SUMMARY
A general approach for modeling the architecture of large and structured RNA molecules is described. The method exploits the modularity and the hierarchical folding of RNA architecture that is viewed as the assembly of preformed double-stranded helices defined by Watson-Crick base pairs and RNA modules maintained by non-Watson-Crick base pairs. Despite the extensive molecular neutrality observed in RNA structures, specificity in RNA folding is achieved through global constraints like lengths of helices, coaxiality of helical stacks, and structures adopted at the junctions of helices. The Assemble integrated suite of computer tools allows for sequence and structure analysis as well as interactive modeling by homology or ab initio assembly with possibilities for fitting within electronic density maps. The local key role of non-Watson-Crick pairs guides RNA architecture formation and offers metrics for assessing the accuracy of three-dimensional models in a more useful way than usual root mean square deviation (RMSD) values.
Large RNA molecules can be modeled computationally as hierarchical assemblies of preformed double-stranded helices and other modules, in which non-Watson-Crick base pairs play a key role.
1. INTRODUCTION
In this article, a general approach for predicting three-dimensional (3D) contacts and modeling the architecture of large and structured RNA molecules on the basis of sequence analysis and sequence alignments is described. The method assumes that the folding is sequential, with modular units being incorporated hierarchically in the final architecture. The aim is the architecture of the assembly and not all the fine atomic details, although the large RNA molecules are assembled using all-atom components. Recently, following the increase in the number of RNA crystal structures, new approaches have been proposed, most of which can be coupled or used in parallel.
The main driving force for RNA architecture is the packing of RNA helices and modules through stacking between terminal base pairs and specific molecular recognition contacts between RNA segments. Structured RNA molecules are able to self-assemble into complex architectural folds because they contain, beyond the Watson-Crick base pairs that maintain the secondary structure, additional tertiary base pairs, often non-Watson-Crick, as well as various types of contacts between segments of the polynucleotide chain. This hierarchical assembly of 3D RNA structures is coupled with the binding of cations (Misra and Draper 2002; Rangan et al. 2003). A reduction in the net charge of the molecule with the initial association of cations induces a collapse of the RNA chain into compact structures (Fang et al. 1999; Heilman-Miller et al. 2001; Sosnick and Pan 2003) that then favor and promote the formation of tertiary interactions. Experimentally, the initial processes that lead to compaction of the RNA are clearly distinguishable from those leading to the formation of the native tertiary structure (Pan and Woodson 1999; Thirumalai et al. 2001). In the following, we consider only the final assembled structure. However, in the modeling approach, it is worthwhile to take into account the possible folding pathways leading to the modeled architecture.
RNA modeling started with attempts at transfer RNAs, first the anticodon loop (Fuller and Hodgson 1967) and later the full tRNA (Levitt 1969; Ninio et al. 1969). Crystallographic data, although reduced to regular RNA helices and tRNA structures, led to a revival in RNA modeling some 20 years after those early attempts (Dock-Bregeon et al. 1989; Krol et al. 1990; Romby et al. 1988; Westhof et al. 1989). Since then, striking progress has occurred in RNA crystallography and the large number of available RNA crystal structures has considerably strengthened our knowledge of RNA structure and folding. It is now apparent that large RNA structures can be parsed into various structural elements: regular RNA helices, junctions between helices, hairpin loops, and RNA modules (Westhof et al. 1996). Several of those structural elements are recurrent and occur in structured RNAs of very diverse origins or functions (Costa and Michel 1995; Leontis and Westhof 2003; Moore 1999). The set of experimental structures forms the basis of all knowledge-based modeling approaches.
2. THE ANALYSIS OF THE SECONDARY STRUCTURE AND THE RNA MODULES
Almost all of the secondary structure information and some of the tertiary structure information can be deduced from comparative sequence analysis (Michel et al. 2000; Pace et al. 1986; Pace et al. 1989). RNA complexity here is linked with topology, which means the content of the RNA sequence that forms branched junctions, terminal and internal loops, pseudoknots, and non-Watson-Crick tertiary contacts. This topological complexity can be translated into internal constraints that can significantly help find the relative spatial positions of the secondary structure elements.
A second critical point in RNA modeling is that, despite six torsion angles along the main chain, only two contribute strongly to the overall folding pathway of the polynucleotide backbone: the two contiguous torsion angles around the phosphate group. They mainly populate three domains (gauche -, gauche +, and trans). The other four torsion angles oscillate around an invariant value most of the time except in particular cases. In addition, the torsion angle between the base and the sugar adopts two conformational domains, syn and anti, with the anti conformation overwhelmingly more frequent. These general rules are still apparent in recent crystal structures (Richardson et al. 2008; Westhof and Fritsch 2000). This ensemble of observations contributed to the prediction power of the RNA modeling method.
This article is intended to describe an overall process of RNA modeling, mainly based on phylogenetic analysis and illustrated with various examples from small to large size assemblies compared, whenever possible, to the available crystal structures. Other approaches relying on automatic folding predictions and at different levels of granulometry have been developed but will not be extensively addressed here (Shapiro et al. 2007).
Energetically, the secondary structure is the main component of an RNA architecture, whereas tertiary structure contributes only minimally to the stability of the native state in terms of Gibbs free energy (Brion and Westhof 1997; Tinoco and Bustamante 1999). Quite naturally, the construction of the tertiary structure of an RNA molecule always assumes and starts from a given secondary structure. Therefore, the determination of the secondary structure is an essential step in the study of the structure–function relationships of an RNA molecule. Among the recent modeling tools based on conformational space searching, three of them (Das and Baker 2007; Ding et al. 2008; Parisien and Major 2008) attempt to predict simultaneously the secondary and tertiary structure, whereas a fourth coarse-grained approach requires the knowledge of the secondary structure (Jonikas et al. 2009). Two main situations need to be considered. In the first case, several sequences of homologous RNAs are known and sequence comparisons can be applied. In the second case, only one sequence is known. Additional information, usually based on chemical and enzymatic probing, is then needed. In this latter case, in which no or few homologs have been reported, the secondary structure can be explored using folding algorithms (Zuker 1989, 2003).
The term “secondary structure” may carry some ambiguity because it includes not only all segments that can build helices formed by any combination of the isosteric Watson-Crick pairings but also, in variable proportions, Watson-Crick as well as non-Watson-Crick pairs involved in tertiary structure (Westhof and Michel 1994). A secondary structure can be broken down into recognizable elementary modules such as the helical regions (stems and pseudoknots) and nonhelical linking elements (hairpin and internal loops, bulges, and multiple junctions). In the secondary structure, a pseudoknot is a specific RNA module that results from standard Watson-Crick pairs involving a single-stranded stretch, located between paired strands, and a distal single-stranded region (Dam et al. 1992; Westhof and Jaeger 1992). The single-stranded regions may belong to a hairpin loop, an internal loop, or a 3′ (or 5′) dangling end; but at least one of them must occur between base‐paired helical strands. When both single-stranded regions are hairpin loops in a single RNA molecule, they are said to form a loop–loop module (Brunel et al. 2002; Lehnert et al. 1996), which is formally equivalent to a pseudoknot. Intermolecular loop–loop interactions between two RNA molecules occur in dimer formation (Bourassa and Major 2002; Ferrandon et al. 1997; Wagner et al. 2004). It is worth noticing that in intramolecular or intermolecular loop–loop motifs, the interactions are not always of the standard Watson-Crick pairing types (Khvorova et al. 2003; Oubridge et al. 2002; Weichenrieder et al. 2000). Formally, the two-dimensional (2D) structure reduces the secondary structure to the set of Watson-Crick base pairs that form a planar graph (i.e., without crossing edges) when the sequence of bases is arranged along a circle and the base pairs are connected by edges. Thus, pseudoknots or loop–loop motifs, which occur in the folding process once one hairpin at least has been formed, belong to the 3D structure and not to the 2D structure.
Although the 2D structure is dominated by Watson-Crick pairs (which represent 60%–70% in a structured RNA), non-Watson-Crick pairs underlie most of the 3D structure. This point was clear already from the structure of tRNAs, where among the tertiary pairs (at least seven), only one is of the standard Watson-Crick type. The x-ray structures that appeared since then have strengthened this conclusion. At the level of the active tertiary structural organization, an architectural module is an arrangement containing a few secondary structure elements associated with a specific geometry and topology. The combination of such substructures leads to compact domains (Batey et al. 1999), which often fold autonomously and independently of the rest of the RNA architecture. An ensemble of observations bears out a view of RNA folding whereby the architecture results from the cooperative compaction of separate and stable substructures, which might undergo only minor and local rearrangement during the process. The introduction of modular units, hierarchically organized and folded, circumvents most of the numerical nightmares inherent to the Levinthal’s paradox of a purely mathematically based prediction of RNA structure at the atomic level even with coarse-grained approximations or ad hoc potentials.
3. COMPARATIVE SEQUENCE ANALYSIS
The comparative approach is based on the assumption that the function has been conserved by the folding architecture during evolution and, consequently, that a consensus secondary structure should be derivable by an alignment of RNA sequences based on a maximization of Watson-Crick covariations (Michel et al. 1982; Pace et al. 1986). An alignment of RNA sequences is, thus, formally equivalent to a secondary structure common to the set of sequences. This approach is the method of choice when a set of RNA sequences with identical biological function is available. The sequences should be arranged in groups and subgroups (ideally of similar size), either according to the phylogenetic classification (Pace et al. 1989) or following a phenotypic parsing (Michel and Westhof 1990). The first step in an alignment consists in the establishment of the paired regions along each sequence and those should be arranged horizontally so that the lengths of the Watson-Crick paired regions juxtapose vertically. In a second step, conservation or semiconservation of bases can be highlighted by a vertical alignment with inclusion of blanks or gaps in a fashion similar to the alignments of protein sequences. Alignments of residues do not necessarily imply structural superimposition but instead a structural correspondence (Brown et al. 2009).
The overall robustness of the approach increases with the diversity of the sequences and the evolutionary distances between them, whereas the accuracy of each prediction depends on the number of covariation events in each group and subgroup (Michel et al. 2000). Within such a scenario, conserved residues, potentially forming Watson-Crick base-pairs, do not display any covariation and, thus, should be regarded as nonproven or with extreme caution. Indeed, conservation of Watson-Crick pairs can reveal either tertiary contacts or an alternative pairing geometry. This can be the case for A-U pairs when they form Hoogsteen/Watson-Crick interactions; for example, the U80A14 pair in tRNAs or the pair U135-A187 in the crystal structure of the P4-P6 domain in the Tetrahymena group I intron (Cate et al. 1996a). One advantage of comparative analysis is that near-Watson-Crick pairs, like the wobble GoU pair or GoA pairs of the Watson-Crick type at the ends of helices, can be noticed and properly assessed. Critically, sequence comparisons allow us to also delimit rather precisely the helical regions and do not intrude into those segments that should form non-Watson-Crick pairs. Another important and nonnegligeable advantage of comparative analysis is that pseudoknotted regions are easily recognized by visual inspection.
4. THE EXTRACTION OF TERTIARY STRUCTURE CONSTRAINTS
The efficiency of the comparative approach stems from the fact that molecular 3D architectures evolve much more slowly than sequences which sample sequence space on a given 3D fold. Global architecture changes extremely slowly as it relies on conserved long-range tertiary interactions. However, phylogenetic methods are fraught with problems related to statistical relevance. With only four bases to choose among, purely coincidental compensatory base changes (or covariations between positions) are bound to occur, leading to ambiguities. Phylogenetically, the level of ambiguity is reduced by new sequences presenting additional covariations. Thus, if the function is identical and the sequences are sufficiently diverse, the noise level (or covariations resulting from historical contingencies) will be decreased by comparisons. The more compensatory base change events there are in the sequences, the more firmly the secondary structure will be established. But the extraction of 3D content from sequences is difficult (Gautheret et al. 1995; Gautheret and Gutell 1997; Michel and Westhof 1990) first because the rules of tertiary interactions are not as well-defined as those contained in the complementarity of the Watson-Crick pairs and, second, because several contacts involve sequence-independent properties like phosphate or ribose hydroxyl groups hydrogen bonding to other chemical groups (Cate et al. 1996a; Zirbel et al. 2009).
5. HOW TO SEARCH FOR RNA MODULES
Here, search is not meant through genomic sequences (Lambert et al. 2004; Macke et al. 2001; Nawrocki et al. 2009) but instead through a common secondary structure deduced from a set of aligned sequences. After alignment and the derivation of a common secondary structure, the RNA parts that do not display Watson-Crick covariations can be suspected to form specific tertiary modules that should be scrutinized for sequence similarities and characteristics to known modules seen in x-ray structures. The compatibility between a new sequence and a given interaction scheme, as provided by a crystal structure, can be checked using isostericity matrices or known variations observed in aligned sequences. The isostericity matrices have been verified for several RNA motifs using structural alignments anchored by crystallographic structures (Leontis et al. 2002b; Lescoute et al. 2005). Still, it is worth noting that sequence analysis of non-Watson-Crick interactions is difficult to perform without a crystallographic structure because many base–base geometries can be accommodated by any base combination (Stombaugh et al. 2009).
RNA–RNA or RNA–protein interactions are mediated by RNA modules, defined recurrent ensembles of ordered non-Watson-Crick base pairs (Leontis et al. 2002a; Leontis and Westhof 2003). A single RNA module comprises a family of sequences all of which can fold into the same 3D structure and can mediate the same types of interaction(s) (Fig. 1). The chemistry and geometry of base pairing constrain the evolution of modules in such a way that random mutations that occur within them are accepted or rejected insofar as they can mediate a similar ordered array of interactions.
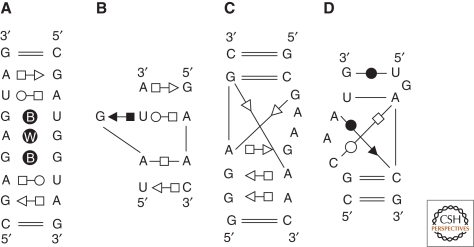
Figure 1.
Some examples of annotated common RNA modules are represented. The annotations use the geometric nomenclature where a circle indicates the Watson-Crick sites, the square the Hoogsteen sites, and the triangle the sugar edge sites (filled black, the bases interact in cis; empty symbols, they interact in trans) (Leontis and Westhof 2001). Using this nomenclature, all base–base pairwise interactions present in nucleic acids have been classified in 12 families in which each family is a 4 × 4 matrix of the bases A, G, C, and U. This classification allows to deduce the isotericity matrices that yield all the possible and geometrically equivalent base pairs in a given family. (A) The double loop E motif of bacterial 5S rRNA. The B within the circle indicates a bifurcated pair and a W a water-mediated interaction. The simple motif occurs in the 16S as well as the 23S rRNAs. (B) The S or bulged G motif in which a base triple occurs. This motif is typical of the eukaryotic loop E of rRNA and of the sarcin/ricin loop in bacterial 23S rRNA. The adenines often interact through their Watson-Crick or sugar-edge sites with other nucleotides (Leontis et al. 2002a). (C) The kink-turn motif (Klein et al. 2001). (D) The C-motif (Leontis and Westhof 2003).
RNA modules (Fig. 1) can be either architectural, forming a bend, e.g., the kink-turn (Klein et al. 2001), or a reorientation within a helix or between helices, or anchors for association as in the tetraloop–tetraloop receptor interaction (Cate et al. 1996b; Costa and Michel 1995, 1997). Up to now, it appears that modules are limited in number and are recurrent, as well as some of the rules of association between them (Leontis and Westhof 2003). Thus, the modules can show sequence variability without impairing their ability to adopt a structure close to the archetype. Surprisingly, the most common RNA–RNA interaction motif, the A-minor motif (Nissen et al. 2001), is also the least specific in its local requirements (Fig. 2). A-minor motifs are mediated by adenines binding into the shallow/minor groove of any combination of stacked and helical Watson-Crick base pairs. Thus, A-minor motifs are mutationally robust and can accommodate many combinations of neutral mutations. RNA nano-objects could successfully be built following some of the above rules (Chworos et al. 2004; Liao and Seeman 2004). New tools are appearing for automatically extracting recurrent modules from databases of x-ray structures (Djelloul and Denise 2008; Sarver et al. 2008).
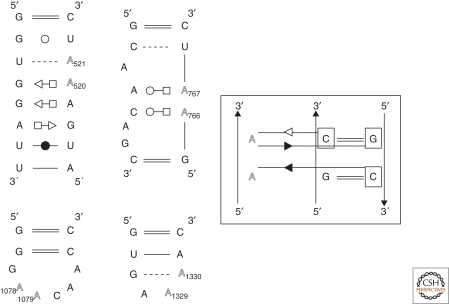
Figure 2.
The figure shows four examples of A-minor interactions (the A residues involved are shown in gray tone). Notice how, despite the similar types of contacts, the two consecutive adenines can belong to very different local environments. At the left, four different types of motifs in which two consecutive adenines form similar types of A-minor interactions (Nissen et al. 2001) with two consecutive base pairs, as shown at the right (first contact at the top, type I A-minor, the adenine H-bonds to both bases of the Watson-Crick pair; second contact below, type II A-minor, the adenine H-bonds to only one base of the Watson-Crick pair). It has been established (Battle and Doudna 2002; Doherty et al. 2001): (1) consecutive adenines recognize without any strong bias any two stacked Watson-Crick pairs; (2) consecutive adenines have a strong preference for complementary Watson-Crick pairs compared to noncomplementary pairs.
6. MODELING LARGE RNA ASSEMBLIES
There are three main categories of tertiary structure interactions, those between two double-stranded helices, those between a helix and a single strand, and those between two single-stranded regions. Sequence analyses together with the growing number of RNA crystal structures have shown that RNA architectures are assembled from modules in a hierarchical manner. The modeling process developed in the laboratory is based on this principle of natural folding processes (Westhof et al. 1996; Westhof and Michel 1994). The process is highly iterative in most cases. The secondary structure is first parsed into modules based on elementary structural elements, the 3D coordinates of which can be generated using appropriate programs (see later). These modules are afterwards assembled to form the RNA architecture. One starts with some module identifications and proceeds to assemble interactively the fragments. During this process, new potential contacts can be identified or suspected. These are then assessed in the set of available aligned sequences. Finally, the geometry of the model is regularized using least-square refinement. In this process, standard hydrogen bonds between nucleotides are used as explicit constraints.
One of the most difficult tasks is the arrangement of the multiple-way junctions between helices. The main problem is the proper choice of helices that stack on each other (Duckett et al. 1995; Hohng et al. 2004; Krol et al. 1990). The natural tendency for right-handedness in RNA strands helps often in the decision process. Thus, the Hoogsteen edge (a purine N7 atom) is more accessible when the purine base is 3′ terminal than when it is either internal or 5′ terminal (Westhof et al. 1989). The avoidance of knot formation should be kept in mind and, in large structures with several long-range loop–loop contacts, like the sense–antisense complex between CopA and CopT, topological criteria have to be considered carefully to avoid knot formation (Kolb et al. 2001). Three-way junctions with two helices approximately coaxially stacked can be divided into three main families depending on the relative lengths of the segments linking the three Watson-Crick helices. Each family has topological characteristics with some conservation in the non-Watson-Crick pairs within the linking segments as well as in the types of contacts between the segments and the helices (Lescoute and Westhof 2006b). The determination of such three-way junctions is only applicable in case of coaxial stacking of two of the three helices, a rather frequent situation.
7. MODELING RNA–PROTEIN COMPLEXES
The intricacies of RNA–protein complexes render the modeling of RNA complexes particularly difficult and challenging. Together with extensive chemical and enzymatic probing, information about some crucial contacts (Romby et al. 1990) and the relative positioning of the interacting surfaces (Caprara et al. 1996; Webb et al. 2001) can be gained, even in the absence of the structures of the binding proteins. However, with the knowledge of the protein crystal structures and additional biochemical evidence, for example cross-linking data, actual docking of protein to the RNA model or directed probing can be attempted (Tsai et al. 2003). Modeling of RNA–protein complexes will constitute a major challenge for the next years.
8. MODELING AND FITTING INTO MEDIUM TO LOW RESOLUTION ELECTRON DENSITY MAPS
The progress in cryoelectron microscopy techniques has led to density maps of large functional objects at resolution around or below 7 Å (Schuler et al. 2006). This allows the fitting of atomic models into the density map (Mitra and Frank 2006). Such models can be assembled from homology models derived from solved crystallographic structures but generally de novo construction based on various RNA modules is required. This approach has been applied to the cricket paralysis virus IRES RNA bound to ribosomes in cryoelectron densities at 7.3 Å (Schuler et al. 2006). The resulting model, obtained before crystallography (Pfingsten et al. 2006), agrees very well with the structure of the IRES RNA structure alone (Pfingsten and Kieft 2008).
9. INTERACTIVE MOLECULAR MODELING WITH ASSEMBLE
Assemble contains a complete set of interactively connected computer tools with web service capabilities dedicated to the analysis of RNA structures, the structural alignments of RNA sequences with or without a known 3D structure, and the modeling with assembly of RNA modules into an RNA architecture. Assemble extends considerably our previous Manip program (Massire and Westhof 1998). Importantly, it can be linked to the S2S application dedicated to RNA alignments (Jossinet and Westhof 2005). As discussed previously, the construction of an RNA 3D model with Assemble starts with the secondary structure. RNA algorithms (Mfold [Zuker 2003] or the RNA Vienna package [Hofacker 2009]) or online repositories (CRW site [Cannone et al. 2002] or RNASTRAND [Andronescu et al. 2008]) provide precomputed secondary structures in CT or BPSEQ files. The secondary structure can also be computed directly from an RNA sequence stored in a FASTA file. Assemble outsources this prediction task to a series of RNA algorithms available as web services (developed in the lab or by third parties). Finally, Assemble can also start the 3D modeling from a solved tertiary structure described in a PDB file. In this latter case, the secondary structure is the result of a 3D annotation process done by RNA algorithms like RNAVIEW (Yang et al. 2003) or MCAnnotate (Lemieux and Major 2002).
The RNA secondary structure is displayed in a 2D panel connected to a 3D one. This 2D panel plays several roles to assist the modeling process. It allows the selection of residues with their visualization in the 3D panel and the definition or edition of 2D structure elements like helices, single-strands, and secondary and tertiary interactions. Helices or single stranded regions defined in the RNA secondary structure can be exported as 3D building blocks with a default regular A-form helical fold. Several options are then available in the 3D panel to adapt the model to the RNA peculiarities. First, the default folding can be altered by modifying the six torsion angles along the sugar-phosphate backbone for any single residue within the 3D structure. The folding of an RNA motif stored in a local repository can be applied to the current 3D selection. The building blocks can be reorganized in the 3D scene to produce the overall shape of the 3D architectural model. If available, electron density maps can be simultaneously displayed for fitting models into density.
Finally, stereochemical and geometrical errors introduced during the modeling process can be fixed using a refinement algorithm embedded into Assemble. The set of base–base interactions defined in the 2D panel are used to deduce the structural and geometrical constraints needed to improve the 3D model. The refinement is achieved by geometrical least-squares using the Konnert-Hendrickson algorithm (Konnert and Hendrickson 1980) implemented in the program Nuclin/Nuclsq (Westhof et al. 1985). The algorithm takes into account bond lengths, valence angles, dihedrals, and has antibump restraints. The resulting function is minimized against a dictionary of distances that have been observed in high-resolution crystal structures of nucleotides and oligonucleotides. Because the refinement program uses the steepest descent algorithm, the conformation of the starting model should not present extremely distorted regions to avoid refinement failure. The refined model can then be collated to the data and the process of interactive modeling and least-square refinement can be looped until the model is satisfactory. A subset of the data should be used as a blind test during the building of the model so as to help validate the model. Further steps of interactive modeling may then be required until a satisfactory solution is reached.
10. COMPARISONS BETWEEN RNA MODELS AND CRYSTAL STRUCTURES
Several RNAs were predicted, sometimes several years, before the x-ray structures became available. This situation is rather unique to the RNA world; this is not the case for example in the protein field where special competitions had to be installed ad hoc (Critical Assessment of Techniques for Protein Structure Prediction; http://predictioncenter.org/). Root mean square deviations between a chosen set of atoms in the structures to be assessed (RMSDs) are usually the main measure for benchmarking prediction tools and comparing predicted structures with experimentally derived structures. For example, Lsqman (Kleywegt 1996) calculates normalized RMSD values according to (Carugo and Pongor 2001). Because RMSDs increase with the size of the molecule, normalization is necessary to allow the comparisons between models of different size. As can be expected, the values for the RMSDs tend to improve with the size of the x-ray crystallographic database on which the modeling is based. However, RMSDs, being based on a least-squares approach, spread errors over the whole molecule so as to minimize the final value. Thus, low values can be obtained for compact structures that may be missing most of the key intramolecular contacts, whereas high values can be observed when the relationship of two domains is wrongly deduced despite the fact that each domain is correctly predicted. An accurate model should present most of the internal interaction contacts maintaining the overall architecture (Lescoute and Westhof 2006a). New metrics have been introduced to calibrate the interaction network fidelity (Parisien et al. 2009). The results show that RMSDs do not provide information about the quality and precision of the base–base interaction networks.
Globally, the interactively modeled architectures with all of the long-range contacts (loop–helix, loop–loop, pseudoknots, …) are in excellent agreement with crystallographic structures, especially for group I introns (Jaeger et al. 1993; Lehnert et al. 1996; Michel and Westhof 1990) and the RNA component of ribonuclease P (Brown et al. 1996; Chen et al. 1998; Haas et al. 1991; Massire et al. 1998). Thus, for the specificity domain of the RNA of ribonuclease P (Fig. 3), the overall RMSD is 11.2 Å but the normalized one is 4.8 Å between the modeled (Massire et al. 1998) and the 3.15 Å resolution x-ray structure (Krasilnikov et al. 2003). Interestingly, the RMSD between the crystal structures of the specificity domains of the two main families of the ribonuclease P RNAs (Krasilnikov et al. 2004) is 1.6 Å. In addition, locally, several motifs were properly identified and inserted into large structures: the ribose zipper in the hairpin ribozyme (Earnshaw et al. 1997; Rupert and Ferre-D’Amare 2001) based on experimental data (Chowrira et al. 1993); the presence of a C-motif in the 5′-UTR of the Thr-aminoacylsynthetase messenger (Caillet et al. 2003; Torres-larios et al. 2002); the presence of a loop-E like structure in domain P7 of subgroup IA2 in group I introns involved in tertiary contacts (Golden et al. 2005; Leontis and Westhof 1998a; Waldsich et al. 2002), or other tertiary contacts like that between P3 and J6/6a in the Azoarcus group I intron (Adams et al. 2004; Rangan et al. 2003). A systematic comparison between phylogenetic (Leontis and Westhof 1998b), chemical probing (Romby et al. 1988), and x-ray data (Correll et al. 1997) led to a refined model of the 5S loop E in spinach chorosplast, which was later strongly supported by NMR data (Vallurupalli and Moore 2003). However, despite a correct architecture with the presence of the proper long-range contacts and the ensuing rather good normalized RMSD between modeled and x-ray structures of the Azoarcus group I intron, 3.85 Å, at the atomic level many contacts are off, sometimes one nucleotide away. Further, a comparison with the crystal structures of group I introns (Adams et al. 2004; Golden et al. 2005; Guo et al. 2004) shows that, although the binding mode of the guanine was correctly predicted (Michel et al. 1989), the adjacent invariant nucleotides were not (Michel and Westhof 1990). A recent analysis between models and crystallographic results of group II introns has been presented (Michel et al. 2009).
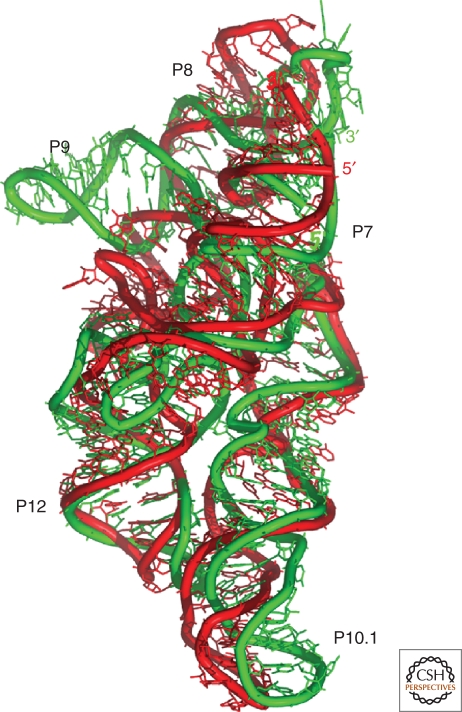
Figure 3.
Models can be compared with x-ray structures to assess their accuracy. One measure, the root mean square deviations (RMSD), is obtained after minimizing the sum of the distances between corresponding atoms. Superimposition of the ribonuclease P specificity domain of Bacillus subtilis the model (in red) (Massire et al. 1998) was assembled 5 years before the x-ray structure (in green) (Krasilnikov et al. 2003).
Up to now, the RNA architectures, assembled using all-atom components, agree very well with the determined x-ray structures despite a rather poor congruence at the atomic level. The interactive 3D modeling of RNA is still the most efficient and reliable method for assembling large structures. The ultimate goal is to model RNA folds with atomic precision as automatically as possible. Despite great progress, new tools based on conformational space searches have not yet produced a novel fold before x-ray determination of major biological impact. Some of them do reproduce known structures with excellent accuracy (Parisien et al. 2009). In the near future, through systematic comparisons between crystal structures and sets of aligned sequences, refined and new key molecular rules will be unraveled, which should lead to an improved accuracy in automatic model prediction.
11. CONCLUSIONS
The modeling approach described here is based on a corpus of observations leading to the paradigm that RNA architecture results from the hierarchical assembly of preformed double-stranded helices defined by Watson-Crick base pairs and RNA modules maintained by non-Watson-Crick base pairs (Fig. 4). Thus, metrics for assessing the accuracy of RNA models should include checks on the number and correctness of non-Watson-Crick pairs in the predicted models (Parisien et al. 2009). The most common long-range RNA–RNA contacts are the A-minor interactions that are mutationally robust and can accommodate many combinations of neutral mutations. This characteristic dilutes the links between RNA sequence and structure. To achieve specificity in RNA folding, global, positional, and orientational, constraints on the native fold must occur upstream in the folding process. Critical parameters are the lengths of the helices, the coaxiality of the helical stacks, and the structure adopted at the junctions of helices (Lescoute and Westhof 2006b). Thus, the molecular neutrality present in the local interactions is partially compensated by global topological criteria, much less accessible to sequence analysis because they are attached to the 3D architecture (Cruz and Westhof 2009).
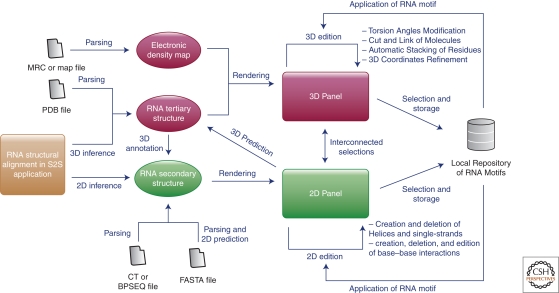
Figure 4.
Integrated sets of computer programs are a necessity for analyzing RNA sequences and structures as well as for modeling RNA structures by the assembly of identified modules. Details of the workflow of the Assemble package (http://bioinformatics.org/assemble). This application is divided into two main components: (1) a 3D panel rendering the 3D model and (2) a 2D panel displaying the secondary structure scaffold of this model. Because of their interconnections, the secondary structure is used as a guide during the modeling process. The secondary structure can be loaded or computed from several file formats or can be inferred from a dedicated tool like the S2S application (Jossinet and Westhof 2005).
Because modeling requires the integration of a vast amount of data at various levels of complexity, the quality of the modeling reflects, in the end, the current understanding of the modeled systems and the quality and usefulness in the integration of knowledge. This present understanding of RNA architecture and modeling bears on the RNA World hypothesis. RNA architecture can be parsed into recurrent modules, limited in number, with defined borders. Modules interact through defined protocols of interaction applicable in many topological situations. Modules can accommodate sequence variations that are neutral within defined geometrical limits. Such hierarchical networks with embedded modularity are typical of self-organizing networks (Ravasz et al. 2002). The physicochemical and structural characteristics of RNA molecules are ideally suited to the evolution of such complex networks. They lead to the appearance of similar modules capable of adaptation. The neutrality of the contacts between the modules allows for a great diversity of architectures despite the use of a limited number of recurrent modular building blocks. For example (Lescoute and Westhof 2006a), it is striking that a central piece of the peptidyl transferase center is made of the most frequent three-way junctions, observed in riboswitches for binding activating ligands or at the active site of the hammerhead ribozymes.
ACKNOWLEDGMENTS
Some of this work was made possible by a grant from the Human Frontier Science Program (RGP0032/2005-C to E.W.).
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA 2004. Crystal structure of a group I intron splicing intermediate. RNA 10: 1867–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronescu M, Bereg V, Hoos HH, Condon A 2008. RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinformatics 9: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA 1999. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl 38: 2326–2343 [DOI] [PubMed] [Google Scholar]
- Battle DJ, Doudna JA 2002. Specificity of RNA-RNA helix recognition. Proc Natl Acad Sci 99: 11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourassa N, Major F 2002. Implication of the prohead RNA in phage phi29 DNA packaging. Biochimie 84: 945–951 [DOI] [PubMed] [Google Scholar]
- Brion P, Westhof E 1997. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct 26: 113–137 [DOI] [PubMed] [Google Scholar]
- Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, et al. 2009. The RNA structure alignment ontology. RNA 15: 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Nolan JM, Haas ES, Rubio MA, Major F, Pace NR 1996. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc Natl Acad Sci 93: 3001–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel C, Marquet R, Romby P, Ehresmann C 2002. RNA loop-loop interactions as dynamic functional motifs. Biochimie 84: 925–944 [DOI] [PubMed] [Google Scholar]
- Caillet J, Nogueira T, Masquida B, Winter F, Graffe M, Dock-Bregeon AC, Torres-Larios A, Sankaranarayanan R, Westhof E, Ehresmann B, et al. 2003. The modular structure of Escherichia coli threonyl-tRNA synthetase as both an enzyme and a regulator of gene expression. Mol Microbiol 47: 961–974 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprara MG, Lehnert V, Lambowitz AM, Westhof E 1996. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell 87: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Carugo O, Pongor S 2001. A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci 10: 1470–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA 1996a. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1684 [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA 1996b. RNA tertiary structure mediation by adenosine platforms. Science 273: 1696–1699 [DOI] [PubMed] [Google Scholar]
- Chen JL, Nolan JM, Harris ME, Pace NR 1998. Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. Embo J 17: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowrira BM, Berzal-Herranz A, Keller CF, Burke JM 1993. Four ribose 2’-hydroxyl groups essential for catalytic function of the hairpin ribozyme. J Biol Chem 268: 19458–19462 [PubMed] [Google Scholar]
- Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L 2004. Building programmable jigsaw puzzles with RNA. Science 306: 2068–2072 [DOI] [PubMed] [Google Scholar]
- Correll CC, Freeborn B, Moore PB, Steitz TA 1997. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell 28: 705–712 [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F 1995. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J 14: 1276–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J 16: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JA, Westhof E 2009. The dynamic landscapes of RNA architecture. Cell 136: 604–609 [DOI] [PubMed] [Google Scholar]
- Dam E, Pleij K, Draper D 1992. Structural and functional aspects of RNA pseudoknots. Biochemistry 31: 11665–11676 [DOI] [PubMed] [Google Scholar]
- Das R, Baker D 2007. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci 104: 14664–14669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV 2008. Ab initio RNA folding by discrete molecular dynamics: From structure prediction to folding mechanisms. RNA 14: 1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djelloul M, Denise A 2008. Automated motif extraction and classification in RNA tertiary structures. RNA 14: 2489–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon AC, Westhof E, Giege R, Moras D 1989. Solution structure of a tRNA with a large variable region: Yeast tRNASer. J Mol Biol 206: 707–722 [DOI] [PubMed] [Google Scholar]
- Doherty EA, Batey RT, Masquida B, Doudna JA 2001. A universal mode of helix packing in RNA. Nat Struct Biol 8: 339–343 [DOI] [PubMed] [Google Scholar]
- Duckett DR, Murchie AI, Lilley DM 1995. The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell 83: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Earnshaw DJ, Masquida B, Muller S, Sigurdsson ST, Eckstein F, Westhof E, Gait MJ 1997. Inter-domain cross-linking and molecular modelling of the hairpin ribozyme. J Mol Biol 274: 197–212 [DOI] [PubMed] [Google Scholar]
- Fang X, Pan T, Sosnick TR 1999. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry 38: 16840–16846 [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C 1997. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3’ UTR-STAUFEN ribonucleoprotein particles. Embo J 16: 1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W, Hodgson A 1967. Conformation of the anticodon loop intRNA. Nature 215: 817–821 [DOI] [PubMed] [Google Scholar]
- Gautheret D, Gutell RR 1997. Inferring the conformation of RNA base pairs and triples from patterns of sequence variation. Nucleic Acids Res 25: 1559–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Damberger SH, Gutell RR 1995. Identification of base-triples in RNA using comparative sequence analysis. J Mol Biol 248: 27–43 [DOI] [PubMed] [Google Scholar]
- Golden BL, Kim H, Chase E 2005. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol 12: 82–89 [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR 2004. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol Cell 16: 351–362 [DOI] [PubMed] [Google Scholar]
- Haas ES, Morse DP, Brown JW, Schmidt FJ, Pace NR 1991. Long-range structure in ribonuclease P RNA. Science 254: 853–856 [DOI] [PubMed] [Google Scholar]
- Heilman-Miller SL, Pan J, Thirumalai D, Woodson SA 2001. Role of counterion condensation in folding of the Tetrahymena ribozyme. II. Counterion-dependence of folding kinetics. J Mol Biol 309: 57–68 [DOI] [PubMed] [Google Scholar]
- Hofacker IL 2009. RNA secondary structure analysis using the Vienna RNA package. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis et al Chapter 12, Unit12 12 [DOI] [PubMed] [Google Scholar]
- Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DM, Ha T 2004. Conformational flexibility of four-way junctions in RNA. J Mol Biol 336: 69–79 [DOI] [PubMed] [Google Scholar]
- Jaeger L, Westhof E, Michel F 1993. Monitoring of the cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3′ terminal intron components. J Mol Biol 234: 331–346 [DOI] [PubMed] [Google Scholar]
- Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB 2009. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA 15: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossinet F, Westhof E 2005. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics 21: 3320–3332 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. Embo J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ 1996. Use of non-crystallographic symmetry in protein structure refinement. Acta Cryst D52: 842–857 [DOI] [PubMed] [Google Scholar]
- Kolb FA, Westhof E, Ehresmann B, Ehresmann C, Wagner EG, Romby P 2001. Four-way junctions in antisense RNA-mRNA complexes involved in plasmid replication control: A common theme? J Mol Biol 309: 605–614 [DOI] [PubMed] [Google Scholar]
- Konnert JH, Hendrickson WA 1980. Restrained parameters thermal factors refinement procedures. Acta Crystallographica A36: 344–349 [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107 [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A 2003. Crystal structure of the specificity domain of ribonuclease P. Nature 421: 760–764 [DOI] [PubMed] [Google Scholar]
- Krol A, Westhof E, Bach M, Luhrmann R, Ebel JP, Carbon P 1990. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res 18: 3803–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A, Fontaine JF, Legendre M, Leclerc F, Permal E, Major F, Putzer H, Delfour O, Michot B, Gautheret D 2004. The ERPIN server: An interface to profile-based RNA motif identification. Nucleic Acids Res 32: W160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert V, Jaeger L, Michel F, Westhof E 1996. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: A complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol 3: 993–1009 [DOI] [PubMed] [Google Scholar]
- Lemieux S, Major F 2002. RNA canonical and non-canonical base pairing types: A recognition method and complete repertoire. Nucleic Acids Res 30: 4250–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E 2002a. Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie 84: 961–973 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E 2002b. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 1998a. A common motif organizes the structure of multi-helix loops in 16S and 23S ribosomal RNAs. J Mol Biol 283: 571–583 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 1998b. The 5S rRNA loop E: Chemical probing and phylogenetic data versus crystal structure. RNA 4: 1134–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2003. Analysis of RNA motifs. Curr Opin Struct Biol 13: 300–308 [DOI] [PubMed] [Google Scholar]
- Lescoute A, Westhof E 2006a. The interaction networks of structured RNAs. Nucleic Acids Res 34: 6587–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E 2006b. Topology of three-way junctions in folded RNAs. RNA 12: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Leontis NB, Massire C, Westhof E 2005. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res 33: 2395–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M 1969. Detailed molecular model for transfer ribonucleic acid. Nature 224: 759–763 [DOI] [PubMed] [Google Scholar]
- Liao S, Seeman NC 2004. Translation of DNA signals into polymer assembly instructions. Science 306: 2072–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29: 4724–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massire C, Westhof E 1998. MANIP: An interactive tool for modelling RNA. J Mol Graph Model 16: 197–205, 255–197 [DOI] [PubMed] [Google Scholar]
- Massire C, Jaeger L, Westhof E 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J Mol Biol 279: 773–793 [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E 1990. Modelling of the three-dimensionnal architecture of group-I catalytic introns based on comparative sequence analysis. J Mol Biol 216: 585–610 [DOI] [PubMed] [Google Scholar]
- Michel F, Costa M, Westhof E 2009. The ribozyme core of group II introns: A structure in want of partners. Trends Biochem Sci 34: 189–199 [DOI] [PubMed] [Google Scholar]
- Michel F, Jaquier A, Dujon B 1982. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie 64: 867–881 [DOI] [PubMed] [Google Scholar]
- Michel F, Costa M, Massire C, Westhof E 2000. Modeling RNA tertiary structure from patterns of sequence variation. Methods Enzymol 317: 491–510 [DOI] [PubMed] [Google Scholar]
- Michel F, Hanna M, Green R, Bartel DP, Szostak JW 1989. The guanosine binding site of the Tetrahymena ribozyme. Nature 342: 391–395 [DOI] [PubMed] [Google Scholar]
- Misra VK, Draper DE 2002. The linkage between magnesium binding and RNA folding. J Mol Biol 317: 507–521 [DOI] [PubMed] [Google Scholar]
- Mitra K, Frank J 2006. Ribosome dynamics: Insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct 35: 299–317 [DOI] [PubMed] [Google Scholar]
- Moore PB 1999. Structural motifs in RNA. Annu Rev Biochem 68: 287–300 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: Inference of RNA alignments. Bioinformatics (Oxford, England) 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J, Favre A, Yaniv M 1969. Molecular model for transfer RNA. Nature 223: 1333–1335 [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci 98: 4899–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Kuglstatter A, Jovine L, Nagai K 2002. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol Cell 9: 1251–1261 [DOI] [PubMed] [Google Scholar]
- Pace NR, Olsen GJ, Woese CR 1986. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell 45: 325–326 [DOI] [PubMed] [Google Scholar]
- Pace NR, Smith DK, Olsen GJ, James BD 1989. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA–a review. Gene 82: 65–75 [DOI] [PubMed] [Google Scholar]
- Pan J, Woodson SA 1999. The effect of long-range loop-loop interactions on folding of the Tetrahymena self-splicing RNA. J Mol Biol 294: 955–965 [DOI] [PubMed] [Google Scholar]
- Parisien M, Major F 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452: 51–55 [DOI] [PubMed] [Google Scholar]
- Parisien M, Cruz JA, Westhof E, Major F 2009. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA 15: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Kieft JS 2008. RNA structure-based ribosome recruitment: Lessons from the Dicistroviridae intergenic region IRESes. RNA 14: 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS 2006. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314: 1450–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Masquida B, Westhof E, Woodson SA 2003. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci 100: 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL 2002. Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, et al. 2008. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution). RNA 14: 465–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romby P, Baudin F, Brunel C, Leal de Stevenson I, Westhof E, Romaniuk PJ, Ehresmann C, Ehresmann B 1990. Ribosomal 5S RNA from Xenopus laevis oocytes: Conformation and interaction with transcription factor IIIA. Biochimie 72: 437–452 [DOI] [PubMed] [Google Scholar]
- Romby P, Westhof E, Toukifimpa R, Mache R, Ebel JP, Ehresmann C, Ehresmann B 1988. Higher order structure of chloroplastic 5S ribosomal RNA from spinach. Biochemistry 27: 4721–4730 [DOI] [PubMed] [Google Scholar]
- Rupert PB, Ferre-D’Amare AR 2001. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410: 780–786 [DOI] [PubMed] [Google Scholar]
- Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB 2008. FR3D: Finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol 56: 215–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM 2006. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Shapiro BA, Yingling YG, Kasprzak W, Bindewald E 2007. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol 17: 157–165 [DOI] [PubMed] [Google Scholar]
- Sosnick TR, Pan T 2003. RNA folding: Models and perspectives. Curr Opin Struct Biol 13: 309–316 [DOI] [PubMed] [Google Scholar]
- Stombaugh J, Zirbel CL, Westhof E, Leontis NB 2009. Frequency and isostericity of RNA base pairs. Nucleic Acids Res 37: 2294–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Lee N, Woodson SA, Klimov D 2001. Early events in RNA folding. Annu Rev Phys Chem 52: 751–762 [DOI] [PubMed] [Google Scholar]
- Tinoco I Jr, Bustamante C 1999. How RNA folds. J Mol Biol 293: 271–281 [DOI] [PubMed] [Google Scholar]
- Torres-Larios A, Dock-Bregeon AC, Romby P, Rees B, Sankaranarayanan R, Caillet J, Springer M, Ehresmann C, Ehresmann B, Moras D 2002. Structural basis of translational control by Escherichia coli threonyl tRNA synthetase. Nat Struct Biol 9: 343–347 [DOI] [PubMed] [Google Scholar]
- Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V 2003. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J Mol Biol 325: 661–675 [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Moore PB 2003. The solution structure of the loop E region of the 5S rRNA from spinach chloroplasts. J Mol Biol 325: 843–856 [DOI] [PubMed] [Google Scholar]
- Wagner C, Ehresmann C, Ehresmann B, Brunel C 2004. Mechanism of dimerization of bicoid mRNA: Initiation and stabilization. J Biol Chem 279: 4560–4569 [DOI] [PubMed] [Google Scholar]
- Waldsich C, Masquida B, Westhof E, Schroeder R 2002. Monitoring intermediate folding states of the td group I intron in vivo. Embo J 21: 5281–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Rose MA, Westhof E, Weeks KM 2001. Protein-dependent transition states for ribonucleoprotein assembly. J Mol Biol 309: 1087–1100 [DOI] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408: 167–173 [DOI] [PubMed] [Google Scholar]
- Westhof E, Jaeger L 1992. RNA Pseudoknots: Structural and functional aspects. Curr Op Struct Biol 2: 327–333 [Google Scholar]
- Westhof E, Fritsch V 2000. RNA folding: Beyond Watson-Crick pairs. Structure Fold Des 8: R55–65 [DOI] [PubMed] [Google Scholar]
- Westhof E, Michel F 1994. Prediction and experimental investigation of RNA secondary and tertiary foldings. In RNA-Proteins interactions: Frontiers in molecular biology IRL Press at Oxford University Press, pp. 25–51 [Google Scholar]
- Westhof E, Dumas P, Moras D 1985. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol 184: 119–145 [DOI] [PubMed] [Google Scholar]
- Westhof E, Masquida B, Jaeger L 1996. RNA tectonics: Towards RNA design. Fold Des 1: R78–88 [DOI] [PubMed] [Google Scholar]
- Westhof E, Romby P, Romaniuk PJ, Ebel JP, Ehresmann C, Ehresmann B 1989. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol 207: 417–431 [DOI] [PubMed] [Google Scholar]
- Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E 2003. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res 31: 3450–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB 2009. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res 37: 4898–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]