Abstract
Hematopoietic development involves the coordinated activity of differentiation and cell cycle regulators. In current models of mammalian cell cycle control, E2f activators (E2f1, E2f2, and E2f3) are portrayed as the ultimate transcriptional effectors that commit cells to enter and progress through S phase. Using conditional gene knock-out strategies, we show that E2f1–3 are not required for the proliferation of early myeloid progenitors. Rather, these E2fs are critical for cell survival and proliferation at two distinct steps of myeloid development. First, E2f1–3 are required as transcriptional repressors for the survival of CD11b+ myeloid progenitors, and then they are required as activators for the proliferation of CD11b+ macrophages. In bone marrow macrophages, we show that E2f1–3 respond to CSF1-Myc mitogenic signals and serve to activate E2f target genes and promote their proliferation. Together, these findings expose dual functions for E2f1–3 at distinct stages of myeloid development in vivo, first as repressors in cell survival and then as activators in cell proliferation. In summary, this work places E2f1–3 in a specific signaling cascade that is critical for myeloid development in vivo.
Keywords: Apoptosis, Cell Cycle, E2f Transcription Factor, Myc, Myeloid Cell, Apoptosis, CSF-1
Introduction
Molecular events regulating hematopoiesis involve coordinated expression of components that regulate the cell cycle (1). Cyclins and cyclin-dependent kinases (CDKs)3 are crucial for the maintenance of hematopoietic stem cells (HSCs) (2, 3). The combined ablation of all three D-type cyclins or their catalytic partners, Cdk4 and Cdk6, results in a decrease in HSC proliferation (4, 5). Conversely, inhibitors of CDKs restrain the ability of HSCs to proliferate. Indeed, mice deficient for p21Cip1 or p27Kip1 have excessive HSC proliferation, rendering HSC pools sensitive to exhaustion (2, 6). Moreover, mice lacking Bmi-1, a repressor of the CDK inhibitor p16Ink4a, show a significant impairment in HSC self-renewal (7).
One consequence of mitogen-mediated activation of CDK activity is the phosphorylation of the retinoblastoma (Rb) tumor suppressor and its two related pocket proteins, p107 and p130 (8). These phosphorylation events lead to the release of E2fs from Rb-E2f complexes and the accumulation of E2f transcriptional activity late in G1 to maximally activate the expression of a wide range of E2f target genes (9). Ablation of murine Rb in adult HSCs leads to increased expression of E2f targets and enhanced proliferation of myeloid lineages, which is significantly exacerbated by the additional loss of p107 and p130 (10, 11). In this classic paradigm of cell cycle control, the three activator E2fs (E2f1, E2f2, and E2f3) are invariably viewed as the final effectors of a transcriptional program that commits cells to enter S phase (12). As cells progress through S phase, E2f1–3 protein levels decrease, and E2f repressors (E2f4, E2f5, E2f6, E2f7, and E2f8) reload on E2f target promoters and down-regulate their expression (13–15). The coordinated oscillatory activation and repression of E2f target gene expression is believed to promote safe passage of cells through cell cycle transitions and provide ample points of control for monitoring appropriate cell proliferation.
Analysis of individual E2f knock-out mice has revealed important roles for E2f1–3 in hematopoiesis. For example, E2f1−/− mice have a slightly increased number of T cells in the thymus due to impaired apoptosis (16), and E2f2−/− mice have defects during erythroid cell maturation and develop autoimmune diseases late in life due to enhanced proliferation of effector/memory T lymphocytes (17). The combined loss of E2f1 and E2f2 leads to impaired maturation of red blood cells, a condition that resembles megaloblastic anemia in humans, and also leads to decreased B-cell differentiation beyond the pre B-cell stage (18). Hematopoiesis has not been carefully evaluated in E2f3−/− mice, partly due to their increased morbidity (19–21). Given the importance of E2f activators in the control of cell proliferation, it is peculiar that mice lacking each of these E2fs have relatively normal cell cycles. One explanation for the absence of proliferative defects in these single or double E2f knock-out mice is the unprecedented functional redundancy that exists among E2f family members during development (20).
Colony-stimulating factor 1 (CSF-1) is a mitogenic ligand required for myeloid cell survival, proliferation, and differentiation (22). Original work by Sherr and colleagues (23–25) demonstrated that binding of CSF-1 to its cognate receptor (CSF-1R) leads to the activation of the Ras/Raf/MAPK pathway and induction of c-Myc expression. Like E2f, c-Myc is a key transcription factor that stimulates cell growth and cell cycle progression (26). Subsequent studies showed that expression of a mutant form of the CSF-1R lacking the ability to transmit a proliferative signal (CSF-1RY809F) failed to induce c-Myc and cyclin D1 in response to CSF-1 stimulation (27, 28). Importantly, enforced expression of these two components was shown to rescue CSF1-mediated proliferation of CSF-1RY809F-expressing cells (25, 27). More recently, Trumpp and colleagues (29, 30) demonstrated that conditional deletion of c-Myc results in the accumulation of HSCs in the bone marrow and depletion of hematopoietic lineages. A link between c-Myc and E2fs has long been speculated to be critical in how these two transcription factor families regulate cell proliferation. Earlier cell culture studies had suggested an intimate relationship linking c-Myc to the regulation of E2fs (31–34); however, in vivo evidence in support of this remains conspicuously absent.
We exploited the well defined signaling pathways of hematopoietic differentiation to rigorously examine the in vivo role of E2f1–3 in cell proliferation. Loss of Rb results in myeloproliferation and the fact that E2f1–3 are regulated by Rb (10). We therefore wanted to investigate whether ablation of E2f1–3 would similarly affect cells of the myeloid lineage. Using Mx-cre transgenic mice (35) and a conditional allele of E2f3 (E2f1−/−E2f2−/−E2f3f/f), we show that E2f1–3 are dispensable for HSC proliferation but are instead required at two distinct steps of myeloid development. We show that E2f1–3 are required for the survival of myeloid progenitors and, subsequently, for the proliferation of committed bone marrow macrophages (BMMs). Interestingly, the survival role of E2fs in myeloid progenitors is associated with a function for these E2fs in transcriptional repression, and the proliferation role in committed BMMs is associated with their function in transcriptional activation. These studies expose and contextualize the dual roles for E2f1–3 in transcription repression and activation in vivo by casting these factors during discrete stages of myeloid differentiation in a defined physiological signaling cascade involving CSF-1 and Myc.
EXPERIMENTAL PROCEDURES
Mice
E2f1−/−, E2f2−/−, and c-Mycf/f mice were a gift from Michael Greenberg, Stuart Orkins, and Andreas Trumpp, respectively. Generation of E2f3f/f mice has been described previously (21). 8–10-week-old mice received five injections of 250 μg of polyinosine-polycytidine (pIpC) every alternate day and mice were sacrificed 24 h after the last injection. All of the experimental as well as control mice were injected with pIpC. Adaptive transfer experiments were performed using cells isolated from bone marrow (BM) of E2f1−/−E2f2−/−E2f3f/f and Mx-cre;E2f1−/−E2f2−/−E2f3f/f mice and injected into lethally irradiated wild type mice via tail vein. Mice were maintained on antibiotic water for 5 weeks after the transplant, followed by five injections of pIpC as described above. Primer sequences for genotyping are listed in supplemental Fig. S5.
Flow Cytometry
Bone marrow cells isolated from the femurs of mice were analyzed by FACS. Briefly, single cell suspensions were prepared from the bone marrow and were counted and stained with a panel of fluorochrome-conjugated antibodies to determine the number of cells in different hematopoietic lineages. For granulocytic macrophage progenitor analysis, the cells were stained as Lin−IL-7Rα−Sca-1−c-Kit+CD34+FcγRII/IIIhigh. After staining, the cells were analyzed using FACSCalibur using Cell Quest Pro software (BD Biosciences). Mice were injected with BrdU 1 h before harvesting of BM and stained with FITC-BrdU antibody using the Cytofix/Cytoperm kit (BD-552598). For analysis of apoptosis, BM cells were stained with FITC-annexin V antibody (BD-556547). BM cells were stained with FITC-conjugated CD11b antibody for sorting CD11b+ myeloid cells from the BM using the FACS Aria system.
Microarray Analysis
BM samples were isolated as described previously. Four independent samples from each genetic group were used for gene expression analysis by Affymetrix microarray. RNA was isolated using TRIzol reagent. RNA was then subjected to purification and processed for hybridization to Affymetrix Mouse Genome 430 2.0 arrays. Genes that increased or decreased at least 1.5-fold (p < 0.001) in Mx-cre samples relative to control samples were used to generate the heat maps illustrated in Fig. 2A. Expression values were normalized and log-transformed using RMAExpress. RMA-normalized data were analyzed using BRB-ArrayTools 3.7.0. The microarray data were deposited with the Gene Expression Omnibus database with accession number GSE25825.
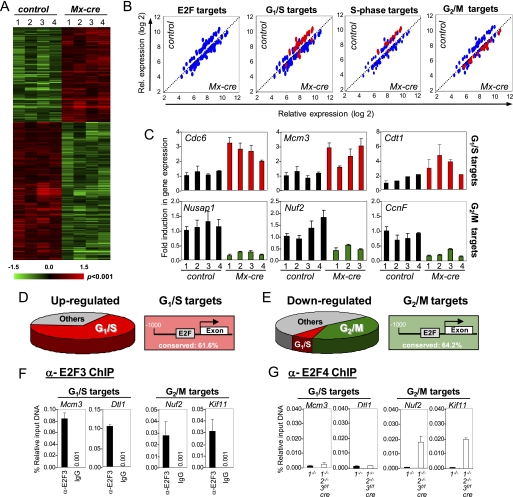
FIGURE 2.
Ablation of E2f1–3 in BM leads to up-regulation of G1/S and down-regulation of G2/M genes in Cd11b+ myeloid cells. A, heat map showing hierarchical clustering analysis of genes differentially expressed (p < 0.001) between Mx-cre;E2f1−/−E2f2−/−E2f3f/f (Mx-cre) and E2f1−/− (control). Four independent samples were analyzed from each genetic group. B, scatter plots comparing E2f target genes between Mx-cre and E2f1−/− (control). Blue dots indicate the total number of genes that were changed. Red dots indicate G1/S and S phase target genes that are up-regulated and/or G2/M target genes that are down-regulated more than 1.5-fold. C, quantitative real-time PCR was performed to compare the relative expression of selected G1/S (top) and G2/M (bottom) E2f target genes in control and Mx-cre. The number on the y axis represents four independent samples from each genetic group. D, pie diagram illustrates that the majority of up-regulated genes in Mx-cre;E2f1−/−E2f2−/−E2f3f/f (Mx-cre) myeloid cells are E2F targets involved in G1/S regulation (left panel). The figure shows the percentage of E2f sites conserved between the human and mouse promoter (right). E, pie diagram illustrates that the majority of down-regulated genes in Mx-cre;E2f1−/−E2f2−/−E2f3f/f (Mx-cre) myeloid cells are E2f targets involved in G2/M regulation (right). F, ChIP assay showing E2f3 recruitment on promoters of G1/S and G2/M genes in wild type myeloid cells. G, ChIP assay in E2f1−/− (closed bars) and Mx-cre;E2f1−/−E2f2−/−E2f3f/f (open bars) myeloid cells showing E2f4 loading on promoters of G1/S and G2/M genes. Error bars, S.D.
Real-time RT-PCR
RNA was isolated using TRIzol reagent from CD11b+ cells and mouse embryo fibroblasts (MEFs) harvested at the indicated time points. To determine the expression of E2fs in different hematopoietic lineages within the bone marrow, cells from bone marrow were isolated and then stained with lineage-specific antibodies (Cd11b, Ter119 and B220) and sorted using FACS. Real time PCR was then performed on RNA isolated from the sorted cells. Reverse transcription of total RNA was performed using Superscript III reverse transcriptase (Invitrogen) and RNase inhibitor (Roche Applied Science) according to the manufacturer's protocol. Real-time PCR was performed using a Bio-Rad iCycler, reactions were performed in triplicate, and relative amounts of cDNA were normalized to RPL4.
ChIP Assays
For ChIP assays, briefly, harvested CD11b+ cells, BMMs, and MEFs were cross-linked, and chromatin was sonicated to an average size of 200–1000 bp. Lysates were subsequently precleared with salmon sperm DNA/Protein G-agarose slurry. Antibodies specific to α-c-Myc (2 μg; SC-262), α-E2f4 (2 μg; SC-1082), or α-E2f3 (2 μg; SC-878) were then added to each sample and incubated overnight at 4 °C. Antibody-protein-DNA complexes were recovered by the addition of 60 μl of salmon sperm DNA/Protein G-agarose slurry and incubation for 1 h at 4 °C. Following extensive washing, the complexes were eluted and decross-linked at 65 °C for 4 h. Finally, samples were treated with Proteinase K (Roche Applied Science) and RNase A (Roche Applied Science) and purified through Qiaquick columns (Qiagen). Real-time PCR quantification of immunoprecipitated DNA was performed using the Bio-Rad iCycler machine with primers specific for the indicated promoter regions. Primer sequences are listed in supplemental Fig. S5.
Isolation and Culture of BMM
Bone marrow cells isolated were grown in RPMI medium containing 50 ng/ml recombinant human CSF-1 (RDI-3025, Fitzgerald) and penicillin/streptomycin antibiotics (Invitrogen) on bacteriological plastic plates for 5 days. For BrdU incorporation and RT-PCR assays, BMMs were synchronized by incubation in RPMI with 0.2% FBS for 20 h. Cells were stimulated by the addition of RPMI supplemented with 50 ng/ml recombinant human CSF-1 and harvested at the indicated time points. BMMs isolated from E2f1+/+2+/+3+/+ and E2f1−/−2−/−3f/f were infected with vector- or cre-expressing retroviruses.
Cell Culture and Retroviral Infection
NIH-3T3 cells expressing the wild type (T56) and mutant (809) human CSF-1R receptor were used in the study (27). Primary MEFs were made from embryonic day 13.5 embryos using standard methods. The c-Mycf/f cell lines were established using the NIH-3T9 protocol (21). All cells were grown in DMEM with 15% fetal bovine serum. For the production of retrovirus, the full-length cDNAs for CSF-1R, cre recombinase and a Myc-tagged E2f3a were subcloned into their respective pBabe-retroviral vectors. High titer retroviruses were produced by transient transfection of retroviral constructs into the Phoenix-Eco packaging cell line as described previously (36). MEFs were infected by incubating the cells with the supernatants containing 4 μg/ml Sequabrene (Sigma) from the transfected cells. Subsequent to infection, cells were grown in selection medium containing either 2.5 μg/ml puromycin (Sigma) or 400 μg/ml hygromycin (Roche Applied Science) or both for 3–5 days. For BrdU incorporation and RT-PCR experiments, subconfluent MEFs were synchronized by incubation in DMEM with 0.2% FBS or serum-free medium for 72 h. Cells were then stimulated to proliferate by the addition of DMEM supplemented with 15% FBS or 100 ng/ml recombinant human CSF-1 and harvested at the indicated time points.
Proliferation and BrdU Assays
Colony formation assays were performed by plating 500 cells/100-mm dish. The colonies were fixed with 70% ethanol and stained with 5 mg/ml crystal violet in 20% methanol. Colonies from three separate plates at the appropriate density were counted, and the mean and S.D. from one representative experiment is reported unless otherwise stated. For BrdU incorporation assays, serum- or CSF-1-stimulated cells were incubated with 50 μm BrdU for the indicated time and subsequently fixed with methanol and acetic acid (1:1). Cells were stained with anti-BrdU antibody (Ab-3, Oncogene) as described previously (37) and counterstained with 4′,6-diamidino-2-phenylinole (DAPI). A total of 500 DAPI-positive nuclei were scored for each time point.
Western Blot
Immunoblot analyses were performed by standard procedures using ECL reagents as described by the manufacturer (Amersham Biosciences). The following commercial antibodies were used: α-E2f1 (SC-193, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), α-E2f2 (SC-633, Santa Cruz Biotechnology, Inc.), α-E2f3 (SC-878, Santa Cruz Biotechnology, Inc.), and α-tubulin (T-9026, Sigma).
Promoter Luciferase Assays
NIH-3T3 cells expressing the wild type CSF-1R, mutant CSR-IR (809) receptors, and c-Mycf/f cells were transfected with the E2f3a and E2f3b luciferase expression vectors, together with Renilla as an internal control, as described previously (32). After transfection, cells were brought to quiescence by serum starvation. Cells were stimulated by the addition of 15% serum or 100 ng/ml CSF-1. Cells were harvested at various times after stimulation, and luciferase activity was measured using a luminometer.
RESULTS
E2f1–3 Are Essential for Myeloid Cell Survival and Development
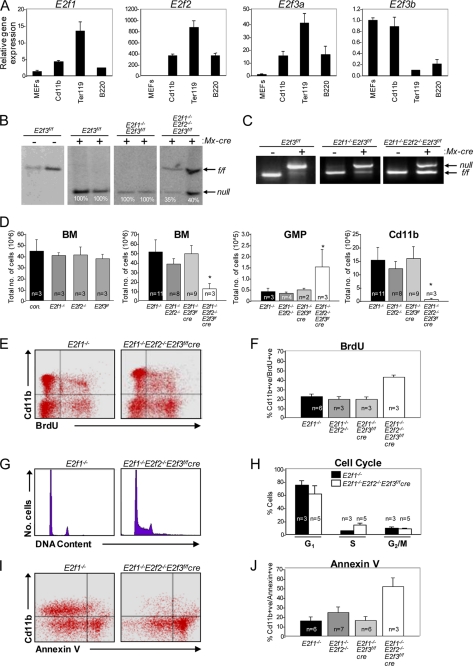
To explore the potential role for the E2f1–3 transcription factors in hematopoietic development, we first examined their expression in the bone marrow of wild type mice. Quantitative RT-PCR assays shown in Fig. 1A illustrate the relative expression of E2f1, E2f2, E2f3a, and E2f3b in mouse embryo fibroblasts (MEFs) and lymphoid, erythroid, and myeloid lineages. This analysis showed that E2f1, E2f2, and E2f3a are highly expressed in all three hematopoietic lineages and that E2f3b expression is significantly restricted to the myeloid lineage. We used null alleles of E2f1 (E2f1−/−) and E2f2 (E2f2−/−) and a conditional allele of E2f3 (E2f3f/f) together with the established Mx-cre transgene to examine the individual and combined roles of the three activator E2fs in vivo. Southern blot and PCR assays showed the efficient Mx-cre-mediated deletion of E2f3f/f in BM cells isolated from pIpC-injected Mx-cre;E2f3f/f and Mx-cre;E2f1−/−E2f3f/f mice (Fig. 1, B and C). The ablation of any single or double combination of activator E2fs had little impact on the total cellularity of BM or individual hematopoietic cell lineages (Fig. 1D and supplemental Fig. S1A). Because of the high degree of functional redundancy among E2fs during embryonic development (20), we examined the consequences of ablating the entire set of activator E2fs (Mx-cre;E2f1−/−E2f2−/−E2f3f/f). Ablation of E2f1–3 resulted in a 4-fold reduction of BM cells and a corresponding decrease in the number of CD11b+ myeloid cells (Fig. 1D and supplemental Fig. S1B). Consistent with a specific defect in the myeloid differentiation cascade, we observed an accumulation of granulocytic macrophage progenitors (Fig. 1D, GMP). Whereas PCR and Southern blot analysis had shown a nearly complete deletion of E2f3f/f in Mx-cre;E2f3f/f and Mx-cre;E2f1−/−E2f3f/f mice, cells isolated from Mx-cre;E2f1−/−E2f2−/−E2f3f/f were only partially deleted (∼40%) (Fig. 1B), indicating a selection against the expansion of cells lacking these three E2fs.
FIGURE 1.
E2f1–3 are essential for myeloid development. Analysis of BM cells isolated from 8–10-week-old mice 2 weeks after pIpC treatment. A, real-time PCR analysis of expression of E2f1, E2f2, E2f3a, and E2f3b in FACS-sorted cells of different hematopoietic lineages. Bone marrow cells were isolated and stained with lineage-specific antibodies (Cd11b, Ter119, and B220) and sorted by FACS. Real-time PCR was then performed on RNA isolated from these sorted cells. B, Southern blot on genomic DNA isolated from BM of Mx-cre E2f3f/f, E2f1−/−E2f3f/f, and E2f1−/−E2f2−/−E2f3f/f mice. A minimum of three samples were analyzed from each genetic group. Two representative samples analyzed from each genetic group are shown. C, E2f3 PCR genotyping on genomic DNA isolated from BM of Mx-cre E2f3f/f, E2f1−/−E2f3f/f, and E2f1−/−E2f2−/−E2f3f/f mice. D, analysis of cells isolated from femur of mice of the indicated genotype showing bone marrow cellularity (p < 0.0003), number of granulocyte-macrophage progenitors (GMP) (p < 0.018), and number of CD11b+ myeloid cells (p < 0.0002). Values are mean ± S.D. (error bars). E, FACS profile of CD11b+ myeloid cells stained with α-BrdU antibody to determine the number of cells in S phase. F, quantification of CD11b+ BrdU-positive cells; values are mean ± S.D. (p < 0.0008). G and H, analysis of DNA content in CD11b+ cells by propidium iodide staining. Values are mean ± S.D. (p < 0.003). I, representative FACS profile of myeloid cells stained with apoptotic marker annexin V. J, quantification of CD11b+ apoptotic cells. Values are mean ± S.D. (p < 0.005).
Given the strict requirement for E2f1–3 in the proliferation of MEFs (21), we analyzed the proliferation of Mx-cre;E2f1−/−E2f2−/−E2f3f/f myeloid cells. Surprisingly, we observed a higher percentage of CD11b+ Mx-cre;E2f1−/−E2f2−/−E2f3f/f cells in S phase when compared with all of the other groups of mice, as determined by BrdU incorporation and flow cytometry (Fig. 1, E–H). In contrast, Annexin V staining revealed massive apoptosis of CD11b+ cells in Mx-cre;E2f1−/−E2f2−/−E2f3f/f mice (Fig. 1, I and J). Transplantation of BM from control and Mx-cre;E2f1−/−E2f2−/−E2f3f/f mice into lethally irradiated wild type mice, followed by the standard pIpC injection regimen, resulted in a similar increase of apoptosis and reduction of myeloid lineages in triply deleted mice (supplemental Fig. S2). Collectively, these results suggest that E2f1–3 are dispensable for mammalian cell proliferation in vivo and are instead required for the survival of myeloid cells through a cell-autonomous mechanism.
E2f1–3 Repress E2f Target Genes in Vivo
To explore the underlying cause for the observed apoptosis in E2f1–3-deficient myeloid cells, we compared global gene expression profiles in control (Mx-cre;E2f1−/−) and Mx-cre;E2f1−/−E2f2−/−E2f3f/f CD11b+-sorted cells. We used an unbiased method similar to gene set enrichment analysis (38) to identify genes that were differentially expressed between the two groups. From the ∼45,000 probes in Affymetrix oligoarrays, 248 genes were up-regulated and 323 were down-regulated >1.5-fold (p < 0.001) in the Mx-cre;E2f1−/−E2f2−/−E2f3f/f samples (Fig. 2A). Gene ontology pathways revealed that most of the differentially expressed genes identified by this method are involved in DNA replication, mitosis, and nucleotide metabolism. Approximately 45 of the 248 up-regulated genes have been reported to be E2f-responsive, and interestingly, 66% of these (30 of 45) have been implicated in promoting the G1/S transition (Fig. 2B and supplemental Table 1), consistent with increased DNA replication in E2f1–3 deficient myeloid cells. Of the 323 down-regulated genes, expression of 69 genes has been shown to be E2f-responsive, and almost half of this group (29 of 69) have been implicated to be involved in G2/M regulation (Fig. 2B and supplemental Tables 1 and 2). Quantitative RT-PCR confirmed the changes in the expression of G1/S- and G2/M-related target genes (Fig. 2C). The majority of promoters in these two classes of G1/S- and G2/M-related gene targets contained E2f consensus binding sites that were conserved between mice and humans (Fig. 2, D and E). Chromatin immunoprecipitation (ChIP) assays on sorted wild type CD11b+ cells showed that E2f3 was specifically loaded onto the proximal promoters of both the G1/S and G2/M gene targets (Fig. 2F and supplemental Fig. S3, A and B). Parallel ChIP assays performed on sorted CD11b+ myeloid cells derived from E2f3a−/− and E2f3b−/− mice suggested that both isoforms participate in the loading of E2f3 on promoters (supplemental Fig. S3, C and D).
We then investigated why loss of E2f1–3 led to an up-regulation of G1/S genes but a down-regulation of G2/M genes. The up-regulation of G1/S genes is consistent with the recent demonstration that E2f1–3 function as repressors in the differentiating retina and small intestine of mice (39, 40). We hypothesized that the down-regulation of G2/M targets in triply deleted myeloid cells may be due to the overcompensation by other “professional” E2f repressors. Expression of the known E2f repressors (E2f4, E2f5, E2f6, E2f7, and E2f8) was not significantly changed in E2f1–3-deficient CD11b+ myeloid cells (supplemental Fig. S3E). Although E2f4 expression was not changed in E2f1–3-deleted BM cells, it remained possible that there might have been a redistribution of E2f4 protein to E2f1–3 target promoters. Indeed, previous studies in E2f1–3-deleted MEFs have shown that E2f4 is inappropriately recruited to the promoters of E2f1–3 target genes and contributes to cell cycle exit in these cells (41). We thus tested this hypothesis by performing E2f4 ChIP on both G1/S and G2/M targets, using E2f4-specific antibodies on sorted CD11b+ cells derived from control and Mx-cre;E2f1−/−E2f2−/−E2f3f/f mice. These assays showed a significant increase in the specific loading of E2f4 onto G2/M target promoters of E2f1–3-deficient CD11b+ cells (Fig. 2G). It would appear that a deficiency of E2f1–3 in myeloid cells can be overcome by the compensatory loading of E2f4 to selected target promoters (G2/M targets), leading to their silencing. How E2f4 is recruited to G2/M-regulated promoters but not to G1/S promoters is not yet clear, but it may involve specific interactions with other factors that co-regulate these G2/M targets and lead to their permanent or more profound repression. We suggest that the up-regulation of G1/S genes and down-regulation of G2/M genes produce conflicting signals that force E2f1–3-deficient cells to replicate DNA and inhibit mitosis, leading to the accumulation of cells in S phase and the initiation of a massive apoptotic response. From these data, we conclude that contrary to their established function in cell culture systems, E2f1–3 function in vivo to repress E2f target gene expression.
E2f1–3 Are Required for the CSF-1-induced Proliferation of BMMs
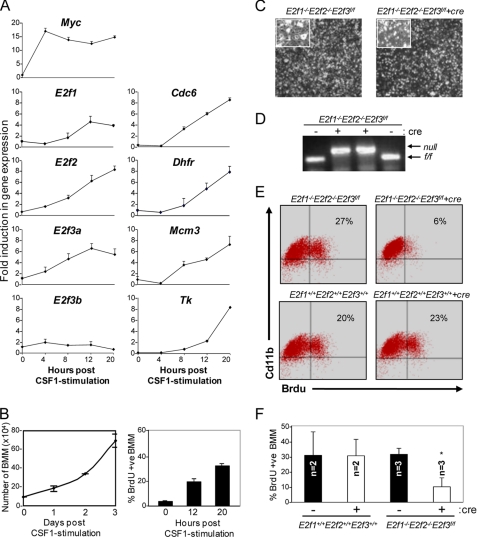
Because Mx-cre-mediated inactivation of E2f1–3 caused an almost complete depletion of the CD11b+ lineage, subsequent differentiation steps potentially regulated by these E2fs could not be investigated further. We thus exploited the ability to differentiate BM cells in culture by the addition of the CSF-1 ligand in order to explore the functions of E2f1–3 in terminally differentiated macrophages (BMMs) (42). To this end, BM cells isolated from the femur of wild type mice were cultured in medium containing CSF-1, allowing the expansion of Cd11b+ cells into differentiated BMMs (43). We then examined the ability of growth factor-starved BMMs to proliferate in response to restimulation with CSF-1. As expected, quantitative RT-PCR assays showed an immediate induction of c-Myc expression that was followed by a marked increase in E2f1, E2f2, and E2f3a expression, a corresponding induction of classic G1/S-regulated E2f target genes (Fig. 3A), and the entry of BMMs into S phase (Fig. 3B). The levels of E2f3b mRNA did not change in response to CSF-1, consistent with its known cell cycle-independent expression pattern (37). The introduction of a cre-expressing retrovirus into E2f1−/−E2f2−/−E2f3f/f BMMs resulted in the efficient deletion of the E2f3f/f allele (Fig. 3D) and a 3-fold reduction in BrdU incorporation (Fig. 3, E and F). Moreover, bright images of E2f1−/−E2f2−/−E2f3f/f-deleted BMMs appeared normal, without any effect on cell survival (Fig. 3C) (data not shown); cre expression had no effect on BMMs containing a wild type E2f3 allele. These results suggest that proliferation of committed BMMs requires E2f1–3 for their ability to activate gene expression and promote their expansion in response to CSF-1-mediated signals.
FIGURE 3.
E2f1–3 are required for BMM proliferation. A, RT-PCR analysis of E2fs and E2f target genes on BMMs at the indicated time points. The values on the y axis represent -fold induction. B, BMMs were plated at a density of 1 × 105 cells/60-mm plate. Cells were grown in a medium containing 50 ng/ml CSF-1 and were harvested and counted every 24 h for 4 days (left). Cells were harvested for BrdU incorporation. Cells were stained with anti-BrdU antibody and counterstained with DAPI, and BrdU-positive cells were counted as described under “Experimental Procedures” (right). C, bright field images of vector- or cre-treated E2f1−/−2−/−3f/f BMMs at 10× magnification. D, E2f3 PCR genotyping on genomic DNA isolated from cre-treated E2f1−/−E2f2−/−E2f3f/f BMMs. E, FACS analysis of BrdU+ Cd11b+ cells of the indicated genotypes. F, BMMs from E2f1+/+2+/+3+/+ and E2f1−/−2−/−3f/f mice were infected with either vector- or cre-expressing retroviruses. BMMs were co-stained with Cd11b and BrdU and analyzed by FACS. The graph depicts the percentage of Cd11b+ cells that are also positive for BrdU. Values are mean ± S.D. (error bars) (p < 0.006).
E2f3a Is a Downstream Target of c-Myc Required for Cell Proliferation
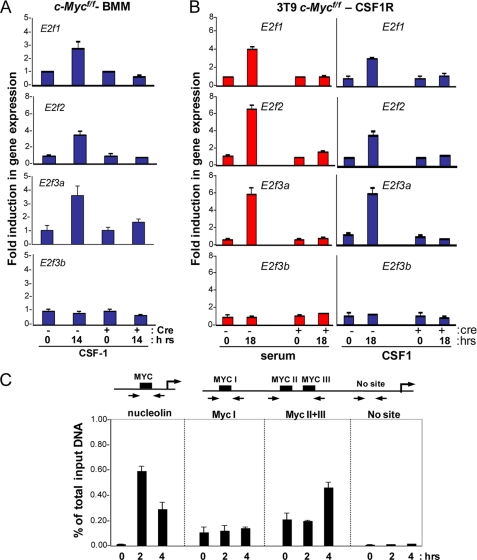
c-Myc is a critical component of the CSF-1 signaling cascade that is necessary for cell proliferation (27). More recent studies have suggested an intimate relationship between c-Myc and the regulation of E2fs (32); however, mechanistic details linking the two transcriptional networks remain unknown. To determine whether the CSF-1 induction of E2f1, E2f2, and E2f3a is mediated by c-Myc, we generated and analyzed c-Myc floxed (Mycf/f) BMMs. We infected Mycf/f BMMs with control or cre-retroviruses, synchronized in G0 and then restimulated with CSF-1. PCR genotyping confirmed the efficient cre-mediated deletion of the conditional c-Mycf/f alleles (supplemental Fig. S4A). Quantitative RT-PCR analysis showed the typical CSF-1 mediated induction of E2f1, E2f2, and E2f3a expression, which was abrogated in c-Myc-deleted BMMs (Fig. 4A). To further corroborate our findings in BMM, we performed similar experiments in c-Myc floxed MEFs overexpressing human CSF-1R (c-Mycf/f-CSF-1R), which are responsive to recombinant human-derived CSF-1 but not endogenous murine CSF-1. This cell system can recapitulate molecular events that follow activation of the CSF-1R and can be exploited to elucidate molecular details of these events. In contrast to control treatment, ablation of c-Myc impaired the ability of cells to enter S phase in response to either serum or CSF-1 (supplemental Fig. S4B). Similarly, no induction of E2f1, E2f2, and E2f3a expression could be observed in c-Myc-deleted cells following CSF-1 stimulation (Fig. 4B).
FIGURE 4.
c-Myc is required for the activation of E2f1, E2f2, and E2f3a. c-Mycf/f BMM or c-Mycf/f-CSF1R cells were infected with either the control or cre-expressing retroviruses and serum-starved. Quiescent cells were then restimulated with medium containing serum or CSF-1 and assessed by RT-PCR analysis for target gene expression at the indicated time points. Total RNA was used to measure the expression of the indicated E2f target genes from c-Mycf/f BMMs (A) and c-Mycf/f-CSF1R cells (B). The y axis represents the average -fold induction in gene expression, where the level at the 0 h time point is equal to 1. C, ChIP assays on lysates from wild type BMMs using antibodies for c-Myc. Immunoprecipitated DNA was measured by real-time PCR using primers flanking the Myc sites on the E2f3a promoter. Results are shown as the percentage of total input at the indicated time upon stimulation of quiescent BMMs with CSF-1. RT-PCR was performed in triplicate, and cycle numbers were normalized to 1% of the input DNA.
Previous studies have shown that the E2f3 locus plays a particularly important role in the control of cell proliferation, and we thus focused our subsequent analysis on the regulation and function of E2f3a (21). To determine whether E2f3a is regulated by c-Myc at the transcriptional level, we analyzed the activity of E2f3a promoter reporter constructs in c-Mycf/f-CSF-1R cells infected with control or cre-expressing retroviruses. These assays revealed that the serum- or CSF-1-mediated increase in E2f3a promoter activity was abolished by the cre-mediated deletion of c-Myc (supplemental Fig. S4C). We then addressed whether enforced expression of E2f3a could restore the proliferation of c-Myc-deleted MEFs. Interestingly, overexpression of E2f3a restored the ability of c-Myc-deleted cells to enter S phase in response to CSF-1 but failed to support mitosis and the continued proliferation of these cells as determined by colony formation assays (supplemental Fig. S5, A–D).
ChIP assays were then used to determine whether c-Myc could directly regulate the expression of E2f3a in BMMs. The E2f3a promoter contains three E-box elements (Myc sites I–III) that can serve as potential binding sites for c-Myc (32). ChIP assays were performed using Myc-specific antibodies, and immunoprecipitated DNA was amplified with primers flanking the E-box sites on the E2f3a promoter (Fig. 4C). As a positive control, we also assayed the nucleolin promoter, an established direct target of c-Myc (44). The results from these ChIP assays showed that c-Myc in BMMs was specifically recruited to Myc I–III sites on the E2f3a promoter but not to flanking irrelevant sequences (No site) in response to CSF-1 (Fig. 4C). Together, these results substantiate our findings that c-Myc is required for the expression of E2f1–3 and is directly involved in CSF-1-mediated activation of E2f3a.
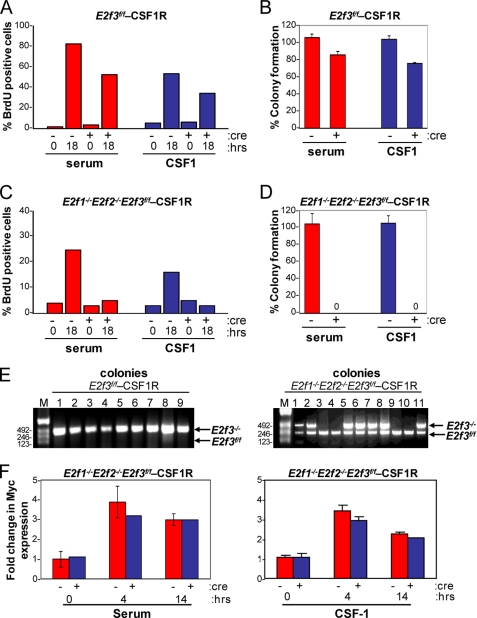
E2f1–3 Are Indispensable for Cell Growth
To evaluate the role of E2f1–3 in CSF-1-mediated cellular proliferation, we used a strategy similar to that described above to generate E2f3f/f and E2f1−/−E2f2−/−E2f3f/f cells overexpressing human CSF-1R (E2f3f/f-CSF-1R and E2f1−/−E2f2−/−E2f3f/f-CSF-1R, respectively). These cells were infected with control or cre-retroviruses as before, synchronized by serum deprivation, and then stimulated by the addition of either serum or CSF-1 to the media. Although the cre-mediated inactivation of E2f3 only marginally reduced the ability of cells to enter S phase, the combined loss of E2f1–3 severely compromised S phase entry in response to either serum or CSF-1 (Fig. 5, A and C). Consistent with a defect in S phase entry, E2f1–3 triply deleted cells failed to form colonies (Fig. 5, B and D). The few colonies that arose in cre-expressing E2f1−/−E2f2−/−E2f3f/f-CSF-1R cells retained at least one intact copy of the E2f3f/f allele (Fig. 5E). Importantly, the normal serum- or CSF-1-mediated induction of c-Myc expression, which occurred ∼4 h post-stimulation, was not affected by the loss of E2f1–3 (Fig. 5F). From these results, we suggest that E2f1–3 are essential components of the CSF-1 mitogenic signaling pathway that function downstream of c-Myc.
FIGURE 5.
E2f1–3 are important for cellular proliferation. The E2f3f/f-CSF-1R and E2f1−/−2−/−3f/f-CSF-1R cell lines were infected with either control- or cre- expressing retroviruses and then used for the following assays. A, BrdU incorporation in E2f3f/f-CSF-1R cell line. Quiescent MEFs with the indicated genotypes were restimulated with medium containing serum or CSF-1 and assessed for BrdU incorporation at the indicated time points. A total of 500 DAPI-stained nuclei from each cell line were counted, and the percentage of BrdU-positive cells is shown. B, E2f3f/f-CSF-1R MEFs were plated for a colony formation assay. Values shown have been corrected for deletion of E2f3 by colony PCR. C, BrdU incorporation of E2f1−/−2−/−3f/f-CSF-1R cell line. The graph shows the percentage of cells positive for BrdU incorporation. D, E2f1−/−2−/−3f/f-CSF-1R MEFs were plated for a colony formation assay. Values shown have been corrected for deletion of E2f3 by colony PCR. E, E2f3 PCR genotyping on genomic DNA from the cre-infected E2f3f/f-CSF1R and E2f1−/−2−/−3f/f-CSF-1R colonies grown in the presence of CSF-1. F, bar graphs showing -fold change in c-Myc expression in control and cre-treated E2f1−/−2−/−3f/f-CSF-1R after stimulation with serum (left) or CSF-1 (right).
E2f1–3 Are Direct Transcriptional Targets of c-Myc
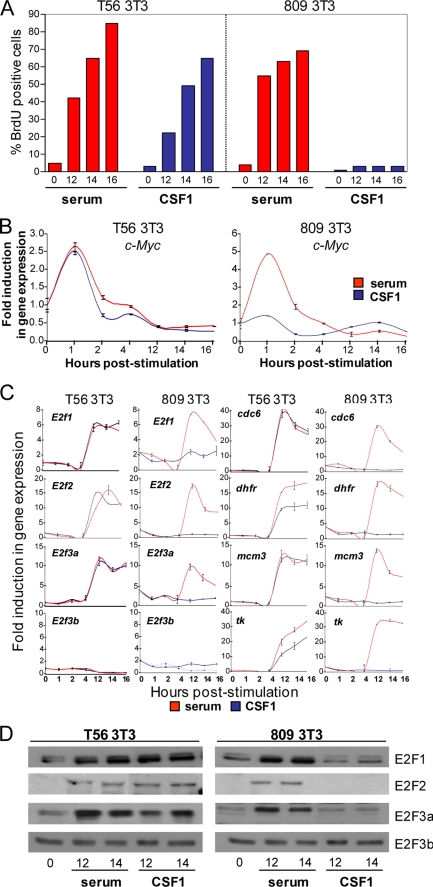
To further explore how CSF1-mediated signaling leads to the activation of E2f3a, we utilized mouse NIH-3T3 cell lines expressing either wild type (T56) or mutant (Y809F) forms of the human CSF-1R receptor that lack the ability to activate the expression of c-Myc and transmit CSF-1-mediated proliferative signals (28). As previously shown, both T56 and mutant Y809F cells entered S phase in response to serum addition, but only T56 3T3 cells responded to CSF-1 (Fig. 6A). Quantitative RT-PCR and Western blot analysis showed that the CSF-1-induced entry of T56 cells into the cell cycle corresponded with the early induction of c-Myc and the subsequent increase of E2f1, E2f2, and E2f3a expression as well as with the increase of E2f target gene expression (Fig. 6C). In contrast, mutant Y809F cells failed to activate the expression of c-Myc, E2f1, E2f2, and E2f3a and classic E2f target genes in response to CSF-1 (Fig. 6, B–D). The expression of E2f3b and other E2f family members was unaffected by CSF-1 stimulation in either T56 or Y809F cells (Fig. 6, C and D) (data not shown).
FIGURE 6.
CSF-1-specific mitogenic signal leads to the activation of E2f1, E2f2, and E2f3a. A, BrdU incorporation (right) in serum-stimulated (red) and CSF-1-stimulated (blue) T56 (left) and 809 (right) NIH-3T3 cells. Quiescent MEFs with the indicated genotypes were stimulated with medium containing serum or CSF-1 and assessed for BrdU incorporation at the indicated time points as described under “Experimental Procedures.” B and C, RT-PCR analysis of c-Myc, E2fs, and E2f target gene expression. Total RNA was harvested from MEFs treated as in A and was used to measure expression of E2f target genes as described under “Experimental Procedures.” The y axis represents the average -fold induction in gene expression, where the level at the 0 h time point is equal to 1. D, cellular lysates from serum- or CSF-1-stimulated cells were used for a Western blot probed with the E2f1, E2f2, and E2f3 antibodies.
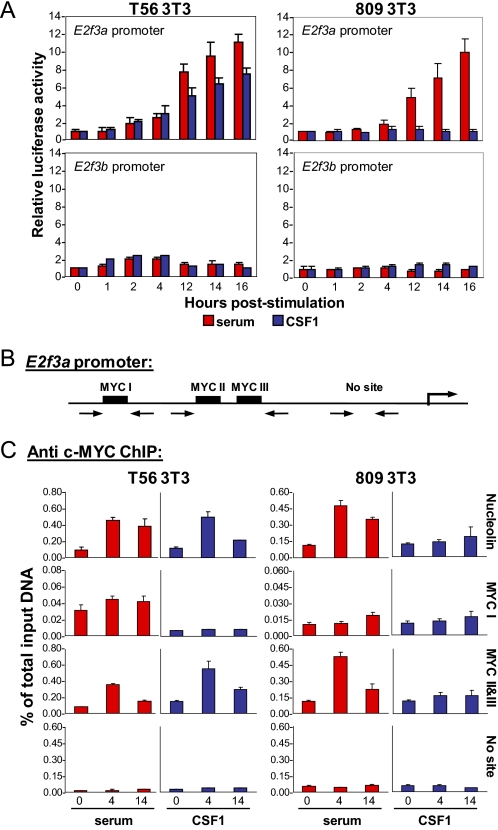
The above results raised the possibility that the induction of E2f1, E2f2, and E2f3a by CSF-1 may be directly dependent on c-Myc function. To test this possibility, we analyzed a series of E2f3a-promoter reporter constructs in both T56 and Y809F cells. CSF-1 stimulation activated the E2f3a reporter in T56 cells but not in Y809F mutant cells (Fig. 7A). The induction of the E2f3a reporter in T56 cells was specific because neither serum nor CSF-1 had any effect on E2f3b reporter activity. These results suggest that the CSF-1 mediated induction of E2f3a mRNA occurs at the transcriptional level. To test for c-Myc binding, we performed ChIP assays in T56 and Y809F cells using c-Myc-specific antibodies. The results from these ChIP assays showed that c-Myc was loaded on Myc I-III sites on the E2f3a promoter in response to both serum and CSF-1 in T56 cells (Fig. 7, B and C). In contrast, no recruitment of c-Myc was observed in Y809F cells following CSF-1 stimulation, consistent with the absence of c-Myc induction in these cells. These results recapitulate our earlier findings in BMMs suggesting that the CSF-1-mediated activation of E2f3a involves the binding of c-Myc to the E2f3a promoter.
FIGURE 7.
The E2f3a promoter, but not the E2f3b promoter, is activated by c-Myc. A, NIH-3T3 cells were transfected with either the E2f3a- or E2f3b-luciferase plasmid, along with a thymidine kinase Renilla luciferase construct as an internal control. Transfected cells were incubated in low serum and then stimulated with either serum (red) or CSF-1 (blue). Cells were harvested at the indicated time points, and luciferase activity was measured. B, schematic representation of the E2f3a promoter showing Myc binding sites. C, ChIP assays on cell lysates from synchronized T56 and 809 MEFs using antibodies for c-Myc. Results are shown as the percentage of total input at the indicated time upon stimulation of quiescent MEFs with serum or CSF-1. RT-PCR was performed in triplicate, and cycle numbers were normalized to 1% of the input DNA.
DISCUSSION
In this study, we evaluated the role of E2fs in hematopoietic development in vivo by analyzing the consequences of conditionally ablating the entire subclass of activator E2fs in HSCs. We show that E2f1–3 regulate cell survival and proliferation at distinct stages during myeloid development through their dual ability to repress and activate transcription, respectively. During early stages of myeloid development, E2f1–3 are dispensable for the proliferation of CD11b+ cells but are instead required for their survival. Once myeloid precursor cells commit to the macrophage lineage, E2f1–3 provide cells with the ability to respond to CSF-1 signals and allow their expansion prior to exit into the periphery. In summary, these findings contextualize the role of E2f1–3 in cell survival and proliferation to specific developmental stages during the life span of a myeloid cell.
The coordinated expression and activity of cell cycle regulators are crucial for proper hematopoietic development (1, 3, 45). HSCs lacking c-Myc accumulate due to their inability to properly differentiate and give rise to hematopoietic lineages (30). Mice lacking D-type cyclins or all three interphase CDKs (Cdk2, Cdk4, and Cdk6) die by middle to late gestation due to defective hematopoiesis attributed to decreased expansion of HSCs (4, 5). In contrast, conditional inactivation of Rb in the hematopoietic system leads to myeloproliferation, but interestingly, Rb-deficient HSCs and progenitor cells did not display any detectable change in the expression of cell cycle-related genes (10). Here, we show that E2f1–3 are not required for HSC proliferation and differentiation in vivo but rather are required for the survival of Cd11b+ myeloid cells. The antagonistic phenotypes observed in myeloid cells deficient for either Rb/p107/p130 or E2f1–3 suggest that myeloid development is tightly regulated by the Rb-E2f pathway.
In the textbook model of mammalian cell cycle regulation, the E2f1–3 proteins are viewed as the ultimate effectors downstream of the CDK/Rb pathway that activate a gene expression program that is critical for S phase entry and cell cycle progression (46). Consistent with this view, loss of E2f1–3 in mouse embryo fibroblasts results in a decrease in the expression of a broad group of E2f targets and a complete cell cycle arrest (21). However, emerging data from mice and other model organisms indicate that E2f function in vivo is complex and probably not restricted to the control of cell cycle progression (46). This study provides additional evidence that E2f1–3, although dispensable for the proliferation of the majority of hematopoietic lineages, are necessary for myeloid cell survival. Global gene expression analysis of E2f1–3-deficient myeloid cells revealed increased expression of G1/S-related E2f target genes but decreased expression of G2/M-related targets. We speculate that this conflict in G1/S and G2/M gene expression programs forces E2f1–3-deficient myeloid cells to inappropriately initiate DNA replication and accumulate in G2/M, resulting in the induction of apoptotic cell death. The molecular signals eliciting this apoptosis remain to be discovered.
In contrast to the rigid view that E2f1–3 proteins function as transcriptional activators, recent in vivo studies demonstrate that these “activators” can also function as “repressors” in specific developmental contexts (40). For instance, in stem cells of the retina and small intestine, E2f1–3 adhere to their canonical role as transcriptional activators whose function is inhibited through association with Rb (40). However, in differentiating epithelial cells of the same tissues, E2f1–3 function in association with Rb as transcriptional repressors (39). From the analysis of tissues deleted for Rb, E2f1–3, or Rb and E2f1-3, it was suggested that phosphorylation of the Rb protein is the molecular event that switches E2f1–3 from functioning as activators in dividing progenitors to repressors in differentiating cells (39). Consistent with studies in the retina and small intestine (39, 40), our results in the hematopoietic system of mice reveal that E2f1–3 function as activators or repressors of G1/S regulated genes in a manner that is dependent on the differentiation stage of the cell. Unlike in the small intestine, retina, and lens, E2f1–3 in myeloid progenitors appear to play a role in the regulation of both G1/S and G2/M genes.
Mice carrying mutant alleles of C/EBPα that lack the ability to bind E2fs fail to support granulocyte differentiation, suggesting that repression of E2f targets by C/EBPα-E2f complexes is essential for myeloid development (47). In the murine liver, C/EBPα is part of a protein complex containing Rb-E2f4 that binds and represses E2f-responsive gene promoters (48), raising the possibility that in E2f1–3-deficient myeloid cells, C/EBPα and Rb-E2f4 complexes might jointly contribute to the observed overcompensatory repression of G2/M targets in these cells.
As Cd11b+ myeloid precursor cells commit to the macrophage lineage, E2f1–3 provides these cells with the ability to expand in response to CSF-1. CSF-1 signaling is complex and difficult to study in vivo. We therefore used the MEFs and NIH 3T3 cells expressing the wild type and mutant (Y809F) forms of human CSF-1R receptor to study mechanism of E2f regulation in CSF-1 signaling pathway and to explore Myc-E2f connection. Using mutant forms of the CSF-1R and conditional alleles of c-Myc, we show that this ability to expand requires the c-Myc-mediated activation of E2f1–3 expression. Engagement of the CSF-1R by its ligand leads to the accumulation of c-Myc protein and its recruitment to canonical Myc-binding sites on the E2f3a promoter. The resulting activation of E2f3a expression provides BMMs with the necessary levels of E2f proteins to activate E2f targets and force cells to enter S phase, divide, and expand prior to exiting into the peripheral organs. In this context of acute growth factor stimulation, as in previous cell culture studies, E2fs conform to their classical role as transcriptional activators that are rate-limiting for cell proliferation.
In summary, our results provide compelling evidence that E2f1–3 are essential for the survival of dividing Cd11b+ precursor cells and the proliferation of terminally differentiated BMMs. Only in the latter circumstance E2f1–3 assume the role of classic transcription activators to stimulate cellular proliferation.
Supplementary Material
Acknowledgments
We thank Bryan Mcelwain and Priya Balasubramanian for technical assistance with flow cytometry. We also thank H. Z. Chen and J. L. Chong for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA85619, R01CA82259, R01HD04470, and P01CA097189 (to G. L.); R01CA053271 and P01CA097189 (to M. C. O.); and CA71907 and CA91765. This work was also supported by the American Lebanese Syrian-Associated Charities of St. Jude Children's Research Hospital (to M. F. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables 1 and 2.
- CDK
- cyclin-dependent kinase
- HSC
- hematopoietic stem cell
- BM
- bone marrow
- BMM
- bone marrow macrophage
- pIpC
- polyinosine-polycytidine
- MEF
- mouse embryo fibroblast.
REFERENCES
- 1. Weissman I. L. (2000) Cell 100, 157–168 [DOI] [PubMed] [Google Scholar]
- 2. Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., Scadden D. T. (2000) Science 287, 1804–1808 [DOI] [PubMed] [Google Scholar]
- 3. Passegué E., Wagers A. J., Giuriato S., Anderson W. C., Weissman I. L. (2005) J. Exp. Med. 202, 1599–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kozar K., Ciemerych M. A., Rebel V. I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R. T., Akashi K., Sicinski P. (2004) Cell 118, 477–491 [DOI] [PubMed] [Google Scholar]
- 5. Malumbres M., Sotillo R., Santamaría D., Galán J., Cerezo A., Ortega S., Dubus P., Barbacid M. (2004) Cell 118, 493–504 [DOI] [PubMed] [Google Scholar]
- 6. Cheng T., Rodrigues N., Dombkowski D., Stier S., Scadden D. T. (2000) Nat. Med. 6, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 7. Park I. K., Qian D., Kiel M., Becker M. W., Pihalja M., Weissman I. L., Morrison S. J., Clarke M. F. (2003) Nature 423, 302–305 [DOI] [PubMed] [Google Scholar]
- 8. Burkhart D. L., Sage J. (2008) Nat. Rev. Cancer 8, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nevins J. R. (2001) Hum. Mol. Genet. 10, 699–703 [DOI] [PubMed] [Google Scholar]
- 10. Walkley C. R., Shea J. M., Sims N. A., Purton L. E., Orkin S. H. (2007) Cell 129, 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Viatour P., Somervaille T. C., Venkatasubrahmanyam S., Kogan S., McLaughlin M. E., Weissman I. L., Butte A. J., Passegué E., Sage J. (2008) Cell Stem Cell 3, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trimarchi J. M., Lees J. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 11–20 [DOI] [PubMed] [Google Scholar]
- 13. Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J. R. (2001) Mol. Cell. Biol. 21, 4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu W., Giangrande P. H., Nevins J. R. (2004) EMBO J. 23, 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giangrande P. H., Zhu W., Schlisio S., Sun X., Mori S., Gaubatz S., Nevins J. R. (2004) Genes Dev. 18, 2941–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Field S. J., Tsai F. Y., Kuo F., Zubiaga A. M., Kaelin W. G., Jr., Livingston D. M., Orkin S. H., Greenberg M. E. (1996) Cell 85, 549–561 [DOI] [PubMed] [Google Scholar]
- 17. Murga M., Fernández-Capetillo O., Field S. J., Moreno B., Borlado L. R., Fujiwara Y., Balomenos D., Vicario A., Carrera A. C., Orkin S. H., Greenberg M. E., Zubiaga A. M. (2001) Immunity 15, 959–970 [DOI] [PubMed] [Google Scholar]
- 18. Li F. X., Zhu J. W., Hogan C. J., DeGregori J. (2003) Mol. Cell. Biol. 23, 3607–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Humbert P. O., Verona R., Trimarchi J. M., Rogers C., Dandapani S., Lees J. A. (2000) Genes Dev. 14, 690–703 [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai S. Y., Opavsky R., Sharma N., Wu L., Naidu S., Nolan E., Feria-Arias E., Timmers C., Opavska J., de Bruin A., Chong J. L., Trikha P., Fernandez S. A., Stromberg P., Rosol T. J., Leone G. (2008) Nature 454, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu L., Timmers C., Maiti B., Saavedra H. I., Sang L., Chong G. T., Nuckolls F., Giangrande P., Wright F. A., Field S. J., Greenberg M. E., Orkin S., Nevins J. R., Robinson M. L., Leone G. (2001) Nature 414, 457–462 [DOI] [PubMed] [Google Scholar]
- 22. Stanley E. R., Guilbert L. J., Tushinski R. J., Bartelmez S. H. (1983) J. Cell. Biochem. 21, 151–159 [DOI] [PubMed] [Google Scholar]
- 23. Matsushime H., Roussel M. F., Ashmun R. A., Sherr C. J. (1991) Cell 65, 701–713 [DOI] [PubMed] [Google Scholar]
- 24. Sherr C. J., Rettenmier C. W., Sacca R., Roussel M. F., Look A. T., Stanley E. R. (1985) Cell 41, 665–676 [DOI] [PubMed] [Google Scholar]
- 25. Roussel M. F., Theodoras A. M., Pagano M., Sherr C. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6837–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Secombe J., Pierce S. B., Eisenman R. N. (2004) Cell 117, 153–156 [DOI] [PubMed] [Google Scholar]
- 27. Roussel M. F., Cleveland J. L., Shurtleff S. A., Sherr C. J. (1991) Nature 353, 361–363 [DOI] [PubMed] [Google Scholar]
- 28. Roussel M. F. (1994) J. Cell Sci. Suppl. 18, 105–108 [DOI] [PubMed] [Google Scholar]
- 29. Laurenti E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W. E., Ehninger A., Knoepfler P. S., Cheng P. F., MacDonald H. R., Eisenman R. N., Bernstein I. D., Trumpp A. (2008) Cell Stem Cell 3, 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson A., Murphy M. J., Oskarsson T., Kaloulis K., Bettess M. D., Oser G. M., Pasche A. C., Knabenhans C., Macdonald H. R., Trumpp A. (2004) Genes Dev. 18, 2747–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leone G., Sears R., Huang E., Rempel R., Nuckolls F., Park C. H., Giangrande P., Wu L., Saavedra H. I., Field S. J., Thompson M. A., Yang H., Fujiwara Y., Greenberg M. E., Orkin S., Smith C., Nevins J. R. (2001) Mol. Cell 8, 105–113 [DOI] [PubMed] [Google Scholar]
- 32. Adams M. R., Sears R., Nuckolls F., Leone G., Nevins J. R. (2000) Mol. Cell. Biol. 20, 3633–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vlach J., Hennecke S., Alevizopoulos K., Conti D., Amati B. (1996) EMBO J. 15, 6595–6604 [PMC free article] [PubMed] [Google Scholar]
- 34. Müller D., Bouchard C., Rudolph B., Steiner P., Stuckmann I., Saffrich R., Ansorge W., Huttner W., Eilers M. (1997) Oncogene 15, 2561–2576 [DOI] [PubMed] [Google Scholar]
- 35. Kühn R., Schwenk F., Aguet M., Rajewsky K. (1995) Science 269, 1427–1429 [DOI] [PubMed] [Google Scholar]
- 36. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leone G., Nuckolls F., Ishida S., Adams M., Sears R., Jakoi L., Miron A., Nevins J. R. (2000) Mol. Cell. Biol. 20, 3626–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 39. Chen D., Pacal M., Wenzel P., Knoepfler P. S., Leone G., Bremner R. (2009) Nature 462, 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chong J. L., Wenzel P. L., Sáenz-Robles M. T., Nair V., Ferrey A., Hagan J. P., Gomez Y. M., Sharma N., Chen H. Z., Ouseph M., Wang S. H., Trikha P., Culp B., Mezache L., Winton D. J., Sansom O. J., Chen D., Bremner R., Cantalupo P. G., Robinson M. L., Pipas J. M., Leone G. (2009) Nature 462, 930–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Timmers C., Sharma N., Opavsky R., Maiti B., Wu L., Wu J., Orringer D., Trikha P., Saavedra H. I., Leone G. (2007) Mol. Cell. Biol. 27, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Himes S. R., Cronau S., Mulford C., Hume D. A. (2005) Oncogene 24, 5278–5286 [DOI] [PubMed] [Google Scholar]
- 43. Tushinski R. J., Stanley E. R. (1985) J. Cell. Physiol. 122, 221–228 [DOI] [PubMed] [Google Scholar]
- 44. Greasley P. J., Bonnard C., Amati B. (2000) Nucleic Acids Res. 28, 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malumbres M., Barbacid M. (2009) Nat. Rev. Cancer 9, 153–166 [DOI] [PubMed] [Google Scholar]
- 46. Chen H. Z., Tsai S. Y., Leone G. (2009) Nat. Rev. Cancer 9, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Porse B. T., Pedersen T. A., Xu X., Lindberg B., Wewer U. M., Friis-Hansen L., Nerlov C. (2001) Cell 107, 247–258 [DOI] [PubMed] [Google Scholar]
- 48. Iakova P., Awad S. S., Timchenko N. A. (2003) Cell 113, 495–506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.