreductase-1 (NQO1), rat and mouse glutathione S-transferase (GST) Ya subunit, and rat GST-P (5). ARE is commonly found in the promoter region of genes encoding phase II detoxification as well as antioxidant enzymes such as NQO1, thioredoxin, thioredoxin reductase, glutathione peroxidase, and hemeoxygenase-1 (6). Analyses of ARE-nuclear protein complexes have identified numerous nuclear transcription factors including c-Jun, Jun-B, Jun-D, c-Fos, Fra1, Nrf1, Nrf2, Nrf3, c-Maf, MafG, MafK, Bach1, Bach2, the Ah (aromatic hydrocarbon) and estrogen receptor. Nrf2 is described as being the most potent inducer of ARE-mediated expression among these transcription factors. Nrf2 associates with small Maf (MafG or MafK) or Jun (c-Jun, Jun-B, and Jun-D) proteins to up-regulate ARE-mediated expression and coordinate the induction of detoxifying enzymes in response to antioxidants and xenobiotics (7–9). Numerous studies support this hypothesis and show that Nrf2 protects various cell types and organs from oxidative stress (10, 11).
Oxidative stress is one main feature in nonalcoholic steatohepatitis and its mouse model DDC feeding (12, 13). Recently, Plum et al. (14) showed a protective role of the IL-6/gp-130 pathway in this cholestatic hepatitis model.
In this study we show that Nrf2 binds to ARE within the promoter region of the IL-6 gene thus highly activating IL-6 transcription. The IL-6 gene appears to be an Nrf2-target. IL-6 expression is induced by Nrf2 stimuli as well as oxidative or electrophilic stress. The results are discussed according to the diverse physiological and pathological functions of IL-6 and Nrf2.
EXPERIMENTAL PROCEDURES
Animals
Nrf2-Knock-out mice were produced by specifically deleting the Nrf2 gene segment (15). WT control mice were littermates of the Nrf2-KO mice. All mice used in this study were 6–8-week-old and maintained in our animal facilities under specific, pathogen-free conditions. The mice were given oral l-sulforaphane ((R)-1-isothiocyanato-4-(methylsulfinyl) butane, 4-methylsulfinylbutyl isothiocyanate, Sigma-Aldrich) at 50 mg/kg body weight/day dissolved in PBS or PBS alone as described in Ref. 16.
For DDC experiments, 6–8-week-old male mice were treated with standard chow containing 0.1% DDC and fed normal drinking water for up to 12 weeks. Liver samples were shock-frozen for homogenization.
Plasmid Construction and Site-directed Mutagenesis
The plasmid pIL6-luc651 containing 651-bp found directly upstream of the transcriptional start site in the human IL-6 gene promoter was fused to a luciferase gene (17). The ARE consensus sequence (positions −289 to −276) of the pIL6-luc651 (5′-GTGACTCAGCA-3′) was altered to pIL6-luc651ΔARE (5′-GTGACTCAAGA-3′) by site-directed mutagenesis using QuickChange II Kit from Stratagene. This mutation was previously shown to inhibit Nrf2 binding (6). All mutant clones were verified by DNA sequencing. The Nrf2 expression plasmid pcDNA-Nrf2 is described in Ref. 18.
Cell Culture and Cotransfection of Reporter and Expression Plasmids
Human hepatoblastoma (Hep-G2) cells were grown in monolayer cultures in Eagle's minimum essential medium (DMEM) plus fetal bovine serum to a final concentration of 10% (all purchased from LGC Promochem, Wesel, Germany).
pIL6-luc651 or pIL6-luc651ΔARE and pRL-TK (4:1 ratio) were mixed with different concentrations of an empty expression plasmid pcDNA3.1 vector (Invitrogen) alone or together with pcDNA-Nrf2 and cotransfected in Hep-G2 cells using Lipofectamine2000 from Invitrogen using procedures supplied by the manufacturer. All experiments were conducted using 96-well plates containing 100 μl Medium and ∼3 × 104 cells per well.
Dual Luciferase Assay
The Dual Luciferase assay was performed as described in Ref. 18. pIL6-luc651 or pIL6-luc651ΔARE reporter plasmids and pRL-TK plasmids (Promega) were cotransfected into HepG2-cells. 24 h after transfection, the cells were seeded to a 96-well plate (3 × 104 cells per well). The activity of both firefly and Renilla luciferases was determined 48 h after transfection with the dual luciferase reporter assay system (Promega). Luciferase activity was measured over 10 s in a luminometer (Promega). The luciferase activities were normalized to the Renilla luciferase activity of the internal control.
IL-6 Quantification
HepG2 cells were grown in the 1.5 ml medium described above using 6-well plates with ∼7 × 107 cells per well. For IL-6 detection within the cell medium (20 μl) we utilized a human IL-6 ELISA from Invitrogen executed in duplicates according to the supplier's recommendation.
Mice were treated as described above. Measurements of liver IL-6 expression were performed in duplicates using microsphere-based Luminex xMAP technology with antibodies directed against mice IL-6 protein (Invitrogen). 30 μg of liver homogenates were analyzed in duplicates using procedures recommended by the manufacturer.
Quantitation of mRNA Concentration
Mice were treated as described above. mRNA concentrations in liver lysates were measured using the QuantiGene multiplex assay (Panomics). 30 μg of liver homogenates were analyzed in duplicates using procedures recommended by the manufacturer. The expression of target-specific RNA molecules was calculated as the mean value from triplicates and corrected to hexose-6-phosphate dehydrogenase.
Histology
Liver samples were fixed in 3.5% formaldehyde, embedded in paraffin, cut, and stained with H&E. Images were captured using light microscopy (Nikon Eclipse 55i) and evaluated using the ImageJ software (11).
Aminotransferase Determination
ALT activities in plasma were determined by an automated enzyme assay (Roche, Mannheim, Germany) as described in Ref. 19.
Statistical Analysis and Bioinformatics
Results were compared using a two-tailed paired Student's t test. Differences were considered significant at p < 0.05. ClustalW was used for multiple sequence alignments.
RESULTS
IL-6 Promoter Alignment
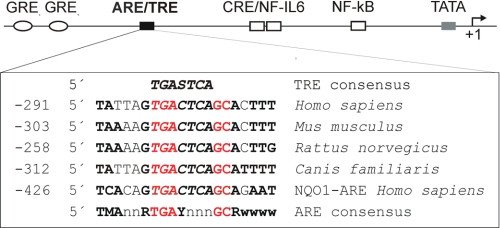
Cross-species conservation of promoter elements is an important functionality indicator. We therefore compared TRE-containing IL-6 promoter regions in humans, mice, rats, and dogs. TRE was embedded in an entire ARE consensus sequence, and this finding was conserved in all selected species (Fig. 1).
FIGURE 1.
Alignment of ARE sequences within the IL-6 promoter. Delineated are the cis-regulatory elements of the IL-6 promoter with the approximate location relative to the transcription start (+1). Below, ARE/TRE sequence of various species and from the human NQO1 promoter are aligned and compared with ARE and TRE consensus sequences. Nucleotides at essential positions for ARE are in red; nucleotides identical to the ARE consensus recognition sequence are marked in bold. The essential TRE sequences are marked in italic. The abbreviations follow standard IUPAC nomenclature (M = A or C, r = A or G, Y = C or T, W = A or T, S = G or C, n = A, T, C or G). All IL-6 promoter sequences are shown as reverse complements.
Effect of Nrf2 on IL-6 Promoter Activity
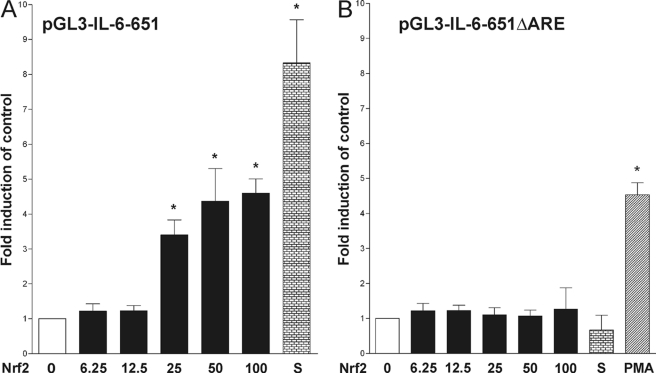
To investigate the functionality of the ARE consensus sequence within the IL-6 promoter dual luciferase reporter gene assays were performed. In the human IL-6 promoter construct, pIL6-luc651 (17), point mutations within the ARE consensus sequence were produced to form an ARE-deficient promoter (pIL6-luc651ΔARE). Previous studies have shown that an over-expression of Nrf2 leads to ARE activation (20). Therefore, we co-transfected HepG2 cells with various amounts of an Nrf2-expression vector along with pIL6-luc651 and pRL-TK. A dose-dependent activity increase was seen in firefly luciferase but not in Renilla luciferase (internal control). The ratio of firefly and Renilla luciferase activity reached a height by a 4-fold induction compared with control (Fig. 2A). Sulforaphane is known for its Nrf2 activation properties (21) and was therefore used as positive control (Fig. 2A). In contrast, Nrf2-expression vector co-transfection with pIL6-luc651ΔARE insufficiently enhanced luciferase activity (Fig. 2B). Stimulation with phorbol 12-myristate 13-acetate (PMA) demonstrated the intact functionality of TRE within the pIL6-luc651ΔARE vector (Fig. 2B). All experiments were conducted on 96-well plates with n = 8.
FIGURE 2.
Effect of Nrf2 on IL-6 promoter activity. Dual-luciferase assay to analyze the effect of Nrf2 expression on pIL6-luc651 (A) and pIL6-luc651ΔARE (B) promoter activity. Nrf2 expression vector pcDNA3.1-Nrf2 (denoted in ng of DNA per well of a 96 well plate) was transiently co-transfected into HepG2 cells along with the pIL6-luc651 and pIL6-luc651ΔARE, respectively, and pRL-TK as described above. Induction of promoter activity by Nrf2 expression, 2 μm sulforaphane (S) or 10 ng/ml PMA was assessed for the respective constructs, corrected for transfection efficiency, and calculated as percentage of induction compared with unstimulated base-line activity. Data represented were obtained from eight independent experiments. Bars represent means and ± S.E. *, significant difference versus control (p < 0.05).
Nrf2-dependent IL-6 mRNA and Protein Synthesis
In vitro and in vivo studies were performed to investigate the effect of common Nrf2 activators on the hepatic expression of IL-6.
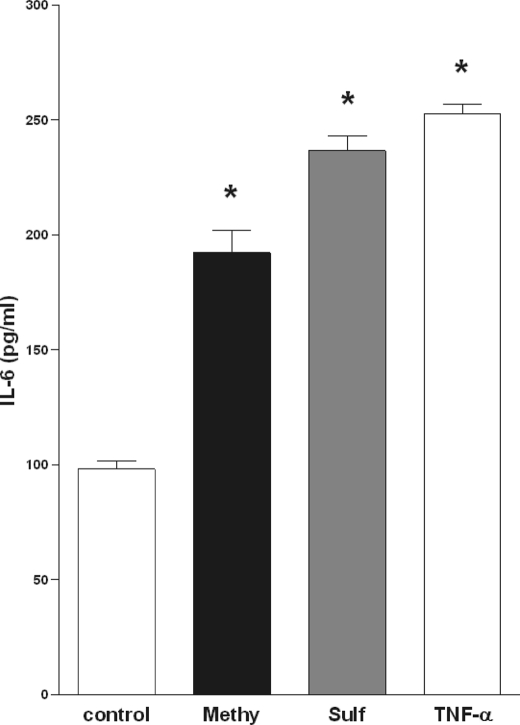
Fig. 3 demonstrates a characteristic IL-6 ELISA from human HepG2 cells treated with 10 μm methysticin or 2 μm sulforaphane for 16 h. Both Nrf2 activators significantly up-regulated IL-6 secretion to a dimension comparable to that seen after TNF-α treatment (Fig. 3). Subsequent in vivo studies utilizing Nrf2 knock-out and wild-type mice further support the in vitro findings and additionally identify Nrf2 as the transcription factor involved.
FIGURE 3.
Stimulation with Nrf2 activators promotes IL-6 production. Production of IL-6 by HepG2 cells treated with vehicle (control), 5 μm Methysticin (Methy.), 2 μm Sulforaphane (Sulf.), or 10 ng/ml TNF-α analyzed by IL-6 ELISA assay. Data represented were obtained from six independent experiments. Bars represent means and ± S.E. *, significant difference versus control (p < 0.05).
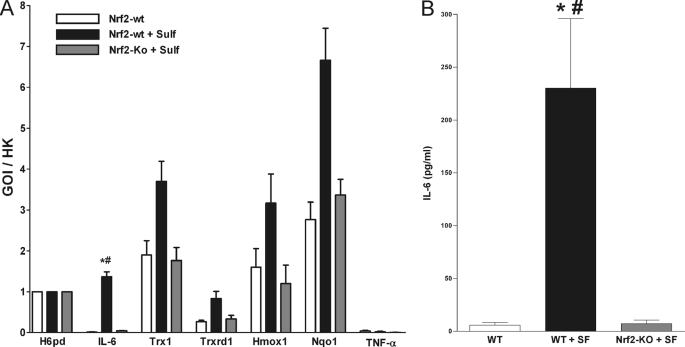
We performed a Quantigene multiplex assay to detect mRNA coding for IL-6, thioredoxin-1 (Trx1), thioredoxin reductase-1 (Trxrd1), hemoxigenase-1 (Hmox1), NAD(P)H:quinine oxidoreductase-1 (Nqo1), TNF-α, and hexose-6-phosphate dehydrogenase (H6pd, used as a housekeeping gene). Serving as an internal control, the measured fluorescence values of the genes of interests (GOI) were divided by the corresponding values of the housekeeping gene (HK). As shown in Fig. 4A, IL-6 mRNA was clearly up-regulated in liver homogenates of wild-type mice treated with sulforaphane for 24 h. In contrast, liver homogenates from sulforaphane treated Nrf2 knock-out mice and untreated control IL-6 mRNA were not detectable (Fig. 4A). As expected, the mRNA of the Nrf2 target genes Trx1, Trxrd1, Hmox1, and Nqo1 were up-regulated in sulforaphane-treated wild-type but not in Nrf2 knock-out mice liver. TNF-α mRNA was not detectable in all three groups (Fig. 4A).
FIGURE 4.
Nrf2-dependent IL-6 mRNA and protein synthesis. To measure the in vivo effect of sulforaphane on both IL-6 mRNA and protein expression fresh homogenized mice liver (30 μg) from untreated WT, sulforaphane-treated WT, and sulforaphane treaded Nrf2-knock-out mice was used. A, to quantify the synthesis of mRNA from IL-6, thioredoxin-1 (Trx1), thioredoxin reductase-1 (Trxrd1), hemoxigenase-1 (Hmox1), NAD(P)H:quinine oxidoreductase-1 (Nqo1), TNF-α, and hexose-6-phosphate dehydrogenase (H6pd, used as a housekeeping gene) a QuantiGene multiplex assay was used. B, to quantify the synthesis of IL-6 protein concentration a microsphere-based Luminex xMAP technology with antibodies raised against the mice IL-6 was used. Bars represent means and ± S.E., n = 3. *, significant difference versus control; #, significant difference versus sulforaphane treated Nrf2-knock-out.
IL-6 mRNA levels of the mice cohorts were then analyzed for their association with IL-6 secretion. We conducted an IL-6 protein Luminex assay using the same liver homogenates used for the Quantigene multiplex assay. IL-6 protein in liver homogenates from WT control was below the detection limit. As expected, we measured a significant increase of IL-6 protein in the homogenates from sulforaphane-treated mice compared with untreated mice. Nrf2 knock-out mice showed no IL-6 up-regulation in response to sulforaphane treatment (Fig. 4B).
DDC Induced in Vivo IL6 Expression
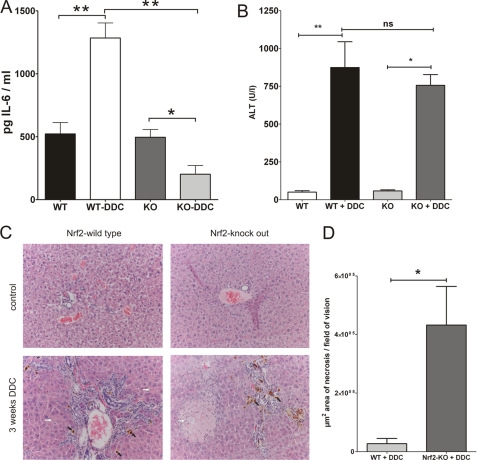
It is known that the IL-6 production is elevated in the liver of DDC-treated mice (14). To explore if Nrf2 is the transcription factor inducing the IL-6 up-regulation under these conditions we treated Nrf2-knock-out and wild type mice with DDC for 3 weeks. We then determined hepatic expression levels of IL-6 by Luminex technique. IL-6 protein expression was significantly induced about 2.5-fold in DDC-treated wild-type mice (1284 pg of IL-6/ml liver homogenate) compared with untreated wild-type mice (522 pg IL-6/ml liver homogenate). However, DDC treated Nrf2-knock-out mice (264 pg of IL-6/ml liver homogenate) failed to up-regulate IL-6 expression compared with untreated Nrf2-knock-out mice (456 pg of IL-6/ml liver homogenate). There were no significant differences between untreated wild type, and Nrf2-knock-out mice (Fig. 5A).
FIGURE 5.
A, Nrf2-knock-out mice fail to up-regulate IL-6 expression in response to DDC-treatment. Hepatic IL-6 protein expression level was determined by luminex technique. Liver homogenate was produced from mice treated with 0.1% DDC for 3 weeks. The pg IL-6 per ml liver homogenate is displayed. B, ALT serum levels were determined 3 weeks after DDC treatment was started. Transaminases increased rapidly in both treatment groups. C, hematoxylin and eosin (H&E) staining was performed on liver sections derived from Nrf2 knock-out and control mice with or without 3 weeks after DDC feeding. Mice develop chronic cholestatic liver injury after DDC feeding (white arrows, necrosis area; black arrow, cholestasis; magnification ×200). D, significantly more liver necrosis was detected in livers of Nrf2 knock-out mice. n = 6; *, p < 0.01; **, p < 0.005.
DDC-caused Liver Injury
ALT level increased after 3 weeks of DDC feeding in both groups (Fig. 5B) with no significant differences between Nrf2-knock-out and wild-type mice.
To better characterize the impact of Nrf2 deletion on liver injury we next performed H&E stainings of liver sections. In Nrf2-knock-out mice significant more necrosis (higher numbers and larger areas) was found 3 weeks after DDC feeding compared with wild-type mice (Fig. 5C). A quantification of necrotic areas showed significant more (and larger) necrotic areas in DDC treated Nrf2-knock-out livers.
DISCUSSION
AREs containing TRE elements mediate high basal transcription of the human NQO1, rat and mouse GST Ya subunit, and rat GST-P and its induction in response to xenobiotics and oxidative stress (5). We found this special promoter element construction in all examined IL-6 promoters (human, mouse, rat, and dog; Fig. 1), thus supporting our hypothesis regarding sequence function.
Utilizing promoter studies in a human hepatic cell line with a cloned IL-6 promoter containing either wild-type or mutated ARE sequences, we experimentally confirmed the ARE consensus sequence function within in the IL-6 promoter. The conducted luciferase assays demonstrated that Nrf2 over-expression induced luciferase activity in wild-type but not in ARE-deficient IL-6 promoter vectors (Fig. 2, A and B). These results demonstrate both the functionality of the ARE sequence as well as the role of Nrf2 as a transcription factor involved.
Further in vitro studies showed that not only an artificial IL-6 promoter construct but also natural IL-6 promoter react to Nrf2 activation. Stimulation of a human hepatic cell line with the well estabished Nrf2 activators methysticin (18) and sulforaphane (21) leads to a significant enhanced IL-6 secretion (Fig. 3).
Subsequently, we brought the study forward into an in vivo mouse model. We demonstrated that sulforaphane administration enhanced IL-6 mRNA expression in wild-type but not in Nrf2-knock-out mice liver (Fig. 4A). We also measured mRNAs of known Nrf2 target genes. As suspected, these genes where up-regulated in wild-type mice but not in Nrf2-knock-out mice and verify the assay integrity (Fig. 4A). We then tested whether an increased IL-6 mRNA production results in an up-regulation of IL-6 protein expression. Our experiment demonstrated an increased IL-6 production in WT but not in Nrf2-knock-out mice treated with sulforaphane (Fig. 4B). We next tested IL-6 expression in a mouse model for steatohepatitis where IL-6 is up-regulated and has a well known protective function (14). As expected, the expression of IL-6 was up-regulated in DDC-treated wild-type mice. In contrast, Nrf2-knock-out mice are not able to up-regulate IL-6 expression in response to DDC treatment (Fig. 5A). These findings further documented an Nrf2, and therefore ARE, driven IL-6 production.
DDC feeding causes cholestatic liver damage and resembles many aspects of certain human chronic liver diseases like necrosis of hepatocytes and elevated blood ALT levels. As expected, DDC treated Nrf2 knock-out mice showed significantly more and larger necrotic areas as DDC treated wild-type mice (Fig. 5, C and D). Further experiments has to be conducted to clarify to which extent the absence of IL-6 expression leads to this enhanced susceptibility to DDC treatment of the Nrf2-knock-out mice. Albeit the levels of ALT increases in both groups to the same extend (Fig. 5B). We speculated that the Nrf2 knock-out and wild-type mice might have comparable initial liver injury and therefore equivalent ALT levels, but the Nrf2 knock-out mice have insufficient mechanisms for liver regeneration leading to extensive necrotic damage (22).
Because IL-6 is neither an antioxidant nor detoxification enzyme, it is surprising to find an entire ARE-consensus sequence contained in the IL-6 gene. However, several studies have shown that IL-6 is redox-regulated and can maintain a protective function in organs subjected to oxidative stress, yet little is known about the molecular mechanisms involved.
Studies regarding possible interactions between IL-6 and Nrf2 are numerous and this research describes one possible relation between Nrf2 and its target gene IL-6.
Hepatic IL-6 expression is enhanced in response to oxidative stress, protects the liver against oxidative injury and reverses hepatic steatosis and alcohol-induced liver injury (23–26). As IL-6-deficient mice, Nrf2-knock-out mice are prone to ethanol-induced hepatic apoptosis and steatosis and displayed a dramatically increased mortality associated with liver failure when fed with ethanol (27).
This similarity in IL-6 and Nrf2 function is also seen in models of hyperoxia-induced lung injury. Both IL-6 and Nrf2 protect pulmonary epithelial cells against oxidative stress (28–30). Several studies have demonstrated a redox-dependent up-regulated IL-6 expression in lung epithelial cells allowing the assumption that Nrf2 may be involved in this process (31, 32).
IL-6 is also found in two groups of cytokines: the myokines and adipocytokines, produced in muscle and visceral fat, respectively. Although the molecular mechanisms underlying IL-6 up-regulation in muscle and fat remain limited, it is generally accepted that oxidative stress causes IL-6 dysregulation. The IL-6 elevation in response to muscle contraction is inhibited by supplementation of the potent antioxidants Vitamin C and E (33, 34). In obesity, oxidative stress in adipose tissue causes an up-regulation of IL-6 and may contribute to obesity-associated metabolic syndrome (35, 36).
Sano and co-workers (37, 38) described a reactive oxygen species-mediated induction of IL-6 in cardiac fibroblasts. Here, IL-6 expression was activated by angiotensin II and inactivated by antioxidants and the ERK1/2 inhibitor PD98059, which are both known to block Nrf2-mediated gene expression. Lee et al. (39) found an oxidative stress mediated, IL-6 induction in vascular smooth muscle cells, which was abolished by modulators of oxidative stress. Both publications demonstrate oxidative stress induced IL-6 expression in the cardiovascular system and imply the participation of Nrf2.
A variety of in vitro and in vivo studies have demonstrated the neuroprotective properties of IL-6 as well as Nrf2 against oxidative damage. As in Nrf2-knock-out mice, IL-6-deficient mice suffer from increased oxidative stress and neurodegeneration after brain injury (40–42). In vitro studies showed that IL-6 protects cultured neuronal cells from apoptosis due to oxidative stress (43, 44). Maeda et al. (45) described a neuroprotective effect of IL6 up-regulation in response to reactive oxygen intermediates. These findings may link neuronal IL-6 up-regulation to Nrf2 activation.
Oxidative stress and a generalized inflammatory state are the main features of preeclampsia. Recently, we described a direct relationship in the etiology of preeclampsia between an Nrf2-mediated fetal defense against oxidative stress and consequent placental toxicity (11). Because IL-6 expression is also elevated in preeclamptic patients (46), one can assume that excessive Nrf2 activity results in a critically enhanced IL-6 expression.
What can be hypothesized about the function of Nrf2 up-regulated IL-6 expression? It is well known that Nrf2 orchestrates the transcription of a battery of antioxidant and detoxifying genes in a variety of cells and organs (10). However, IL-6 activates the transcription of another category of protective genes such as Bcl-2, Bcl-xL, Mcl-1, FLIP, Ref-1, cyclin D1, and c-Myc, via STAT3 activation (47). It may therefore be speculated that Nrf2 broadens its protection by interacting with the IL-6/STAT3 pathway.
An Nrf2-dependent IL-6 up-regulation is described in the study at hand. In addition to Nrf2, ARE can be bound by various other factors, functioning as both an activator and inhibitor of transcription. Yang et al. (48) reported that only IL-6, rapidly up-regulates c-Maf transcription. Considering that c-Maf homodimers are negative regulators of ARE-driven transcription (49), this may be indicative of a negative feedback loop.
However, our present findings are limited to hepatic Nrf2 driven IL-6 expression. They provide a first step in understanding the contribution of the ARE-system in IL-6 expression. Nrf2 may have an important function in the regulation of inflammatory processes and may indicate the possible role of IL-6 in oxidative stress defense.
Acknowledgments
We thank Christiane Jaeschke, Michaela Nicolau, Lian Chen, Sonja Seiter, Inka Geuring, and Angela Rüben for the excellent technical assistance.
We acknowledge funding from the Deutsche Forschungsgemeinschaft Pu 214/3-2, Pu 214/4-2, and Pu 214/5-2, and SFB617. This research project was also supported by the START-Program of the Faculty of Medicine, RWTH Aachen.
- ARE
- antioxidant responsive element
- l-sulforaphane
- (R)-1-isothiocyanato-4-(methylsulfinyl)butane, 4-methylsulfinylbutyl isothiocyanate
- DDC
- 1,4-dihydro-3,5-dicarbethxycollidine
- PMA
- phorbol 12-myristate 13-acetate
- GOI
- gene of interest
- HK
- housekeeping
- ALT
- alanine aminotransferase.
REFERENCES
- 1. Nishimoto N., Kishimoto T. (2006) Nat. Clin. Practice 2, 619–626 [DOI] [PubMed] [Google Scholar]
- 2. Dendorfer U., Oettgen P., Libermann T. A. (1994) Mol. Cell Biol. 14, 4443–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rushmore T. H., Morton M. R., Pickett C. B. (1991) J. Biol. Chem. 266, 11632–11639 [PubMed] [Google Scholar]
- 4. Jaiswal A. K. (1994) Biochem. Pharmacol 48, 439–444 [DOI] [PubMed] [Google Scholar]
- 5. Xie T., Belinsky M., Xu Y., Jaiswal A. K. (1995) J. Biol. Chem. 270, 6894–6900 [DOI] [PubMed] [Google Scholar]
- 6. Wasserman W. W., Fahl W. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhakshinamoorthy S., Long D. J., 2nd, Jaiswal A. K. (2000) Curr. Top. Cell Regul. 36, 201–216 [DOI] [PubMed] [Google Scholar]
- 8. Jaiswal A. K. (2004) Free Radic Biol. Med. 36, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 9. Venugopal R., Jaiswal A. K. (1998) Oncogene 17, 3145–3156 [DOI] [PubMed] [Google Scholar]
- 10. Lee J. M., Li J., Johnson D. A., Stein T. D., Kraft A. D., Calkins M. J., Jakel R. J., Johnson J. A. (2005) Faseb J. 19, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 11. Wruck C. J., Huppertz B., Bose P., Brandenburg L. O., Pufe T., Kadyrov M. (2009) Histopathology 55, 102–106 [DOI] [PubMed] [Google Scholar]
- 12. Farrell G. C., Larter C. Z. (2006) Hepatology 43, S99-S112 [DOI] [PubMed] [Google Scholar]
- 13. Stumptner C., Fuchsbichler A., Heid H., Zatloukal K., Denk H. (2002) Hepatology 35, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 14. Plum W., Tschaharganeh D. F., Kroy D. C., Corsten E., Erschfeld S., Dierssen U., Wasmuth H., Trautwein C., Streetz K. L. (2010) Am. J. Pathol. 176, 2236–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan K., Lu R., Chang J. C., Kan Y. W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13943–13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kong L., Tanito M., Huang Z., Li F., Zhou X., Zaharia A., Yodoi J., McGinnis J. F., Cao W. (2007) J. Neurochem. 101, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 17. Eickelberg O., Pansky A., Mussmann R., Bihl M., Tamm M., Hildebrand P., Perruchoud A. P., Roth M. (1999) J. Biol. Chem. 274, 12933–12938 [DOI] [PubMed] [Google Scholar]
- 18. Wruck C. J., Götz M. E., Herdegen T., Varoga D., Brandenburg L. O., Pufe T. (2008) Mol. Pharmacol. 73, 1785–1795 [DOI] [PubMed] [Google Scholar]
- 19. Streetz K. L., Wustefeld T., Klein C., Kallen K. J., Tronche F., Betz U. A., Schütz G., Manns M. P., Muller W., Trautwein C. (2003) Gastroenterology 125, 532–543 [DOI] [PubMed] [Google Scholar]
- 20. Venugopal R., Jaiswal A. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMahon M., Itoh K., Yamamoto M., Chanas S. A., Henderson C. J., McLellan L. I., Wolf C. R., Cavin C., Hayes J. D. (2001) Cancer Res. 61, 3299–3307 [PubMed] [Google Scholar]
- 22. Beyer T. A., Xu W., Teupser D., auf dem Keller U., Bugnon P., Hildt E., Thiery J., Kan Y. W., Werner S. (2008) EMBO J. 27, 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cressman D. E., Greenbaum L. E., DeAngelis R. A., Ciliberto G., Furth E. E., Poli V., Taub R. (1996) Science 274, 1379–1383 [DOI] [PubMed] [Google Scholar]
- 24. Tacchini L., Cairo G., De Ponti C., Massip M., Rosellò-Catafau J., Peralta C. (2006) Free Radic Res. 40, 1206–1217 [DOI] [PubMed] [Google Scholar]
- 25. El-Assal O., Hong F., Kim W. H., Radaeva S., Gao B. (2004) Cell Mol. Immunol. 1, 205–211 [PubMed] [Google Scholar]
- 26. Jin X., Zhang Z., Beer-Stolz D., Zimmers T. A., Koniaris L. G. (2007) Hepatology 46, 802–812 [DOI] [PubMed] [Google Scholar]
- 27. Lamlé J., Marhenke S., Borlak J., von Wasielewski R., Eriksson C. J., Geffers R., Manns M. P., Yamamoto M., Vogel A. (2008) Gastroenterology 134, 1159–1168 [DOI] [PubMed] [Google Scholar]
- 28. McGrath-Morrow S., Lauer T., Yee M., Neptune E., Podowski M., Thimmulappa R. K., O'Reilly M., Biswal S. (2009) Am. J. Physiol. Lung Cell Mol. Physiol. 296, L565–L573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waxman A. B., Kolliputi N. (2009) Am. J. Respir. Cell Mol. Biol. 41, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolliputi N., Waxman A. B. (2009) Am. J. Physiol. Lung Cell Mol. Physiol. 297, L6–L16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simeonova P. P., Toriumi W., Kommineni C., Erkan M., Munson A. E., Rom W. N., Luster M. I. (1997) J. Immunol. 159, 3921–3928 [PubMed] [Google Scholar]
- 32. Kida H., Yoshida M., Hoshino S., Inoue K., Yano Y., Yanagita M., Kumagai T., Osaki T., Tachibana I., Saeki Y., Kawase I. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 288, L342–L349 [DOI] [PubMed] [Google Scholar]
- 33. Fischer C. P., Hiscock N. J., Penkowa M., Basu S., Vessby B., Kallner A., Sjöberg L. B., Pedersen B. K. (2004) J. Physiol. 558, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vassilakopoulos T., Karatza M. H., Katsaounou P., Kollintza A., Zakynthinos S., Roussos C. (2003) J. Appl. Physiol. 94, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 35. Higdon J. V., Frei B. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 365–367 [DOI] [PubMed] [Google Scholar]
- 36. Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y., Nakayama O., Makishima M., Matsuda M., Shimomura I. (2004) J. Clin. Invest. 114, 1752–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sano M., Fukuda K., Sato T., Kawaguchi H., Suematsu M., Matsuda S., Koyasu S., Matsui H., Yamauchi-Takihara K., Harada M., Saito Y., Ogawa S. (2001) Circ. Res. 89, 661–669 [DOI] [PubMed] [Google Scholar]
- 38. Wruck C. J., Claussen M., Fuhrmann G., Romer L., Schulz A., Pufe T., Waetzig V., Peipp M., Herdegen T., Gotz M. E. (2007) J. Neural. Transm. Suppl. 72, 57–67 [DOI] [PubMed] [Google Scholar]
- 39. Lee P. C., Ho I. C., Lee T. C. (2005) Toxicol. Sci. 85, 541–550 [DOI] [PubMed] [Google Scholar]
- 40. Li J., Calkins M. J., Johnson D. A., Johnson J. A. (2007) Methods Mol. Biol. 399, 67–78 [DOI] [PubMed] [Google Scholar]
- 41. de Araujo E. G., da Silva G. M., Dos Santos A. A. (2009) Ann. N.Y. Acad. Sci. 1153, 57–64 [DOI] [PubMed] [Google Scholar]
- 42. Penkowa M., Giralt M., Carrasco J., Hadberg H., Hidalgo J. (2000) Glia 32, 271–285 [DOI] [PubMed] [Google Scholar]
- 43. Nakajima A., Yamada K., Zou L. B., Yan Y., Mizuno M., Nabeshima T. (2002) Free Radic Biol. Med. 32, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 44. Bissonnette C. J., Klegeris A., McGeer P. L., McGeer E. G. (2004) Neurosci. Lett. 361, 40–43 [DOI] [PubMed] [Google Scholar]
- 45. Maeda Y., Matsumoto M., Hori O., Kuwabara K., Ogawa S., Yan S. D., Ohtsuki T., Kinoshita T., Kamada T., Stern D. M. (1994) J. Exp. Med. 180, 2297–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernardi F., Guolo F., Bortolin T., Petronilho F., Dal-Pizzol F. (2008) J. Obstet. Gynaecol. Res. 34, 948–951 [DOI] [PubMed] [Google Scholar]
- 47. Taub R. (2003) J. Clin. Invest. 112, 978–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Y., Ochando J., Yopp A., Bromberg J. S., Ding Y. (2005) J. Immunol. 174, 2720–2729 [DOI] [PubMed] [Google Scholar]
- 49. Dhakshinamoorthy S., Jaiswal A. K. (2002) Oncogene 21, 5301–5312 [DOI] [PubMed] [Google Scholar]