Abstract
When dsDNA polymers containing the identical number of base pairs were electrophoresed through a nanopore in a voltage biased silicon nitride membrane, the measured time integral of blocked ionic current (the event charge deficit, ecd [1]) for each translocation event was the same regardless of whether the molecules were in a linear, circular relaxed, or supercoiled form. Conversely, when DNA polymers containing different numbers of base pairs were electrophoresed through a nanopore, the ecd depended strongly on, and predicted the value of, the molecule’s number of base pairs. Measurements showed that the magnitude of the current blockages was strongly affected by a molecule’s form. The current blockages exhibited characteristic differences that distinguished between single stranded linear, double stranded linear, circular relaxed, and supercoiled forms. Because the data that establish ecd are usually determined concomitantly with current blockade measurements, our results show that a single nanopore assay can simultaneously determine both DNA conformation and base number.
Keywords: nanopore, DNA conformation, DNA base number
Introduction
A DNA molecule’s conformation and configuration as well as its number of bases and sequence are important characteristics that affect its functional properties. It is well known that chromosomal DNA may have different degrees of supercoiling and that plasmids can exist in linear, relaxed-circular, or supercoiled forms. Because essential cellular processes, such as DNA packaging, transcription, replication, recombination, response to environmental stress, and protein binding are all dependent on DNA topology [2-8], a simple method that can simultaneously determine DNA conformation and base number would be advantageous.
Although electrophoresis in a gel is one of the most common methods used to study DNA’s conformation, the electrophoretic mobility of a DNA molecule is a function of both its conformation and its length [9-12]. Consequently, it is usually not possible to determine both the conformations of DNA molecules and their lengths from a single electrophoretic run. Here we show that data gathered in a few minutes from a single nanopore experiment can concurrently identify both DNA conformation and approximate base number by sensing changes of ionic current as DNA molecules are driven through a biased nanopore in a thin silicon nitride membrane separating two ionic solution-filled chambers,
As initially recognized in a voltage biased protein nanopore [13-15] and subsequently in solid state nanopores [16-19], a nucleic acid polymer molecule that translocates through such a pore partially blocks its ionic conductivity. Analyses of blockage amplitudes and blockage durations can reveal information about polymer length and conformation [14, 17, 20-22], base pair unzipping kinetics [23, 24] and can, in some special cases, even discriminate between single bases in a polymer [25-27]. Our results now show that while the blockage amplitude alone can be an excellent indicator of molecule conformation, the event charge deficit (ecd) [1], which is the measured time integral of obstructed ionic current or the integrated event area, serves as a powerful measure of DNA length or molecular weight.
Materials and methods
Nanopore chip fabrication
Our nanopores were fabricated in a free standing 280 nm thick low stress silicone nitride membrane supported by a 380 μm thick silicone substrate using a combination of focus ion beam milling and feedback controlled ion beam sculpting [16, 28]. We found that 10-20 nm diameter pores are the most convenient for determining DNA conformation and base pair number. The ion beam sculpting technique produces these nanopores with high yield (gt;90%). Depending on the DNA samples and solution conditions used, these pores can last for hours and even days before they become unusable because they become irreversibly clogged. Although TEM images show the projected view of the nanopore’s perimeter, because the diameter of the pore is not the same throughout its unknown length, its precise contours cannot be fully ascertained. Hence, the absolute value of the current blockages caused by identical DNA molecules can vary from one nanopore to the next by as much as 20% even for similar diameter nanopores. To minimize effects due to pore-to-pore variation in thickness, diameter, shape, or surface charge, a single nanopore was used for the full set of experiments needed to generate any one single figure shown here.
Nanopore DNA analysis setup
The DNA translocation experimental setup was previously described in detail [17, 29]. Briefly, a chip with a single nanopore separated two ionic solution filled chambers whose only ionic connection was via the solution inside the nanopore. Both chambers were filled with buffered KCl solution (1M to 2M, as indicated) containing 10 mM Tris, 1 mM EDTA, pH 7.0, and, where indicated, 20% glycerol to increase solution viscosity [29] so as to slow DNA translocation. Except where otherwise stated, only a single DNA sample was added at a time to the cis chamber. A 120 mV bias applied to Ag/AgCl electrodes embedded in each chamber’s solution drove negatively charged DNA molecules through the nanopore from the negatively biased cis chamber to the positively biased trans chamber. As each DNA molecule translocated through the nanopore, it caused a detectable ionic current blockage that was recorded using an Axopatch integrated system (Axopatch 200B, Axon Instruments) with a low pass Bessel filter set at 100 kHz. This current blockage event was characterized by its average current drop (ΔIb = Iopen pore Itranslocating DNA), translocation duration (td), and the event charge deficit, . The voltage, buffer chemical composition, and the molecule itself modulate both ΔIb and td [29]. After each use, and between samples, the chambers were extensively flushed with fresh KCl solution to remove residual DNA. The chambers were considered clean only when no event was recorded from freshly added buffered KCl solution during at least 10 minutes. For each DNA data set the number of recorded events was between 3,000 and 50,000.
Gel Electrophoresis
Electrophoresis was performed in 1% agarose, TAE1X buffer, 6V/cm electric field for 2 hours and the gel was subsequently stained with EtBr and visualized using an image acquisition work station (BioDoc-It, UVP).
DNA
Except where otherwise indicated, all DNA and restriction enzymes were purchased from New England Biolabs. To break apart annealed sticky ends of dsDNAs or base-paired complementary regions of ssDNA, all ssDNA and the ladder DNA samples were heated to 65° C for 5 minutes just prior to being stirred into the cis chamber. 5.4 kbp ΦX174 was purchased as either the supercoiled form (RFI) or the nicked, circular relaxed form (RFII). Linearized ΦX174 double stranded DNA was produce from the RFI form by digestion with Ssp I restriction enzyme followed by electrophoretic purification in a 0.6% agarose gel and recovered by electro elution. Linearized ΦX174 single stranded DNA was produced by annealing the circular single stranded Virion DNA with an oligonucleotide (5′-GAGCACGAGAGCGGTCAGTAGC-3′); Midland Certified Reagents, Midland TX) to create an efficiently digested site that was subsequently cleaved by BsrBI restriction enzyme. The linearized BsrBI digested ΦX174 DNA was purified on a 0.8% agarose gel and recovered by electro elution. The DNA ladder (Lambda hind III phage DNA) was used as purchased.
Analysis of translocation events
The average blockade current, ΔIb, and the translocation time, td, were extracted from the recorded data using custom Matlab® routines. The start of a translocation event was defined as one that caused the nanopore current to drop monotonically below two thresholds; the end of a translocation event was signaled by the current trace climbing monotonically back to the open channel current past both of these thresholds. The event duration, td, was defined by the time between the current drop across the first threshold (~40pA below the baseline) and its rise through the first threshold to the open channel current. The first threshold value was set to be beyond the baseline noise level and about the half peak height of the ΔIb for linear dsDNA which is ~40pA. The second threshold was set to be at 70% of the maximum ΔIb value for linear unfolded dsDNA. Thus short current spikes were not counted as current blockage events. The arithmetic mean of the current blockage value was calculated from the current range between the downward and upward crossings of the second threshold. The event charge deficit, ecd, was defined as the integral of obstructed ionic current over the duration of an event, , in units of kilo electron charge (ke). To correct for baseline drift during an experiment, the average current during 500 microseconds immediately before the current dropped below the first threshold was used to define the open channel current, or baseline, for each event. The errors to the values estimated in this paper are the standard deviations.
Results and Discussion
To verify that a nanopore assay can determine a molecule’s contour length regardless of its conformation, 5.4 kbp ΦX174 DNA was purified in linear, circular relaxed, and supercoiled form (Fig. 1, lanes 1-4); each of the three forms was assayed by adding the DNA to the solution on the cis side of the nanopore. Prior to adding DNA, the nanopore exhibited a stable open pore current. After adding DNA (final concentration ~10 nM), transient decreases of the open pore current were evident. About 10,000 events (~25 min.) were recorded for each form, after which both chambers and the nanopore were washed and similar assays were repeated for each of the two remaining DNA forms (Fig. 2a-c) and finally for a mixture of equal concentrations of all three forms together (Fig. 2d).
Figure 1.

Electrophoresis of DNAs used for the nanopore assays of figures 2. Lanes 1 - 4, 5.4 kbp ΦX174 in (1) supercoiled, (2) linear, and (3) circular relaxed forms; (4) a mixture of supercoiled, linear, and circular relaxed.
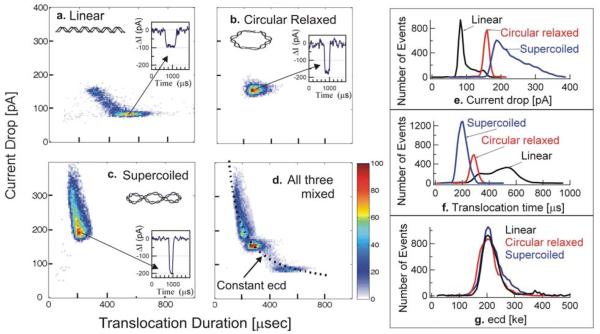
Figure 2.
(a-d) Event plots for the 5.4 kbp DNA at different forms: linear dsDNA (a), circular relaxed (b), supercoiled (c), and a mixture of the three molecular forms (d). The highest density of events in each panel is normalized to 100 and colored red. (e-g) Histograms for the parameters extracted in a-d.: the average blockade current (e), the translocation times (f), and the ecd (g). All measurements were performed using the same 18 nm diameter nanopore in buffered 1M KCl with 20% glycerol added.
Inspection of the distribution of events in the event plots (Fig. 2a-d) showed a clustering of the events. The cluster shapes differed for each conformation. Linear DNA (Fig. 2a) events were clustered within a horizontally distributed group with a broadly distributed td and a narrowly distributed ΔIb.(Fig. 2e, f), and a tailing group extending toward smaller td and higher ΔIb. The former horizontally distributed group represents molecules that were unfolded during translocation, whereas the latter tail represent molecules that were variously folded during translocation [1]. Circular relaxed DNA events were narrowly distributed in a unique, almost circular pattern (Fig. 2b), indicating a narrow distribution of both td and ΔIb (Fig. 2e, f). Supercoiled DNA events were clustered within an almost vertically distributed group (Fig. 2c), with a narrowly distributed td and a broadly distributed ΔIb (Fig. 2e, f).
Whereas the characteristic features of the event plots (Fig. 2a-d) provided qualitative identification of each DNA conformation. Histograms of the events provided clear quantitative data showing that the most probable current blockades produced by each of the 3 conformations clearly differed from each other, with ΔIb = 85 pA for the linear form, 165pA for the circular relaxed form, and 190pA for the supercoiled form (Fig. 2e). But, because the translocation duration time data (Fig. 2f) for each of these forms compliments the current blockade magnitudes to establish the measured time integral of obstructed ionic current, or ecd, this ecd value was the same (~204 ke) for the three identical length 5.4 kbp DNAs regardless of their different conformations (Figs. 2d and 2g). The insensitivity of the ecd to DNA conformation strongly suggested that the ecd value could provide an unambiguous estimate of contour length.
To test the idea that the ecd can measure DNA contour length, a DNA ladder containing a mixture of seven different known lengths of linear double stranded DNAs was translocated through a nanopore. A histogram of the calculated ecd from about 17,500 translocation events showed six ecd peaks, each containing ~2,500 events, except the second peak that contained ~5,000 events (Fig. 3a). This single peak was presumed to represent both the 2,027 bp DNA and the 2,322 bp DNA, whose ecd values were apparently too close to resolve as individual peaks. Plotting the peak values of the ecd as a function of DNA length (Fig. 3b), a nonlinear relation was observed in which ecd = CLβ where L is the DNA length in base pairs, C a fitting constant, and the exponent β was 1.34±0.15. This value is in good agreement with the analogous exponent observed by Storm et al. [22]. More importantly, this experiment demonstrated that ecd can be used as a measure of DNA contour length.
Figure 3.
(a) Histogram of the ecd observed when the DNA ladder, a mixture of 500, 2,027, 2,322, 4,361, 6,557, 9,416, and 23,130 bp linear double stranded DNA was translocated through a nanopore. The experiment was performed in 1.6M buffered KCl and 20% glycerol in a 12 nm diameter pore. (b) ecd as a function of DNA length L(bp). The solid curve is a fit of ecd = C Lβ.
Experiments that compared a linear 5.4 kbp dsDNA and its linear 5.4 kb single stranded analogue demonstrated how ssDNA can be distinguished from dsDNA and how both the number of bases in each form as well as the form of each DNA can be extracted from a single nanopore assay. Because the mean blockade current for the ssDNA was significantly less than the mean blockade current of any dsDNA form (ΔIb(ss)/ΔIb(ds) ≈ 157 pA/228 pA ≈ 0.69) (Fig. 4), this characteristically low blockade current immediately indicated the presence of ssDNA (see also [1]). Consequently, the ecd for the linear 5.4 kb ssDNA was about half that of the 5.4 kbp dsDNA (ecdss/ecdds = 0.49). Thus, when the characteristically low current blockades indicate the presence of ssDNA, the contour length of this ssDNA can be estimated from the ecd value by bearing in mind that this value will be 1/2 that of equal length dsDNA.
Figure 4.

5.4 kb ds and ssDNA measured through a 12 nm pore in 2M buffered KCl. (a) The event plot. (b) The ecd.
To confirm that the ecd can be used as a measure of DNA base number even when several different length unknown DNAs are in different conformations, blind assays were performed on samples prepared by the Harvard University based investigators and shipped, labeled only as samples A through D, but without further descriptions, to the University of Arkansas, where the samples were translocated through several nanopores to determine their conformation and length using the kind of data and analyses depicted in Figs. 2 - 5. The results of these experiments (Table 1) show that all of the assays performed were able to determine the conformation of the unknown DNAs from blockade current measurements and their distribution in event plots, while simultaneously providing approximate estimates of length from ecd measurements.
Figure 5.

Conformation and length determination of an unknown DNA sample “D” from a nanopore experiment. a) The event plot for the DNAs measured. b) ecd of the DNA ladder (●) and sample D (X). The solid curve is a fit of ecd = C Lβ for the DNA ladder. The experiment was performed in 1.6 M buffered KCl solution containing 20% glycerol.
Table 1.
Nanopore-based determination of conformation and contour length by translocation through a 16 nm diameter nanopore; the experiment has been performed in 1.6M KCl at pH 7. The errors represent the standard deviations of the values calculated.
| Sample | Known Conformation and Length | Nanopore determined Conformation and Length |
|---|---|---|
| A | Linear dsDNA, 2.6 kbp | Linear dsDNA, 2.7±0.2 kbp |
| B | Circular relaxed, dsDNA, 10 kbp | Circular relaxed, dsDNA 8.0±3.8 kbp |
| C | Linear ssDNA, 7.2 kb | Linear ssDNA, 6.0±1.7 kb |
| D | Circular relaxed, dsDNA, 7.2 kbp | Circular relaxed, dsDNA, 6.9±0.6 kbp |
As an example of our procedure, a preparation of 7.2 kbp circular relaxed dsDNA, labeled only as “D”, was shipped from Harvard University to the authors in Arkansas for characterization. The sample was characterized (Table 1) by translocation through a 12 nm pore which was calibrated using a sample containing the DNA ladder shown in Figure 3. After recordin~7,000 events from the DNA ladder, the chambers were washed and the unknown sample D was added. After recording ~9.000 sample D events, the recorded events from the calibrating ladder and the unknown sample D were superimposed to produce the event plot of Figure 5a. The event plot shows clusters of events corresponding to the calibrating DNA ladder at ~2.17 kbp (a mixture of 2,027 and 2,322 kbp), 4.36, 6.56, 9.42, and 23 kbp (not shown) and from the unknown DNA sample labelled“D”.
The conformation of sample D as circular relaxed dsDNA was evident from the pattern formed by the sample D events (which cluster into a nearly circular pattern) (Fig. 5a) and from the blockade current measurements (which clustered at a ΔIb value nearly double that of the linear dsDNA used to calibrate the nanopore). To estimate the length of sample D, the ecd data calculated from the DNA ladder measurements were plotted vs. the known lengths of the ladder samples to generate the nonlinear relation showing ecd = CLβ (Fig. 5b), from which C = 26.4±0.7 and β = 1.4±0.1 were obtained. The corresponding ecd peak value for the unknown sample D was ecdD = 392±47 ke from which L for sample D can be calculated as follows:
Discussion and conclusions
Our results show that a nanopore assay can simultaneously evaluate a DNA molecule’s conformation and its base number based on two parameters extracted from the ionic current blockage induced by each DNA molecule translocation through the nanopore: the average blockade current drop or blockade amplitude, ΔIb, and the ecd. The mean current blockage amplitude ΔIb is proportional to the average hydrodynamic cross section of DNA traversing the nanopore. It provides information about the number of DNA strands that are simultaneously traversing the nanopore and the manner in which these strands are folded, as illustrated in Fig. 6. On the other hand, the ecd represents the excluded charge during a translocation event and is the integral of blocked ionic current during the time the entire contour length of the molecule translocates through the nanopore. Thus, observations of blockade amplitude can be used to determine conformation whereas the integral of obstructed ionic current, or ecd, can be used to estimate the number of bases, or the contour length of the molecule.
Figure 6.

Idealized schematic drawings of DNA molecules in different conformations in a nanopore.
When DNA molecules are translocating through a nanopore, if the mean current blockage for a linear (ln) dsDNA is ΔIb(ln), it is expected to be ΔIb(cr) = 2 ΔIb(ln) for a circular relaxed (cr) DNA since its cross section in the pore is twice that (Fig. 6b) of the linear form (Fig. 6a). For a supercoiled DNA (sc), as illustrate in Fig. 6c, ΔIb(sc) is expected to be larger than the ΔIb(cr) and its amplitude depends on the number of superhelical turns. The number of superhelical turns of a DNA can be estimated by comparing the most probable value of ΔIb(sc) with ΔIb(cr). For example, for the 5.4 kbp supercoiled DNA in Fig.2c, the peak value of ΔIb(sc) is 190 pA, it is ΔIb(cr)=165 pA for the circular relaxed DNA in Fig. 2b. Assuming the arc length S of a supercoiled DNA is half the DNA contour length L, S=L/2 = 2.7 kbp ~ 918 nm = S0N, where N is the number of superhelical turns. The diameter of a dsDNA is d = 2.2 nm, and the pitch of a superhelical turn is P (Fig. 6c). For a DNA translocation through a nanopore, ΔIb*td=constant [1], thus the ratio of ΔIb(sc)/ ΔIb(cr)=td(cr)/td(sc). The greater the number of superhelical turns, the shorter are the pitch, P (Fig. 6c), and the translocation time, td. That is, td(sc) ∝ P, or td(cr)/td(sc)=So/P= ΔIb(sc)/ ΔIb(cr). Therefore, we have, S0 / P = 190 pA/165pA = 1.15 . Assuming an idealized geometry as shown in Fig. 6c, , solving for P, P =12.2 nm was calculated. Thus, S0=14.03 nm, so the estimated number of superhelical turns in the 5.4 kbp supercoiled DNA was: N ~ 918/14.03 = 65.4 turns. Using this method, we can estimate the number of superhelical turns for a supercoiled DNA. This estimation needs to be verified in future experiments with a DNA containing a known number of superhelical turns.
Our results show that a nanopore assay can reveal both conformation and base number simultaneously and can, furthermore, do so over an unprecedented dynamic range of DNA contour lengths. Since ecd vs DNA contour length follows a power law, (ecd ~ Lβ), the ca. 1.4 value of β suggests that the resolution of the nanopore method will be better for large molecules, especially for L> 40 kbp, where electrophoresis in gels begins to fail. Nanopore technology is a one-step experiment, much faster (0.5 hour per sample) than electrophoresis, very reliable for discriminating ln, cr and sc DNA (100% reliability). But compared with gel electrophoresis or mass spectrometry, a nanopore provides only a rough estimation of DNA contour length and, as shown in Figure 3, could not resolve between a 2,027 kbp molecule and a 2,322 kbp molecule. Although 2-D gel electrophoresis has much higher resolution in determining the number of superhelical turns [30-32] compared to the nanopore technique, linear dsDNA would migrate together with certain sc isomers in a 2-D electrophoresis [31]. Furthermore, 2-D gels involve a two-step process requiring 24-48 hours to whereas contour length and an estimate of contour length from a nanopore experiment can be available within 30 minutes.
At least two factors could contribute the poor base number estimation. One is the time scale of the DNA length measurement. In gel electrophoresis, the contour length of a DNA is measured in about 103 seconds. In the nanopore method, the length of a DNA is measured for each molecule in less than a second, usually in less than 10−3 seconds. The other factor is that in gel electrophoresis the DNA molecules are in the confining spaces within a gel during the entire measurement time (103 seconds). In a nanopore analysis, less than about a 10 nm length of the DNA is linearized in the confined space of the nanopore channel at any moment in time, whereas the rest of the molecule is entropically undefined and free to adopt any of many conformations.
In this context, it will be of interest in future nanopore experiments to determine if the base pair number of circular DNA molecules -- either supercoiled or relaxed – are better resolved than the contour lengths of linear molecules.
Acknowledgements
We thank Bradley Ledden for nanopore fabrication, Professor Jene A. Golovchenko for assisting FIB hole fabrication. This work is supported by NSF/MRSEC 0080054, ABI-PT06, and NIH 1R21HG003290-03.
References
- [1].Fologea D, Gershow M, Ledden B, McNabb DS, et al. Nano Letters. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rippe K, Hippel P. H. v., Langowski J. TIBS. 1995 December;20:500–506. [Google Scholar]
- [3].Wang JC, Lynch SA. In: Regulation of Gene Expression in Escherichia Coli. Lin ECC, Lynch SA, editors. R. G. Landes Company; Austin, TX: 1996. [Google Scholar]
- [4].Dorman CJ. Trends in Microbiology. 1996;4:214–216. doi: 10.1016/0966-842X(96)30015-2. [DOI] [PubMed] [Google Scholar]
- [5].Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. PNAS. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tomanee P, Hsu JT. Journal of Liquid Chromatography & Related Technologies. 2004;27:1483–1490. [Google Scholar]
- [7].Peter BJ, Arsuaga J, Breier AM, Khodursky AB, et al. Genome Biology. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bae S-H, Yun SH, Sun D, Lim HM, Choi B-S. Nucleic Acids Research. 2006;34:254–261. doi: 10.1093/nar/gkj428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dingman CW, Fisher MP, Kafekuda T. Biochemistry. 1972;11:1242–1250. doi: 10.1021/bi00757a020. [DOI] [PubMed] [Google Scholar]
- [10].Depew RE, Wang JC. PNAS. 1975;72:4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Keller W. PNAS. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Serwer P, Allen JL. Biochemistry. 1984;23:922–927. doi: 10.1021/bi00300a020. [DOI] [PubMed] [Google Scholar]
- [13].Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys. J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc. Natl. Acad. Sci. USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li J, Stein D, McMullan C, Branton D, et al. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- [17].Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. Nature Materials. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- [18].Siwy Z, Dobrev D, Neumann R, Trautmann C, Voss K. Appl. Phys. A. 2003;76:781–785. [Google Scholar]
- [19].Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nature Materials. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- [20].Chen P, Gu J, Brandin E, Kim Y-R, et al. Nano Letters. 2004;4:2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Heng JB, Ho C, Kim T, Timp R, et al. Biophysical Journal. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Storm AJ, Chen JH, Zandbergen HW, Dekker C. Physical Review E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- [23].Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Phys. Rev. Lett. 2003;90:238101–238104. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- [24].Mathe J, Visram H, Viasnoff V, Robin Y, Meller A. Biophys. J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Howorka S, Cheley S, Bayley H. Nature Biotechnology. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- [26].Vercoutere W,S, Winters-Hilt HO,D, Deamer DH, Akeson M. Nature Biotechnology. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- [27].Ashkenasy N, Sanchez-Queasada J, Bayley H, Ghadiri MR. Angew. Chem. Int. Ed. 2005;44:2–5. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stein DM, McMullan CJ, Jiali Li, Golovchenko JA. Review of Scientific Instruments. 2004;75:900–905. [Google Scholar]
- [29].Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hanai R, Roca J. In: DNA Topoisomerase Protocols. Osheroff M.-A. B. a. N., editor. Humana Press; Totowa, New Jersey: 1999. [Google Scholar]
- [31].Lee C-H, Mizusawa H, Kakefuda T. PNAS. 1981:78, 2838–2842. doi: 10.1073/pnas.78.5.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vologodskaia MY, Vologodskii AV. J. Mol. Biol. 1999;289:851–859. doi: 10.1006/jmbi.1999.2811. [DOI] [PubMed] [Google Scholar]