Abstract
Cells lacking the exosome-associated protein Rrp47 show similar defects in stable RNA processing to those observed in the absence of the catalytic subunit Rrp6, but the precise mechanism(s) by which Rrp47 functions together with Rrp6 remains unclear. Deletion complementation analyses defined an N-terminal region of Rrp47, largely coincident with the bioinformatically defined Sas10/C1D domain, which was sufficient for protein function in vivo. In vitro protein interaction studies demonstrated that this domain of Rrp47 binds the PMC2NT domain of Rrp6. Expression of the N-terminal domain of Rrp47 in yeast complemented most RNA-processing defects associated with the rrp47Δ mutant but failed to complement the defect observed in 3′-end maturation of box C/D small nucleolar RNAs. Consistent with these results, protein capture assays revealed an interaction between the C-terminal region of Rrp47 and the small nucleolar ribonucleoproteins Nop56 and Nop58. Filter binding assays demonstrated that deletion of the lysine-rich sequence at the C terminus of Rrp47 blocked RNA binding in vitro. Furthermore, a protein mutated both at the C terminus and within the N-terminal domain showed a synergistic defect in RNA binding without impacting on its ability to interact with Rrp6. These studies provide evidence for a role of Rrp47 in registering a small nucleolar ribonucleoprotein particle assembly, functionally characterize the Sas10/C1D domain of Rrp47, and show that both the C terminus of Rrp47 and the N-terminal domain contribute to its RNA-binding activity.
Keywords: Protein Domains, Protein-Protein Interactions, Ribonuclease, RNA, RNA-binding Protein, RNA Processing, Small Nucleolar RNA (snoRNA), Yeast Genetics
Introduction
The exosome ribonuclease complex was initially characterized through its function in the processing of 3′-extended precursors to stable RNAs such as 5.8 S rRNA, small nucleolar RNAs (snoRNAs),5 and small nuclear RNAs (snRNAs) and in the degradation of excised RNA fragments that are released during such RNA-processing reactions (1, 2). It has since become clear that the exosome also plays a crucial role in nuclear RNA surveillance mechanisms that recognize and degrade transcripts that are incorrectly processed, that do not receive the appropriate post-transcriptional modification, or that are not correctly assembled into ribonucleoprotein particles (3–5). More recently, studies have shown that the exosome is responsible for the degradation of short-lived transcripts that are the result of pervasive transcription throughout the genome (6–9). In the cytoplasm, the exosome also functions in normal mRNA turnover and in translation-coupled RNA surveillance mechanisms. The role of the exosome in RNA-processing and surveillance pathways has been the subject of a number of recent reviews (3, 4, 10–14).
Catalytic activity of the eukaryotic exosome is attributed to two subunits, Rrp44/Dis3 and Rrp6 (15, 16). Rrp44 is a member of the RNase II family of 3′ → 5′ exoribonucleases but also has an endonucleolytic activity associated with its N-terminal PIN domain (17–19). Rrp6 is related to another bacterial 3′ → 5′ exoribonuclease, RNase D (20–22). These enzymes have differing substrate preferences in vitro, and yeast strains mutant for Rrp44 or Rrp6 show distinct stable RNA-processing phenotypes, with rrp6 mutants accumulating characteristic precursor species, including a 3′-extended form of 5.8 S rRNA and adenylated forms of snoRNAs and U6 snRNA (1, 2, 6, 23, 24). RNA analyses of yeast mutants expressing a C-terminally truncated Rrp6 mutant that cannot bind stably to the exosome suggest that this enzyme might carry out some of its functions independently of the exosome complex (25).
Yeast strains lacking the small nuclear protein Rrp47 (also known as Lrp1) show comparable RNA-processing phenotypes to those observed in the absence of Rrp6, and Rrp47 is found associated with exosome complexes that contain Rrp6 (26–29). We have previously shown that Rrp47 interacts directly with the N-terminal region of Rrp6 (30) comprising the PMC2NT domain (31). Like other exosome factors, Rrp47 is conserved throughout eukaryotes, and the homologous human protein, C1D, functions in stable RNA-processing pathways and interacts with the Rrp6 homologous protein PM/Scl-100 (32). Both Rrp47 and C1D have RNA- and DNA-binding activity, with an apparent specificity for double-stranded DNA and structured RNA substrates (30, 32, 33). Rrp47 has been shown to bind both Rrp6 and nucleic acid concomitantly, suggesting that it might promote Rrp6 activity by enabling the enzyme to bind structural elements within RNA (30).
Rrp47 lacks previously characterized domains that are associated with nucleic acid- or protein-binding activity. In the absence of available structural data for Rrp47, we initiated a mutagenesis study to map the regions of the protein that are required for its characterized RNA-processing functions in vivo and for its Rrp6- and RNA-binding activities in vitro. Mutually supportive data from biochemical and genetic experiments define a domain within the N-terminal portion of Rrp47 that can interact with Rrp6 and that is sufficient for Rrp47 function in yeast. In contrast, stable binding of Rrp47 to RNA in vitro required the full-length protein. Finally, RNA analyses of rrp47 mutants and protein interaction studies in vitro suggest an unexpected role for Rrp47 in sensing the assembly of box C/D small nucleolar ribonucleoprotein (snoRNP) particles.
EXPERIMENTAL PROCEDURES
Bioinformatics
The secondary structure of Rrp47 was predicted using the web server Phyre (34). Rrp47 homologous sequences were identified by WU-BLAST2 using the EBI server (35). Multiple sequence alignments were created using ClustalW and viewed using Jalview (36).
Construction and Growth of Strains
Details of plasmid constructions, primer sequences, and strain descriptions are provided in the supplemental “Experimental Procedures.” Plasmid constructs were generated by standard restriction digestion cloning techniques or site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). Yeast strains carrying plasmids were grown under standard selective conditions, whereas strains expressing genomically encoded tandem affinity purification-tagged proteins (Open Biosystems) were grown in YPD medium (yeast extract, peptone, and dextrose).
Expression and Purification of Recombinant Proteins
GST, GST-22, GST-C, GST-PMC2NT, and His6-Rrp47 proteins were induced in BL21(DE3)LysS cells, and cell lysates were made in 20 mm HEPES, pH 7.6, 300 mm NaCl, 10 mm imidazole, and 1 mm PMSF as described previously (30). Full-length His6-Rrp47 and the N* sextuple point mutant (E79A/R82G/K84I/Y86S/K89M/K91I) were purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and ion exchange chromatography over SP-Sepharose as described (30). For the ΔC, S100X, I162X, V176X, G181X, and N*G181X mutant Rrp47 proteins, the Ni-NTA eluates were dialyzed against 20 mm HEPES, pH 7.6, and 150 mm NaCl and passed over SP-Sepharose beads, and the nonbound fraction was retained. Proteins were analyzed by SDS-PAGE and Western blotting using the anti-pentahistidine monoclonal antibody (Qiagen) or anti-GST antiserum (Sigma).
Expression of the heteromeric His9-Nrd1·MBP-Nab3 complex was performed as described (37). This expression construct does not encode the C-terminal 26 residues of Nrd1, 191 residues from the N terminus of Nab3, and 238 residues from the C terminus of Nab3. The Nrd1·Nab3 complex was purified by Ni-NTA affinity chromatography and detected in pulldown assays by Western blot analyses using monoclonal antibodies against the His epitope (Qiagen) or MBP (Sigma).
Protein and RNA Analyses
The SL-AU RNA (ggguuuccuucucaaacauucuguuugguagugaguaauuaaaaaugaauu) was generated by in vitro transcription of EcoRI-linearized plasmid p299 using T7 RNA polymerase. Transcription products were dephosphorylated by incubation with alkaline phosphatase, gel-purified, and 5′-32P-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Yeast tRNAPhe (Sigma) was dephosphorylated and 5′-labeled as described above. RNA-binding experiments were performed using a modified double-filter binding assay (38). Purified recombinant proteins were serially diluted in RNA-binding buffer (10 mm Tris-HCl, pH 8, 150 mm NaCl, 2 mm MgCl2, 0.1 mm EDTA, and 10% glycerol) and 20-μl aliquots were mixed with 5 μl of 32P-labeled RNA substrate (final concentration, 10 nm). After incubation on ice for 10 min, the reaction mixtures were filtered through a double membrane consisting of Hybond N+ overlaid with Hybond C (GE Healthcare) using a Minifold II slot blot apparatus (Schleicher & Schüll) and washed three times with 0.5 ml of binding buffer. RNA retained on each membrane was visualized using a Storm PhosphorImager (GE Healthcare), and the data were quantified using ImageQuant software (GE Healthcare). Triplicate assays were performed in parallel.
Pulldown experiments using extracts from Escherichia coli cells expressing recombinant GST-PMC2NT and Rrp47 proteins were performed essentially as described (30). Yeast cell extracts were made in lysis buffer comprising 10 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 5 mm MgCl2. Protein capture assays were performed by adding 0.5 ml of cell extract from yeast strains expressing epitope-tagged proteins to 20 μl of Ni-NTA beads or glutathione-Sepharose beads precharged with recombinant protein. Binding reactions were carried out for 2 h at 4 °C. After washing five times with 1 ml of cell lysis buffer containing 0.1% Nonidet P-40 for 10 min, the beads were routinely incubated with 1 μg of RNase A for 30 min at 20 °C and then rewashed. Retained protein was eluted in 50 μl of elution buffer (20 mm HEPES, pH 7.6, and 300 mm NaCl containing 250 mm imidazole or 25 mm reduced glutathione). In the experiment shown in supplemental Fig. S3, proteins from yeast cell extracts that were retained on Rrp47-charged beads were treated with DNase I, micrococcal nuclease, or RNase A before rewashing and elution with imidazole-containing buffer. Samples were resolved by SDS-PAGE and analyzed by Western blotting using the peroxidase/anti-peroxidase antibody (Sigma).
Total cellular RNA was resolved through 8% polyacrylamide gels containing 50% urea, transferred to a Hybond N+ membrane, and hybridized with 5′-32P-labeled oligonucleotide probes as described (25). The probes used were as follows: 5.8 S rRNA (o236), gcgttgttcatcgatgc; 5′-external transcribed sequence (ETS; o274), cgctgctcaccaatgg; snR38 (o272), gagaggttacctattattacccattcagacagggataactg; snR50 (o494), ctgctgcaaattgctacctc; snR52 (o318), gtatcagagattgttcacgctaatg; U6 (o517), atctctgtattgtttcaaattgaccaa; and SCR1 (o242), aaggacccagaactaccttg.
RESULTS
The N-terminal Sas10/C1D Domain of Rrp47 Is Sufficient for Normal Growth
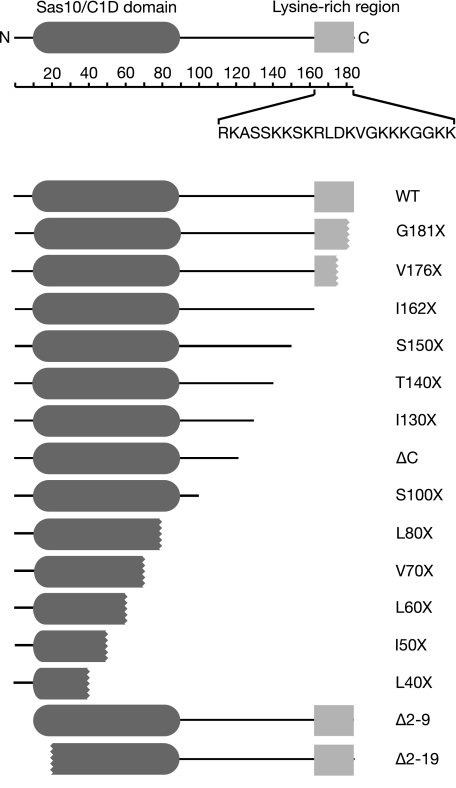
Yeast strains lacking Rrp47 exhibit a moderate slow growth phenotype in selective medium (30), whereas strains lacking both RRP47 and REX1 genes are nonviable (27). To determine the region of Rrp47 required for function in vivo, plasmids encoding N- or C-terminal deletion mutants were assayed for their ability to complement the synthetic lethality of rrp47Δ rex1Δ double mutants in a plasmid shuffle assay using a host strain that also allowed visualization of plasmid loss through a red/white colony-sectoring phenotype (39) and for their ability to rescue the slow growth phenotype of rrp47Δ single mutants. The deletion mutants generated in this study are depicted in Fig. 1.
FIGURE 1.
Mutations generated in Rrp47. The position of the Sas10/C1D domain within the Rrp47 protein and the sequence of the lysine-rich C-terminal peptide are indicated. The scale indicates the position along the length of the protein in amino acid residues. The coding regions present in each of the C- or N-terminal truncation mutants is indicated; truncations that encroach into the Sas10/C1D domain or the lysine-rich C-terminal peptide are shown by serrated lines.
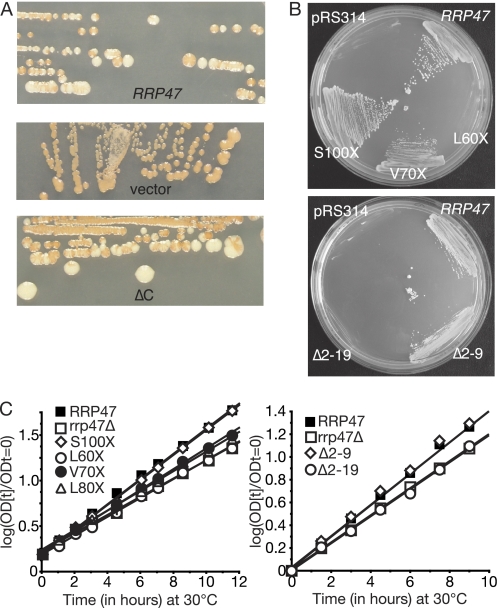
Protein structure prediction analyses suggested that the N-terminal two-thirds of Rrp47 are largely α-helical in nature (supplemental Fig. S1). We therefore initially generated and tested a mutant encoding the N-terminal 120 residues, denoted ΔC. This mutant fully complemented the synthetic lethality of the rrp47Δ rex1Δ double mutant in the colony-sectoring assay and plasmid shuffle assay (Fig. 2A and data not shown), demonstrating that the C-terminal region of Rrp47 is not required for the essential function of Rrp47 in vivo in the absence of Rex1. Further consideration of the predicted secondary structure suggested possible looped regions within the Rrp47 structure around residues 95–100 and 68–74. Truncation at Ser-100 generated a construct that complemented the synthetic lethality of the double mutant, with the colonies obtained on medium containing 5′-fluoroorotic acid for the rrp47Δ rex1Δ double mutant that expressed the S100X rrp47 allele being of similar size to the wild-type control (Fig. 2B). Truncation at Val-70 also complemented the rrp47Δ rex1Δ double mutant, but the colonies obtained were significantly smaller. A slightly larger C-terminal truncation (L60X) failed to support growth (Fig. 2B). Consistent with these data, rrp47Δ transformants expressing the S100X mutant grew comparably to cells expressing the wild-type allele, whereas V70X and L80X mutants only partially restored the slow growth phenotype of the rrp47Δ strain, and the L60X mutant grew at a rate comparable with the vector-transformed control (Fig. 2C). A short deletion (Δ2–9) at the N terminus allowed complementation in the plasmid shuffle assay and rescued the slow growth phenotype, whereas the larger Δ2–19 deletion did not (Fig. 2, B and C). The region of Rrp47 that is required for function in vivo, as defined by these genetic complementation assays, closely maps to residues 10–89, which constitute the bioinformatically predicted Sas10/C1D domain (Pfam accession number PF04000) (40, 41) (see Fig. 1). We conclude from these data that residues 10–100 of Rrp47, encompassing the Sas10/C1D domain, are critical for protein function in vivo.
FIGURE 2.
The N-terminal Sas10/C1D domain of Rrp47 is required for protein function. A, colony-sectoring assay. An rrp47Δ rex1Δ double mutant harboring a plasmid-borne copy of the RRP47 gene was transformed with plasmids encoding the wild-type RRP47 gene, the ΔC mutant, and a vector control. Loss of the original plasmid through growth on YPD was followed using the red/white colony-sectoring assay. B, plasmid shuffle assay. The rrp47Δ rex1Δ double mutant was transformed with plasmids encoding S100X, V70X, L60X, Δ2–9, and Δ2–19 rrp47 truncations and then grown on solid medium containing 5′-fluoroorotic acid to purge the cells of the original RRP47 plasmid. C, growth rate analyses. An rrp47Δ strain was transformed with plasmids encoding rrp47 truncation mutants, the wild-type RRP47 gene, or the vector, and the growth rates of the resultant transformants were measured in selective medium. Growth assays were performed in duplicate with the average values plotted. The r2 values for all individual data sets lay between 0.995 and 0.999.
The Sas10/C1D Domain of Rrp47 Binds Rrp6
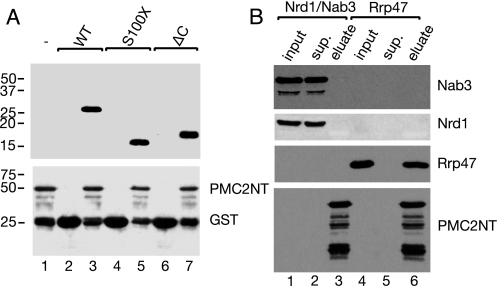
We had previously shown that recombinant Rrp47 binds to the N-terminal PMC2NT domain of Rrp6 in E. coli cell lysates (30). To test whether the Sas10/C1D domain of Rrp47 is sufficient for this interaction with Rrp6, purified His-tagged full-length protein and the ΔC mutant were added to beads charged with GST or GST-PMC2NT fusion protein. Both purified full-length Rrp47 and the ΔC construct bound to the GST-tagged PMC2NT domain, whereas neither protein was retained on beads charged with GST (Fig. 3A). As observed previously (30), a number of polypeptides shorter than the ∼50-kDa GST fusion of the Rrp6 N-terminal domain were detected upon Western blot analysis, which presumably arise through proteolytic degradation. The S100X truncated Rrp47 protein was stably expressed and also bound to the GST-PMC2NT fusion protein. In control reactions, the unrelated Nrd1·Nab3 protein complex failed to purify with the tagged Rrp6 N-terminal domain (Fig. 3B). Similar results were obtained when pulldown assays were performed on nonfractionated cell lysates containing the recombinant proteins (data not shown). Proteins with more extensive C-terminal truncations (L80X and V70X) showed a specific but progressively decreasing level of binding to the GST-PMC2NT fusion protein but were not expressed at sufficient levels to allow a direct comparative analysis using the binding assay (data not shown).
FIGURE 3.
The N-terminal domain of Rrp47 binds to Rrp6. A, pulldown assays of His-tagged full-length (WT), S100X, and ΔC Rrp47 proteins with a GST fusion of the PMC2NT domain of Rrp6 (lanes 1, 3, 5, and 7) and the GST control (lanes 2, 4, and 6). Glutathione-Sepharose beads charged with ∼100 μg of GST or GST fusion protein were washed and then incubated with ∼50 μg of purified recombinant Rrp47. Eluates were resolved by SDS-PAGE and analyzed by Western blotting using antibodies against the His6 epitope (upper panel) or GST (lower panel). The electrophoretic mobility of bait proteins and molecular mass markers (in kDa) are indicated on the right and left of the panels, respectively. B, pulldown assays of Rrp47 and the Nrd1·Nab3 complex control with the GST-PMC2NT fusion protein. Equivalent amounts of input, supernatant (sup.) and eluate fractions were analyzed by Western blotting using epitope tag-specific antibodies. Proteins detected in the Western blot analyses are indicated. Multiple bands were detected for the GST-PMC2NT fusion protein in both panels because of proteolytic degradation.
Both N- and C-terminal Regions of Rrp47 Contribute to RNA Binding
We then asked whether the N-terminal region of Rrp47 has RNA-binding activity. Filter binding assays were performed on full-length Rrp47 or C-terminally truncated mutants using either yeast tRNAPhe or a model-structured RNA (denoted SL-AU) comprising the stem-loop at the 5′-end of the yeast internal transcribed sequence 2 (42) linked to a 3′-A/U-rich sequence.
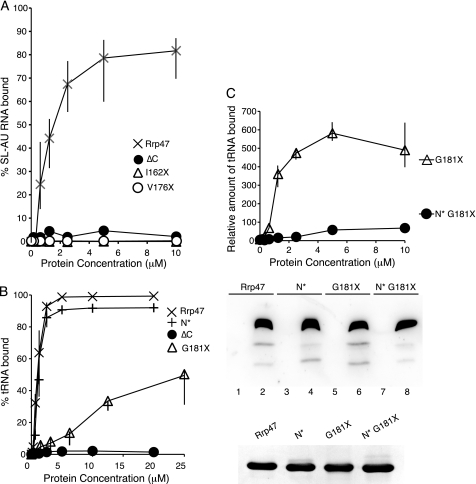
Consistent with previous gel shift assays (30), full-length Rrp47 bound to tRNAPhe and the SL-AU RNA substrates with an apparent dissociation constant of ∼1 μm (as calculated using the concentration of the monomeric protein) (Fig. 4, A and B) and showed no binding activity with a control substrate transcribed from the polylinker of a cloning vector (data not shown). This selective binding probably reflects the degree of structure in the RNAs assayed because Rrp47 has previously been shown to bind structured RNAs but not poly(A) (30). In contrast to the wild-type protein, the ΔC mutant showed no significant binding to either tRNAPhe or the SL-AU RNA substrates (Fig. 4, A and B). The C-terminal region of Rrp47 and its homologous proteins are typically very basic, with 12 of the final 22 amino acids in the protein from Saccharomyces cerevisiae being either lysine or arginine residues (shown schematically in Fig. 1; for a sequence alignment, see supplemental Fig. S2). Mutant proteins lacking the C-terminal 22 residues (denoted I162X) or nine residues (denoted V176X) showed no detectable RNA-binding activity, whereas a mutant lacking only the last four residues (denoted G181X) exhibited a substantial decrease in RNA-binding activity (Fig. 4, A and B). A GST fusion of the C-terminal 22 residues showed weak nonspecific binding activity to all RNA substrates tested, including the polylinker transcript (data not shown). Thus, the basic C terminus of Rrp47 is required for stable interaction with RNA but is, in itself, not sufficient for the stability and specificity of binding.
FIGURE 4.
RNA binding of Rrp47. A and B, filter binding saturation curves obtained for wild-type or mutant Rrp47 proteins with the SL-AU and tRNAPhe substrates. All assays were performed in triplicate; the vertical bars indicate the range of values. The sextuple mutant (N*) carries the point mutations E79A, R82G, K84I, Y86S, K89M, and K91I. C, upper panel, parallel filter binding assays of the G181X truncation mutant and the N*G181X mutant. The amount of radiolabeled tRNA retained is plotted against the concentration of Rrp47 protein. Assays were performed in triplicate; the vertical bars indicate the ranges of values. Center panel, pulldown assays of the purified proteins used in the RNA-binding assays. Purified Rrp47 proteins were incubated with beads charged with GST control (lanes 1, 3, 5, and 7) or the GST-PMC2NT fusion protein (lanes 2, 4, 6, and 8), and the eluates were analyzed by SDS-PAGE and Western blotting using the anti-His antibody. Lower panel, Coomassie Blue-stained SDS-polyacrylamide gel of the purified recombinant Rrp47 proteins used in these assays.
There are several well conserved, charged, or aromatic amino acid residues within the N-terminal region of Rrp47 that lie clustered within the primary sequence, namely Glu-79, Arg-82, Lys-84, Tyr-86, Lys-89, and Lys-91 (supplemental Fig. S2). We mutated these residues, singularly and in combination, and tested the resultant mutant proteins for their ability to interact with RNA. The single-point mutants E79A, R82A, K84A, Y86A, K89A, and K91A showed no defect in RNA-binding activity in vitro (data not shown), and neither did a sextuple mutant wherein each of the above residues was mutated (denoted N*) (Fig. 4B). However, combining the sextuple mutant with the compromised G181X mutant (denoted N*G181X) caused an essentially complete loss of RNA-binding activity in the resultant protein (Fig. 4C). This effect does not reflect a gross structural alteration of the N-terminal region in the N*G181X mutant protein because the N*, G181X, and N*G181X mutants bound to the PMC2NT region of Rrp6 in a comparable manner to the wild-type protein in pulldown assays (Fig. 4C). These results demonstrate that the N-terminal region and the extreme C terminus both contribute to the RNA-binding activity of Rrp47.
The C-terminal Region of Rrp47 Has a Specific Function in snoRNA Maturation
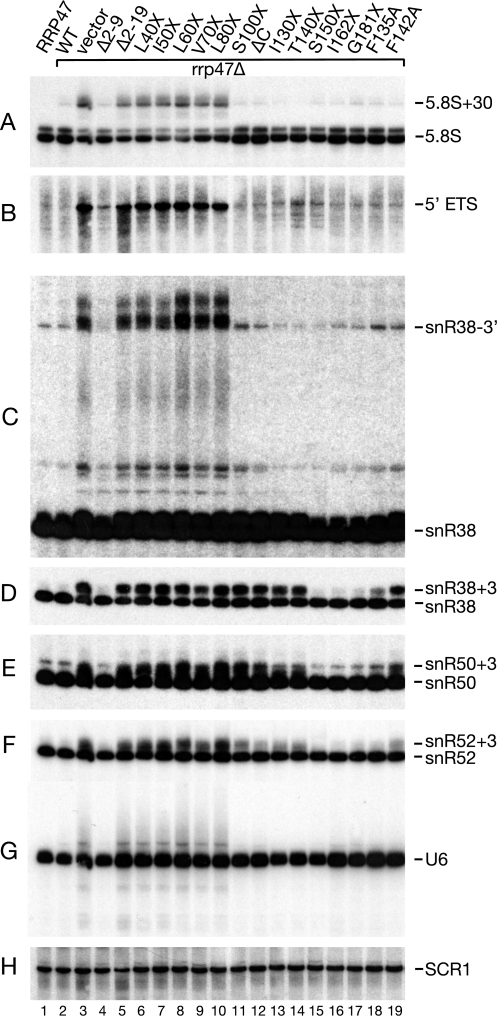
Yeast rrp47Δ strains accumulate 3′-extended forms of 5.8 S rRNA and box C/D snoRNAs and are defective in the degradation of the excised 5′-ETS pre-rRNA fragment (27, 28). To determine which regions of the Rrp47 protein are required for its function in these processes, Northern blot hybridization analyses were performed on total cellular RNA from a wild-type strain, an rrp47Δ strain, and a series of N- and C-terminal rrp47 truncation mutants.
As reported previously (27, 28), the rrp47Δ mutant accumulated the 3′-extended 5.8 S + 30 species to high levels, with a corresponding decrease in the level of mature 5.8 S rRNA, and accumulated readily detectable amounts of the 5′-ETS fragment (Fig. 5). Consistent with previous data (28, 30), this mutant also showed significantly increased levels of a 3′-unprocessed form of snR38 and expressed an snR38 species slightly longer than the mature form of wild-type strains (labeled snR38–3′ and snR38+3, respectively, in Fig. 5, C and D) and accumulated both discrete and diffuse snR38 species of intermediate length. The rrp47Δ mutant also accumulated extended U6 snRNA species. The diffuse extended forms observed for U6 and snR38 are consistent with polyadenylated forms of these RNAs previously observed in rrp6Δ mutants (1, 2, 6, 24).
FIGURE 5.
RNA Analyses of rrp47 mutants. RNA was resolved through an 8% denaturing polyacrylamide gel and analyzed by Northern blot hybridization. Hybridization patterns were visualized by autoradiography. Lane 1, RNA from a wild-type strain; lanes 2–19, RNA from an rrp47Δ strain transformed with plasmids encoding rrp47 alleles, as indicated. Probes used were complementary to the following RNAs: 5.8 S rRNA (A), 5′-ETS (B), snR38 (C and D), snR50 (E), snR52 (F), U6 (G), and SCR1 (H). The major species detected are indicated on the right. D shows a shorter exposure than C that reveals the relative levels of snR38 and the 3′-extended snR38+3 species. SCR1 served as a loading control.
Mutant rrp47 alleles encoding proteins that do not encompass the complete Sas10/C1D domain (Δ2–19, L40X, I50X, L60X, V70X, L80X) exhibited RNA-processing phenotypes that are characteristic of rrp47Δ mutants (Fig. 5, lanes 5–10). These mutants either failed to complement the rrp47Δ allele genetically or exhibited slow growth (Fig. 2 and data not shown), and so their expression levels may well be compromised. In contrast, the S100X mutant showed no defect in 5.8 S rRNA processing or 5′-ETS degradation and did not accumulate the extended forms of snR38 or U6 (Fig. 5, compare lanes 10 and 11). Similarly, the Δ2–9 mutant showed no detectable defect in 5.8 S rRNA processing or in the maturation of box C/D snoRNAs and did not accumulate extended U6 snRNA species. These results correlate closely with the genetic complementation analyses (Fig. 2) and suggest that N- or C-terminal deletions that encroach into the N-terminal Sas10/C1D domain either impact on Rrp47 expression levels or inhibit Rrp47 function in stable RNA-processing and degradation pathways in vivo.
Intriguingly, strains expressing the S100X, ΔC, I130X, and T140X rrp47 mutants accumulated the snR38+3 species but not the longer extended forms of snR38 (Fig. 5, C and D, lanes 11–14), showed no defect in 5.8 S rRNA processing or 5′-ETS degradation, and did not accumulate 3′-extended forms of U6 snRNA. To test whether this effect was specific to intron-encoded snoRNAs, the blots were also hybridized with probes complementary to sequences within the independently transcribed snoRNAs snR50 and snR52. Slightly extended forms of snR50 and snR52 also accumulated in the S100X, ΔC, I130X, and T140X mutants, albeit to a lesser extent in the case of snR52 (Fig. 5). Comparable phenotypes were observed for other intron-encoded (snR39) and independently transcribed (snR54 and snR13) snoRNAs (data not shown).
In contrast, strains expressing the longer S150X, I162X, and G181X mutant proteins showed little or no accumulation of aberrant forms of any of the RNAs tested (Fig. 5, lanes 15–17). Inspection of a multiple alignment of Rrp47 sequences (supplemental Fig. S2) suggested that a short sequence showing a very high degree of conservation across yeast species (amino acids 128–144 in S. cerevisiae, consensus yeast sequence PAISKSNFQGKHTKFED) maps to the region around position 140, with Ile-130, Ser-131, Phe-135, His-139, and Phe-142 being highly conserved. The two phenylalanine at positions 135 and 142 were individually mutated to alanine residues, and RNA was analyzed from strains expressing the mutant proteins. The F142A mutant showed a specific RNA-processing defect in snoRNA 3′-end maturation, with a marked accumulation of the +3 species for snR38, snR50, and snR52, whereas the F135A mutant had a considerably weaker effect (Fig. 5, D–F, lanes 18 and 19). These data support a specific role for residues within the C-terminal region of Rrp47 in the final maturation of box C/D snoRNAs and demonstrate that the basic C terminus, although required for stable RNA binding in vitro, is dispensable for RNA-processing functions in vivo.
The C-terminal Region of Rrp47 Interacts with snoRNP Proteins
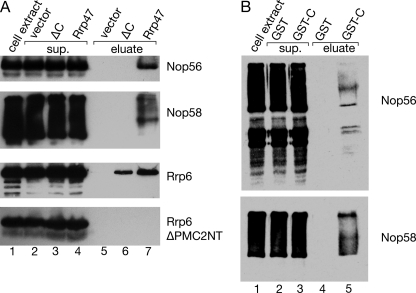
To test whether the C-terminal region of Rrp47 mediates an interaction between the exosome and snoRNP particles, we assayed immobilized recombinant Rrp47 for its ability to bind epitope-tagged forms of the snoRNP proteins Nop56 and Nop58 in yeast cell lysates. Both Nop56 and Nop58 were readily detected in the eluate fractions from beads charged with full-length Rrp47 but not in eluates from beads charged with the ΔC mutant (Fig. 6A). In control reactions, the full-length and truncated Rrp47 proteins bound full-length Rrp6, and both failed to retain a truncated Rrp6 mutant lacking the N-terminal PMC2NT domain. Both the C-terminally tagged Nop56 and Nop58 proteins consistently migrated on SDS-polyacrylamide gels as one major band with additional, slower migrating, diffuse bands. The reason for this is not clear but presumably reflects a common structural feature in these proteins, such as the C-terminal KK(D/E) repeats (43). The beads were routinely incubated with RNase A prior to elution of the retained material, and incubation with micrococcal nuclease or DNase I also failed to release Nop56 from the Rrp47-charged beads (supplemental Fig. S3). To address whether the C-terminal region of Rrp47 is sufficient for this interaction, residues 121–184 were expressed as a GST fusion (denoted GST-CT), and the resultant protein was assayed for Nop56 and Nop58 binding as described above. The GST-CT fusion protein bound to both Nop56 and Nop58 present in cell extracts, whereas no binding was observed for the GST control to either snoRNP protein (Fig. 6B). These experiments show that Rrp47 can bind to box C/D snoRNP proteins in an RNase- and DNase-insensitive manner and that the C-terminal region of Rrp47 is necessary and sufficient for this interaction.
FIGURE 6.
Rrp47 interacts with Nop56 and Nop58. Cell extracts from yeast strains (∼5 mg of total protein) expressing tandem affinity purification-tagged proteins were incubated with beads precharged with ∼500 μg of recombinant proteins. After binding, the beads were washed and then incubated with RNase A prior to rewashing and elution of the retained protein. Equivalent fractions of cell extract and supernatant (sup.) and a 100-fold equivalent of the eluate fractions were then resolved by SDS-PAGE and analyzed by Western blotting. The epitope-tagged protein expressed in yeast is indicated on the right. A, pulldown assays with full-length Rrp47, the ΔC mutant, and a vector control. Lysates from strains expressing epitope-tagged full-length Rrp6 or an Rrp6 mutant protein lacking the PMC2NT domain were used as positive and negative controls, respectively. B, pulldown assays with a GST fusion of the C-terminal region of Rrp47 (GST-CT) or GST alone.
DISCUSSION
Genetic complementation assays and Northern blot analyses of cellular RNAs from rrp47 mutants provide independent corroborative data sets demonstrating that the N-terminal region of Rrp47 encompassing residues 10–100 comprises a domain that is critical for its function. Biochemical protein binding studies demonstrated that the N-terminal domain of Rrp47 interacts directly with the PMC2NT domain of Rrp6. These data are consistent with a key biochemical activity of Rrp47 being its ability to interact with Rrp6. Indeed, previous studies from our laboratory showed that the interaction with Rrp6 is required for normal expression levels of Rrp47 (30). This empirically determined N-terminal domain correlates with the bioinformatically defined Sas10/C1D domain (residues 10–89) (40) that is also found in the 18 S rRNA synthesis factors Sas10/Utp3 and Lcp5 (44, 45) and in the metazoan protein neuroguidin, a factor involved in translational regulation during neuronal development (41, 46). We have found no evidence for an interaction between Sas10 or Lcp5 and Rrp6,6 but it remains feasible that Sas10/C1D family members interact with PMC2NT-related domains within other proteins.
An alternative but not mutually exclusive hypothesis is that the Sas10/C1D family may represent a group of RNA-binding proteins. Both Sas10/Utp3 and Lcp5 are found associated with U3 snoRNA, whereas neuroguidin is associated with the mRNA cap-binding protein eIF4E and the mRNA polyadenylation element-binding protein CPEB and is therefore presumably bound to mRNP particles. Our studies reported here show that the RNA-binding activity of Rrp47 is dependent upon both basic residues at the extreme C terminus of the protein and residues within the N-terminal domain. Notably, Sas10, Lcp5, and neuroguidin also have clusters of basic residues close to their C termini (45–47), and basic residues are conserved across the Sas10/C1D domain family at positions equivalent to Lys-84 and Lys-89 of Rrp47 (339 and 290 residues of all 382 available sequences being either lysine or arginine at these positions, respectively) (Pfam accession number PF04000) (41).
Given the RNA-binding activity of Rrp47 and the requirement for this protein in stable RNA-processing events, an unexpected observation was that short truncations from the C terminus of Rrp47 that prevented stable RNA binding in vitro (e.g. the I162X mutant) had no discernible effect on Rrp47-dependent RNA-processing events in vivo. One interpretation of these data would be that the RNA-binding activity of Rrp47 is not strictly required for its function in cellular stable RNA-processing pathways, perhaps because of redundancy with other RNA-binding proteins. Alternatively, interactions with other protein factors may enable the RNA-binding activity of the Sas10/C1D domain of Rrp47 to be sufficient for protein function in vivo.
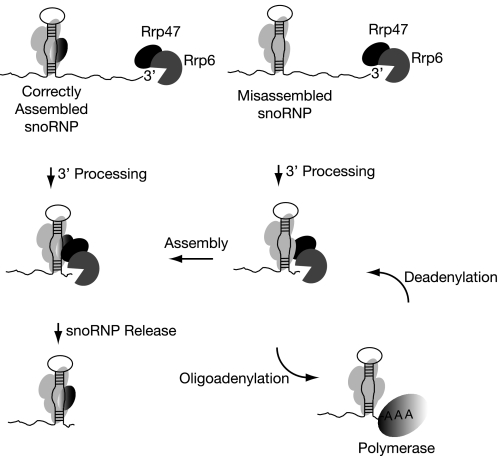
Our data reveal specific interactions between the C-terminal region of Rrp47 and proteins of box C/D snoRNP complexes and demonstrate that rrp47 mutants lacking the C-terminal region have specific defects in final 3′-maturation of box C/D snoRNAs. Previous studies have revealed similar snoRNA-processing phenotypes for conditional mutants of the snoRNP proteins Nop58 and Nop1 (48, 49). The 3′-extended snoRNAs have been shown to be generated by post-transcriptional oligoadenylation (24). Together, these data suggest a model (Fig. 7) wherein, during normal snoRNP assembly, Rrp47 interacts with the RNP particle during 3′-maturation and allows release of the mature complex. Conversely, if assembly of the snoRNP particle is delayed, the lack of interaction between Rrp47 and the snoRNP particle promotes snoRNA oligoadenylation and/or impedes the removal of the oligoadenylate tail by Rrp6. Oligoadenylated snoRNAs can be detected at low levels in wild-type cells, and transcriptional pulse-chase analyses have shown that oligoadenylated products are processed to mature snoRNAs in wild-type cells (24). Thus, oligoadenylation represents a step in a productive processing pathway for at least a fraction of the snoRNA transcripts under normal physiological conditions. Our results point to an important novel function of Rrp47 as a sensor for RNP assembly during 3′-end maturation of snoRNAs.
FIGURE 7.
Model for Rrp47 function in snoRNP assembly. snoRNP particles assemble cotranscriptionally through binding of core proteins to the snoRNA. Correct 3′-end formation of snoRNAs requires the 3′-exonuclease Rrp6. Correct assembly is sensed through interactions between Rrp47 and proteins of the snoRNP particle, allowing release of the mature snoRNP complex. In the absence of the Rrp47-snoRNP interaction, the snoRNA is adenylated by TRAMP (Trf4/Air2/Mtr4 polyadenylation complex) and maintained in an oligoadenylated form by the competing action of polymerase and exonuclease activities. This provides an extended period for assembly to be completed. Other exonucleases can substitute for Rrp6 in trimming of the pre-snoRNA but not complete removal of the oligoadenylate tail. For simplicity, Rrp6, rather than the complete exosome complex, is shown.
Delays in RNA-processing or assembly pathways can facilitate targeting of RNA substrates to quality control systems (50–52). In many cases, adenylation of RNA substrates is an initial step in such RNA degradation mechanisms, some of which involve Rrp6. Our data are consistent with the model (24) that snoRNA adenylation stalls the fate of the RNA and provides a kinetic opportunity for the delayed assembly of RNP particles to be completed (Fig. 7). Indeed, adenylation of stable RNAs during productive processing appears to be quite widespread (2, 24, 53–55). Similar interactions to those described here between Rrp47 and snoRNP proteins might also underlie the coupling of RNP assembly and processing of other RNAs.
To date, Rrp47 has been envisaged to function as an RNA-binding protein that allows Rrp6 to degrade structured RNA (13, 30). The data presented here reveal an unsuspected role for Rrp47 in RNP substrate recognition during final 3′-end maturation of box C/D snoRNAs. It is tempting to speculate that polyadenylated species that accumulate in rrp47Δ and rrp6Δ mutants, such as primary snoRNA transcripts (24), cryptic unstable transcripts, or extended forms of U6 snRNA (6) (see Fig. 5), arise because of the absence of similar interactions between the Sas10/C1D domain of Rrp47 and other factors involved in RNA transcription termination or processing.
Supplementary Material
Acknowledgment
We thank Jeff Corden (John Hopkins School of Medicine, Baltimore, MD) for providing the Nrd1/Nab3 expression construct.
This work was supported by Biotechnology and Biological Sciences Research Council Research Grant BB/D001161 (to J. A. S.), by Yorkshire Cancer Research and Wellcome Trust Research Grant 088306/Z/09/Z (to M. F.), by a Biotechnology and Biological Sciences Research Council Ph.D. studentship (to J. L. C.), and by a University of Sheffield Ph.D. studentship (to R. M. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S3, Table 1, and additional references.
M. Turner and P. Mitchell, unpublished data.
- snoRNA
- small nucleolar RNA
- snRNA
- small nuclear RNA
- snoRNP
- small nucleolar ribonucleoprotein
- Ni-NTA
- nickel-nitrilotriacetic acid
- ETS
- external transcribed sequence
- MBP
- maltose-binding protein.
REFERENCES
- 1. Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. (1999) EMBO J. 18, 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Hoof A., Lennertz P., Parker R. (2000) Mol. Cell. Biol. 20, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vanacova S., Stefl R. (2007) EMBO Rep. 8, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Houseley J., Tollervey D. (2008) Biochim. Biophys. Acta 1779, 239–246 [DOI] [PubMed] [Google Scholar]
- 5. Anderson J. T., Wang X. (2009) Crit. Rev. Biochem. Mol. Biol. 44, 16–24 [DOI] [PubMed] [Google Scholar]
- 6. Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Libri D., Jacquier A. (2005) Cell 121, 725–737 [DOI] [PubMed] [Google Scholar]
- 7. Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., Mapendano C. K., Schierup M. H., Jensen T. H. (2008) Science 322, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 8. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L. M. (2009) Nature 457, 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neil H., Malabat C., d'Aubenton-Carafa Y., Xu Z., Steinmetz L. M., Jacquier A. (2009) Nature 457, 1038–1042 [DOI] [PubMed] [Google Scholar]
- 10. Lebreton A., Séraphin B. (2008) Biochim. Biophys. Acta 1779, 558–565 [DOI] [PubMed] [Google Scholar]
- 11. Schmid M., Jensen T. H. (2008) Trends Biochem. Sci. 33, 501–510 [DOI] [PubMed] [Google Scholar]
- 12. Belostotsky D. (2009) Curr. Opin. Cell Biol. 21, 352–358 [DOI] [PubMed] [Google Scholar]
- 13. Lykke-Andersen S., Brodersen D. E., Jensen T. H. (2009) J. Cell Sci. 122, 1487–1494 [DOI] [PubMed] [Google Scholar]
- 14. Tomecki R., Drazkowska K., Dziembowski A. (2010) ChemBioChem 11, 938–945 [DOI] [PubMed] [Google Scholar]
- 15. Liu Q., Greimann J. C., Lima C. D. (2006) Cell 127, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 16. Dziembowski A., Lorentzen E., Conti E., Séraphin B. (2007) Nat. Struct. Mol. Biol. 14, 15–22 [DOI] [PubMed] [Google Scholar]
- 17. Lebreton A., Tomecki R., Dziembowski A., Séraphin B. (2008) Nature 456, 993–996 [DOI] [PubMed] [Google Scholar]
- 18. Schaeffer D., Tsanova B., Barbas A., Reis F. P., Dastidar E. G., Sanchez-Rotunno M., Arraiano C. M., van Hoof A. (2009) Nat. Struct. Mol. Biol. 16, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schneider C., Leung E., Brown J., Tollervey D. (2009) Nucleic Acids Res. 37, 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mian I. S. (1997) Nucleic Acids Res. 25, 3187–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burkard K. T., Butler J. S. (2000) Mol. Cell. Biol. 20, 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Midtgaard S. F., Assenholt J., Jonstrup A. T., Van L. B., Jensen T. H., Brodersen D. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11898–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Briggs M. W., Burkard K. T., Butler J. S. (1998) J. Biol. Chem. 273, 13255–13263 [DOI] [PubMed] [Google Scholar]
- 24. Grzechnik P., Kufel J. (2008) Mol. Cell 32, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Callahan K. P., Butler J. S. (2008) Nucleic Acids Res. 36, 6645–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 27. Peng W. T., Robinson M. D., Mnaimneh S., Krogan N. J., Cagney G., Morris Q., Davierwala A. P., Grigull J., Yang X., Zhang W., Mitsakakis N., Ryan O. W., Datta N., Jojic V., Pal C., Canadien V., Richards D., Beattie B., Wu L. F., Altschuler S. J., Roweis S., Frey B. J., Emili A., Greenblatt J. F., Hughes T. R. (2003) Cell 113, 919–933 [DOI] [PubMed] [Google Scholar]
- 28. Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., Tollervey D. (2003) Mol. Cell. Biol. 23, 6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Synowsky S. A., van Wijk M., Raijmakers R., Heck A. J. (2009) J. Mol. Biol. 385, 1300–1313 [DOI] [PubMed] [Google Scholar]
- 30. Stead J. A., Costello J. L., Livingstone M. J., Mitchell P. (2007) Nucleic Acids Res. 35, 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staub E., Fiziev P., Rosenthal A., Hinzmann B. (2004) BioEssays 26, 567–581 [DOI] [PubMed] [Google Scholar]
- 32. Schilders G., van Dijk E., Pruijn G. J. (2007) Nucleic Acids Res. 35, 2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nehls P., Keck T., Greferath R., Spiess E., Glaser T., Rothbarth K., Stammer H., Werner D. (1998) Nucleic Acids Res. 26, 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 35. Lopez R., Silventoinen V., Robinson S., Kibria A., Gish W. (2003) Nucleic Acids Res. 31, 3795–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clamp M., Cuff J., Searle S. M., Barton G. J. (2004) Bioinformatics 20, 426–427 [DOI] [PubMed] [Google Scholar]
- 37. Carroll K. L., Ghirlando R., Ames J. M., Corden J. L. (2007) RNA 13, 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong I., Lohman T. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5428–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kranz J. E., Holm C. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6629–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitchell P. (2010) Biochem. Soc. Trans. 38, 1088–1092 [DOI] [PubMed] [Google Scholar]
- 42. Yeh L. C., Lee J. C. (1990) J. Mol. Biol. 211, 699–712 [DOI] [PubMed] [Google Scholar]
- 43. Gautier T., Bergès T., Tollervey D., Hurt E. (1997) Mol. Cell. Biol. 17, 7088–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y., Beyer A. L., Hunt D. F., Baserga S. J. (2002) Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wiederkehr T., Prétôt R. F., Minvielle-Sebastia L. (1998) RNA 4, 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jung M. Y., Lorenz L., Richter J. D. (2006) Mol. Cell. Biol. 26, 4277–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kamakaka R. T., Rine J. (1998) Genetics 149, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lafontaine D. L., Tollervey D. (1999) RNA 5, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lafontaine D. L., Tollervey D. (2000) Mol. Cell. Biol. 20, 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reinisch K. M., Wolin S. L. (2007) Curr. Opin. Struct. Biol. 17, 209–214 [DOI] [PubMed] [Google Scholar]
- 51. Doma M. K., Parker R. (2007) Cell 131, 660–668 [DOI] [PubMed] [Google Scholar]
- 52. Houseley J., Tollervey D. (2009) Cell 136, 763–776 [DOI] [PubMed] [Google Scholar]
- 53. Piper P. W., Bellatin J. A., Lockheart A. (1983) EMBO J. 2, 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chapon C., Cech T. R., Zaug A. J. (1997) RNA 3, 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- 55. Egecioglu D. E., Henras A. K., Chanfreau G. F. (2006) RNA 12, 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.