Abstract
By detailed NMR analysis of a human telomere repeating unit, d(CCCTAA), we have found that three distinct tetramers, each of which consists of four symmetric single-strands, slowly exchange in a slightly acidic solution. Our new finding is a novel i-motif topology (T-form) where T4 is intercalated between C1 and C2 of the other duplex. The other two tetramers have a topology where C1 is intercalated between C2 and C3 of the other parallel duplex, resulting in the non-stacking T4 residues (R-form), and a topology where C1 is stacked between C3 and T4 of the other duplex (S-form). From the NMR denaturation profile, the R-form is the most stable of the three structures in the temperature range of 15–50°C, the S-form the second and the T-form the least stable. The thermodynamic parameters indicate that the T-form is the most enthalpically driven and entropically opposed, and its population is increased with decreasing temperature. The T-form structure determined by restrained molecular dynamics calculation suggests that inter-strand van der Waals contacts in the narrow grooves should contribute to the enthalpic stabilization of the T-form.
INTRODUCTION
Telomeres, which exist at the ends of chromosomes, have a large number of repetitive sequences in which one DNA strand typically has G-rich 3′ termini and the counter-strand has C-rich 5′ termini (1). These consecutive G and C clusters are interspersed with short sequences containing adenosine and thymidine residues. For example, in human somatic cells, telomeres have 500–3000 repeats of [5′-d(TTAGGG)-3′]·[5′-d(CCCTAA)-3′] with 130–210 base G overhangs, and the repetitive sequences gradually shorten with age in vivo and in vitro (2). G-rich telomere DNA oligomers take a four-stranded structure with four guanines hydrogen-bonded in the Hoogsteen manner. C-rich telomere sequences also take a unique four-stranded DNA structure, the so-called i-motif (3,4). In a slightly acidic solution, two parallel-stranded duplexes are associated by forming C·C+ base pairs (Fig. 1A), their base pairs are fully intercalated, and the relative orientation of the duplexes is anti-parallel. The i-motif structure is characterized by unusual inter-strand sugar–sugar NOE cross-peaks (H1′–H1′, H1′–H2″, H1′–H4′) which are not detected for a conventional DNA duplex (3,4). Both an inter-molecularly- and an intra-molecularly-folded i-motif were reported for several C-rich oligodeoxynucleotides (5–8), and these intra-molecular i-motifs were found even at neutral pH. In order to investigate the biological role of the C-rich strands in telomeres, protein-binding studies of the C-rich strands have been carried out (9,10), and recently, several nuclear factors have been found to recognize the C-rich strand of the human repeating unit, d(CCCTAA)n (11), suggesting some biological roles of the C-rich repeating unit.
Figure 1.
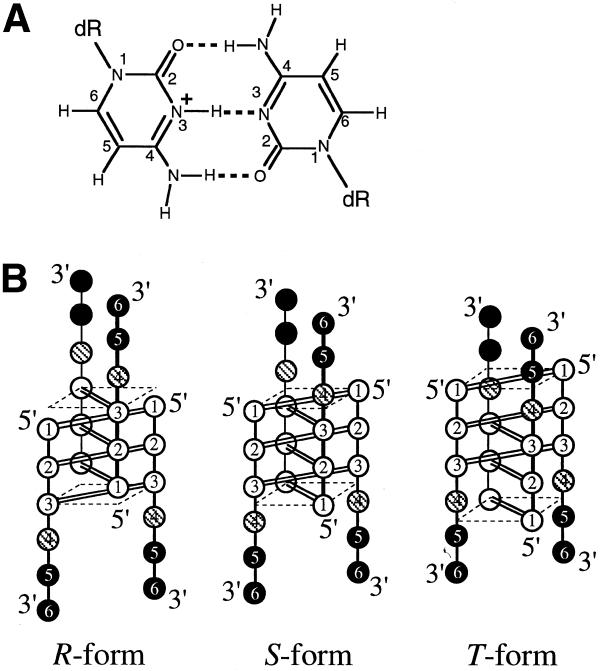
(A) Chemical structure of the C·C+ base pair. dR indicates a deoxyribose moiety. (B) Schematic drawings of NMR-observable i-motif topologies of d(CCCTAA): R-form (left), S-form (middle) and T-form (right). Cytidine, thymidine and adenosine residues are symbolized by open, shaded and closed circles, respectively. C·C+ base pairs are formed in the parallel duplexes for all the topologies, indicated by double lines between open circles, and the plane formed by the outermost C1·C1+ pair is indicated by broken lines.
Detailed i-motif structures have been extensively studied for human telomere repeating units and their analogous sequence. Crystallographic data on d(TAACCC) confirmed the four-stranded folding with a stable loop formed by the 5′ thymidine Hoogsteen-base paired to the third adenosine residue (12). For an analogous sequence, d(CCCAAT), two arrangements of intermolecular A·A·T base triplets were found, involving both asymmetric and symmetric A·A base pairs joined to thymidine residues by Watson–Crick and reverse Hoogsteen base pairing, respectively (13). These structural data suggest that the interaction between the adenosine and thymidine residues attached to consecutive cytidine residues affects the folding topology and stability of the i-motif. Recent NMR studies on intra- or bi-molecular i-motif folding topology showed that i-motif folding highly depends on the loop sequence consisting of A and T residues (14,15).
We have previously reported that short d(CnT) (n = 3 and 4) sequences exhibit two distinct tetramers (each fully symmetric) exchanging slowly on the NMR time-scale although d(TCn) takes a single tetramer topology (16). One tetramer takes an intercalated conformation where the 3′-end thymidine residue is stacked on the C1·C1+ pair of the other duplex (S-form, analogous to the middle structure in Fig. 1B). The other is a tetramer in which one of the duplexes is shifted by one nucleotide unit (R-form, analogous to the structure on the left in Fig. 1B), resulting in non-stacking 3′-end thymidine residues and an equal number of stacked C·C+ pairs to that of the S-form. It is worth noting that the i-motif structure potentially possesses structural diversity and heterogeneity, and that only one thymidine residue at the 5′- or 3′-end changes the i-motif topology and stability. The existence of at least two intra-molecular i-motif isomers was also demonstrated by NMR spectroscopy for a human telomere four-repeat sequence, d[(CCCTAA)3CCC] (6), whose complementary G-rich sequence can fold into an intra-molecularly four-stranded structure (17,18). Investigation of the isomerization provides us with further knowledge about the inter- and intra-molecular i-motif folding.
In the present study, we have explored the i-motif structure of the human telomere repeating sequence, d(CCCTAA), by NMR spectroscopy to examine whether isomeric i-motif conformations can exist for the oligomer and to elucidate an effect of the adenosine residues on the conformational isomerization of the i-motif structure of d(CCCTAA). We have found that the repeating unit of the human telomere can take a new tetrad conformation besides the two conformations observed for d(CCCT) mentioned above, and have discussed the relationship between the i-motif structure and thermal stability.
MATERIALS AND METHODS
Sample preparation
The DNA oligomer, d(CCCTAA), was purchased from Bex (Tokyo, Japan) and purified by reversed-phase liquid chromatography (RPLC) as previously described (19). The purity of the oligomers was checked by RPLC and 1H NMR. The strand concentration used in the following NMR measurements was estimated using extinction coefficients at 260 nm, which were calculated by the nearest-neighbor method (20).
NMR measurements
NMR samples of d(CCCTAA) were prepared in 20 mM acetate buffer (pH 4.5) containing 100 mM NaCl. The strand concentration was 10 mM. The sample was pre-melted at 80°C in an NMR tube prior to measurement and then stored at various temperatures. Attainment of the equilibrium among single- and four-stranded components was confirmed by measuring the time-course of one-dimensional (1D) 1H NMR spectra. Even at low temperatures (∼10°C), the equilibrium was reached within 30 min, and the 1D spectra were unchanged during several two-dimensional (2D) NMR measurements. The equilibrium was also reversible against temperature. The NMR spectra were recorded on Bruker ARX-500 and AMX-750 spectrometers. For the resonance assignment, 2D NMR spectra, DQF-COSY (21,22), HOHAHA (mixing time 40 and 80 ms) (23,24), NOESY (mixing times 100, 200 and 350 ms) (25,26) and 1H-31P HMQC (27) were measured in the temperature range of 10–40°C. For the structure calculation of the T-form, a NOESY spectrum was acquired by applying a short mixing time of 50 ms at 20°C to evaluate accurate NOE cross-peak intensity. Exchangeable protons were assigned in 90% H2O/10% D2O by jump-and-return NOESY (28) at 5–20°C. 1H and 31P chemical shifts were referred to internal sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4 and external 85% H3PO4, respectively.
Restrained molecular dynamics calculation
The distance restraints were obtained from the 50 ms NOESY cross-peak intensities by counting the number of contours, and classified to three classes: strong (1.8–2.9 Å), medium (1.8–3.7 Å) and weak (3.0–4.5 Å). The NOE cross-peaks that could only be observed in the NOESY spectra after longer mixing times were classified as very weak (4.0–6.5 Å), taking account of the spin diffusion effect (3,29). Partly overlapped signals were classified as weak or very weak NOEs. Since some distance restraints could not be obtained from NOESY spectra due to the completely overlapped signals of the C2 and C3 residues, distance restraints of H1′–H1′ and amino–H2′/2″ derived from the typical i-motif structure (29) were introduced into the calculation to maintain the i-motif structure around these central cytidine residues. The force constants for the upper and lower boundaries of the NOE restraints were 30 kcal/mol/Å. The initial structure of the T-form was generated from four B-form single-strands with the T-form topology (structure on the right in Fig. 1B), using the INSIGHT molecular modeling program (Biosym Inc.). Restrained molecular dynamics (rMD) and energy minimization (rEM) calculations included 18 hydrogen-bond distances between cytosine base pairs. The hydrogen bonds were restrained by the length: O4–N4, N4–O4 and N3–N3 distances were set to 2.8 ± 0.15 Å (29). Asymmetric centers of the deoxyriboses were restrained by 72 chirality restraints (S or R) per tetramer besides the NOE restraints. No dihedral restraints were employed in the calculation. A dielectric constant ɛ = 4rij was used for simulating the solvent environment. First, rEM of 2000 steps were carried out, followed by simulated rMD calculations of 6 ps at 500 K (Discover module-Amber force field, Biosym Inc.) with reduced scale factors of some energy terms (0.01 for non-bond, 0.4 for valence angles, 0.1 for torsion angles and 0.1 for out-of-planes) to search conformational space for structures consistent with the NMR restraints. The temperature was gradually reduced from 500 to 300 K in the subsequent cooling step of 6 ps with increasing the scale factors mentioned above up to 1, and then rMD of 4 ps at 300 K and a final rEM calculation of 2000 steps were carried out. The seven lowest total energy structures out of 30 calculated structures were analyzed using the program Curves 5.3 (30). The helical parameters were obtained by determining a global axis for residues of C1–A5.
Analysis of thermodynamic parameters
Concentrations of single- and four-stranded components were determined by integrating corresponding peak areas of 1D spectra measured with a 45° pulse (5 µs), a long relaxation delay of 10 s and 32 scans. Uncertainty in the integration of peak areas was estimated to be ±5–10% based on repetitions of experiments. The equilibrium constant ([tetramer]/[single]4) can be written as Ki = θi/(4C03θu4). [θi and θu are the fraction of a folded oligodeoxynucleotide (i = R-, S- or T-form) and an unfolded single-stranded species, respectively. C0 is the total strand concentration.] The Ki value for each tetramer was determined from the denaturation profiles in the temperature range where both θi and θu values were >0.1. The linearity (correlation coefficient ≥ 0.998) was obtained between the logarithm of the obtained equilibrium constant and the inverse of the temperature value, conforming to the two-state transition model where each tetramer is dissociated to four single-strands. Enthalpy and entropy values were determined for each tetramer by a slope and a y-intercept of the plot of ln(Ki) and 1/T on the basis of the general equation: ln(K) = –(ΔH/R)·(1/T) + (ΔS/R).
RESULTS
Observation of three i-motif conformations of d(CCCTAA)
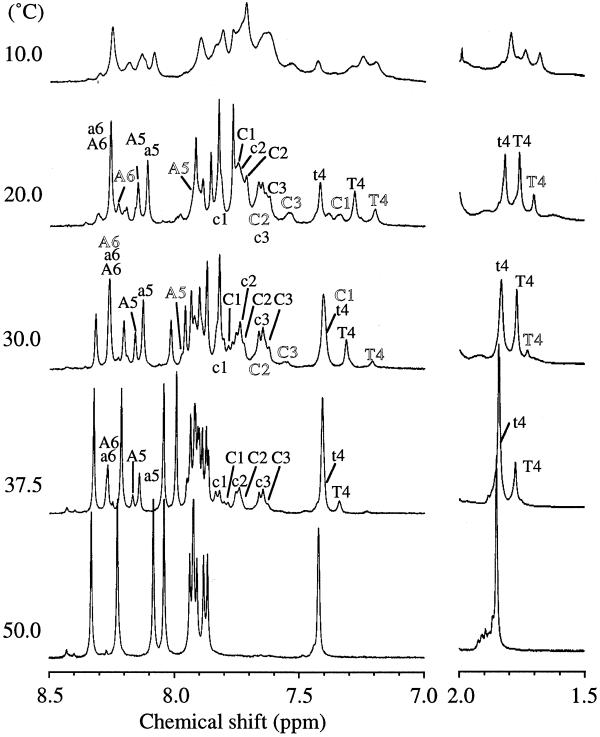
At varied temperatures, 1H NMR measurements were carried out for d(CCCTAA) at the strand concentration of 10 mM, pH 4.5, and the resulting 1D spectra for the aromatic proton region (7.0–8.5 p.p.m.) are depicted in Figure 2 with H8/H6 proton assignment conducted by 2D NMR as described shortly. Each 1D spectrum was collected after the attainment of the equilibration. At temperatures >50°C, only the single-stranded component was observed. With a decrease in temperature, signals arising from folded species appeared besides the single-stranded peaks. At ~40°C, two kinds of signal were clearly observed for all the protons, labeled with lowercase and capital letters with sequence numbers in Figure 2. These two peak sets will, hereafter, be called Peaks-1 and Peaks-2, labeled with lowercase and capital letters, respectively. The population of Peaks-1 was significantly larger than that of Peaks-2. An additional third peak set appeared at <30°C and is labeled with outlined letters in Figure 2. This third peak set will be called Peaks-3. With a further decrease in temperature, these three peak sets showed almost equal intensity.
Figure 2.
Temperature dependence of 1D 1H NMR spectra of the aromatic proton region of d(CCCTAA) in D2O. Conditions: strand concentration, 10 mM, pH 4.5. Lowercase, capital and outlined letters represent residues of R-, S- and T-form, respectively.
Resonance assignment of three i-motif conformations of d(CCCTAA)
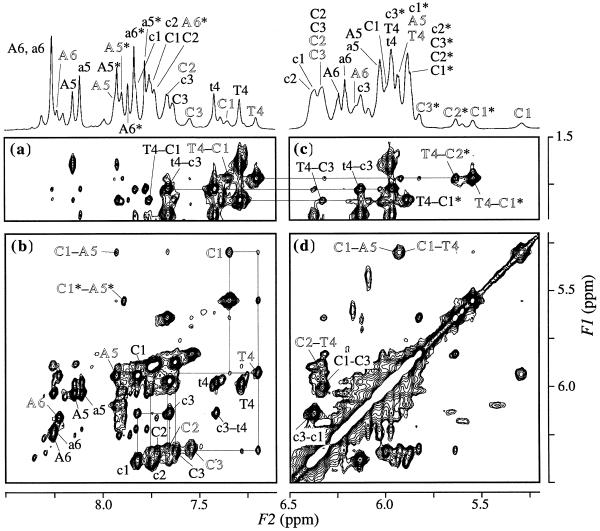
To assign all the signals of single-stranded peaks and the three peak sets, 2D 1H NMR spectra (DQF-COSY, HOHAHA and NOESY) were measured at various temperatures between 10 and 40°C. At first, the single-stranded signals were assigned at 40–50°C and then Peaks-1 and -2 were identified by 2D measurements over the temperature range of 30–40°C. The NOESY spectra obtained at 20°C are shown in Figure 3, where the small population of single-stranded species and the three peak sets are clearly evident. The chemical shifts of the single-strand are listed in Table S1 available as Supplementary Material at NAR Online. The H1′–H1′ NOEs characteristic of i-motif were observed for Peaks-1, -2 and -3 (Fig. 3d), and NOE cross-peaks of amino–H2′/2″ protons were also observed (data not shown), indicating that the peak sets originate from i-motif structures. In addition, no NOE cross-peak was observed among the peak sets even with a long mixing time of 350 ms. These results indicate that the three peak sets arise from three distinct tetramers, each of which consists of four symmetric single-strands, and that these tetramers slowly exchange in solution on the NMR time-scale. The slow exchange of i-motif conformations whose four strands are symmetric was previously reported for the two tetramers of d(CnT) and d(5mCCT) (5mC, 5-methylcytidine) (16,29).
Figure 3.
The 200 ms NOESY spectra for d(CCCTAA) measured at 20°C and pH 4.5 in D2O. The regions of (a) T4 CH3–H8/H6/H2, (b) H1′/H5–H8/H6, (c) T4 CH3–H1′/H5 and (d) H1′/H5–H1′/H5 are displayed with 1D spectra with 1H assignment. Lowercase, capital and outlined letters represent residues of R-, S- and T-form respectively. Asterisks (e.g. C1*, A1*) indicate a cytidine H5 and adenosine H2 proton. Intra-residue NOE connectivities between H1′ and H8/H6 in (B) are indicated by residue names, and inter-residue NOEs are indicated by hyphenated residue names. The rectangular patterns are indicated in (B).
Peaks of T4 CH3 at 1.82, 1.76 and 1.71 p.p.m. were connected to H6 signals at 7.43, 7.28 and 7.20 by NOE cross-peaks, respectively (Fig. 3a). Hence, these H6 peaks in the aromatic region were assigned to T4 H6 protons, which was also confirmed by a weak four-bond cross peak between T4 H6 and CH3 in the HOHAHA and DQF-COSY spectra (data not shown). The H6 signal at 7.43 p.p.m. was completely overlapped with the single-stranded T4 H6 signal in the temperature range of 30–40°C (Fig. 2). By analysis of the NOESY and HOHAHA spectra, each deoxyribose proton of T4 was found to show three distinct peaks. Thus, all the protons of the T4 residue could be classified into the three peak sets.
For the cytidine residues, the resonance assignment of the aromatic protons and H1′ protons is shown in the 1D spectra of Figure 3. The assignment of C1 deoxyribose and base protons started with the most upfield-shifted C1 H5′/H5″ protons, which were confirmed by the absence of the cross-peaks in the 1H-31P HMQC experiments (Fig. S1, see Supplementary Material available at NAR Online), and was unambiguously accomplished by using DQF-COSY, HOHAHA and NOESY spectra. The assignment of C2 and C3 protons was carried out on the basis of the inter-strand NOE cross-peaks with the C1 and T4 protons by assuming the three distinct conformations in Figure 1B. This assignment will be described in detail later. For the adenosine residues, the assignment was carried out based on sequential NOEs of T4 H1′/H2′/H2″–A5 H8 and A5 H1′/H2′/2″–A6 H8. The chemical shift assignment of all the non-labile protons is summarized in Table S1. Consequently, each proton of d(CCCTAA) exhibited three distinct peaks, and it was confirmed that the three peak sets (Peaks-1, -2 and -3) originate from different i-motif conformations. The most prominently shifted peaks were observed for the C1 residues of Peaks-3. The chemical shift values of the C1 H6/H5/H1′ protons of Peaks-3 considerably differed from those of Peaks-1 and -2, both of which are rather similar to each other, suggesting that the i-motif conformation of Peaks-3 is different from that of Peaks-1 and -2.
Tetramer conformation of Peaks-1
For Peaks-1 (lowercase letters in Figs 2 and 3), neither inter-residue H1′–H1′ NOEs nor H1′–H6 NOE cross-peaks in both directions (5′ to 3′ and 3′ to 5′), the so-called ‘rectangular’ pattern, were observed between T4 and any cytidine residues. Strong sequential NOE cross-peaks as seen in conventional DNA duplexes were observed between C3 and T4, such as C3 H1′/H2′/H2″–T4 H6, C3 H6/H5–T4 CH3 (Fig. 3a–c). Furthermore, the H1′–H1′ and rectangular pattern NOEs were observed between the C1 and C3 residues (Fig. 3b and d). The same NOE pattern was reported for the R-form of d(CCCT) (16). In addition to these NOE cross-peaks, the cytidine H1′ and H6 chemical shift (δ) patterns for Peaks-1 were also similar to those observed for the R-form of d(CCCT): δ(C1) ≅ δ(C2) > δ(C3) for H1′ and δ(C1) > δ(C2) > δ(C3) for H6 (16). In particular, the H1′ chemical shifts indicate that the C1 and C2 residues are in a similar environment while C3 is in a different one. In the R-form, the C1·C1+ and C2·C2+ pairs are stacked and buried in the i-motif structure and the C3·C3+ pair is stacked outside of the cluster of the C·C+ pairs, which coincides with the H1′ chemical shift pattern. The fact that the chemical shift of T4 H6 of Peaks-1 was completely degenerated with that of the single-strand in the temperature range of 30–40°C supports the R-form whose T4 residues are not involved in the i-motif. Thus, the topology for Peaks-1 was determined to be the R-form (left in Fig. 1B) whose thymidine residues are not stacked on the cluster of the C·C+ pairs.
Tetramer conformation of Peaks-2
For Peaks-2, the NOESY cross-peak pattern showed the inter-residue H1′–H1′ cross-peaks (Fig. 3d) and rectangular pattern (Fig. 3b) between the C1 and C3 residues. A strong NOE was observed for T4 CH3 and C1 H5, and weak NOEs were observed for T4 CH3–C1/C3 H6 (Fig. 3A). These NOE cross-peaks were previously reported for the S-form defined for d(CCCT) (16). The cytidine H1′ and H6 chemical shift pattern of Peaks-2 [δ(C1) < δ(C2) ≅ δ(C3) for H1′ and δ(C1) > δ(C2) > δ(C3) for H6] was also identical to that observed for the S-form of d(CCCT). It should be noted that the H1′ chemical shift values of the C2 and C3 residues of Peaks-2 are equal to those of the C1 and C2 residues of Peaks-1, as mentioned above. In other words, the C2 and C3 residues of Peaks-2 would be stacked inside the C·C+ cluster. Thus the i-motif topology for Peaks-2 is the S-form where the T4·T4 base pair is stacked on the C1·C1+ pair of the other parallel duplex. Weak sequential NOE cross-peaks were observed for C3 H1′–T4 H6 of the S-form in the NOESY spectra obtained with the long mixing times of 200 and 350 ms (Fig. 3b). Such a weak sequential cross-peak was also previously reported for d(CnT) as well as d(TC5) (3,16). These sequential cross-peaks should be partly due to spin diffusion effects because they were not observed in the NOESY spectra obtained with a shorter mixing time.
Tetramer conformation of Peaks-3
For Peaks-3, the NOESY spectra showed three inter-residue H1′–H1′ cross-peaks of C1–T4, C2–T4 and C1–A5 (Fig. 3d) accompanied by a weak NOE of C1 H1′–T4 H4′. Weak rectangular patterns were also observed for these residue pairs (Fig. 3b). A strong NOE of C1 H5–T4 CH3 and a weak NOE of C2 H5–T4 CH3 were observed (Fig. 3c). These cross-peaks indicate that the T4·T4 base pair is intercalated between covalently bonded C1 and C2 residues of the other duplex, and that Peaks-3 arises from a ‘thymidine’-intercalating i-motif, T-form (right in Fig. 1B). Additional NOE cross-peaks of C1 H5–A5 H2 and C1 H1′–A5 H8, as shown in Figure 3b, demonstrate that the adenine ring of A5 is stacked on the cytosine ring of C1. The chemical shift pattern of the cytidine H1′ and H6 also supports that Peaks-3 comes from the topology of T-form as follows. The C1 H5, H6 and H1′ protons of Peaks-3 were largely shifted upfield by >0.4 p.p.m., as compared to the corresponding protons of the other two peak sets. In particular, the C1 H1′ proton of the T-form was shifted to the upfield by 1.09 and 0.71 p.p.m. as compared to that of Peaks-1 (R-form) and Peaks-2 (S-form), respectively. On the other hand, the H1′ chemical shifts of C2 and C3 of Peaks-3 were almost identical both to those of C1 and C2 for the R-form and to those of C2 and C3 for the S-form. The chemical shift information indicates that the C1 residue of Peaks-3 is located in a considerably different environment from the C2 and C3 residues, which should be stacked inside the i-motif. All the NMR data manifest that Peaks-3 arises from the T-form. The large upfield shifts of the C1 protons should be caused by the large ring current field of the A5 residue, which is supposedly stacked on the C1 residue in the T-form. These largely deviated chemical shift values of the T-form could be an indicator of the formation of the T-form for longer repetitive telomere sequences.
Exchangeable proton assignment
The imino proton of T4 showed a strong broad peak at 11.35 p.p.m. and a weak sharp peak at 11.02 p.p.m. at 10°C, and the former area was almost twice as large as the latter one. A weak NOE cross-peak with the T4 CH3 signal at 1.82 p.p.m. (S-form) was observed for the former peak, and the latter was connected by an NOE cross-peak with the T4 CH3 signal at 1.71 p.p.m. (T-form) (data not shown). No NOE cross-peak was observed for the T4 imino proton of the R-form probably because of fast exchange of the imino proton signal with a water signal. Consequently, the broad peak was assigned to T4 imino protons of the R- and S-form, and the sharp peak was assigned to that of the T-form. For imino protons of C·C+ pairs, the signals of all the three tetramers were severely overlapped in the narrow chemical shift range of 15.50–15.65 p.p.m. The inter-strand cytidine amino–H2′/2″ NOEs are characteristic of i-motif structure, and those characteristic amino–H2′/2″ NOEs were observed in the jump-and-return NOESY spectra of d(CCCTAA). The amino protons of R-form C3 and T-form C1/C2 were unambiguously assigned on the basis of NOE cross-peaks with the separately observed H5 chemical shifts, and these chemical shift values are included in Table S1. However, the internal and external amino protons of the other cytidine residues were overlapped in the narrow ranges of 9.0–9.4 and 8.0–8.3 p.p.m., respectively.
NOE analysis and structure calculation of T-form
The dispersion of the chemical shifts of the T-form enabled us to investigate local structure around the thymidine residue by analyzing the NOESY cross-peaks of the T-form. The schematic representation of inter-residue distances of the T-form from NOESY spectra is shown in Figure S2. As mentioned above, the H1′–H1′ cross-peaks were observed for C1–T4, C1–A5 and C2–T4, and NOESY spectra obtained with the longer mixing times showed the rectangular pattern for these residue pairs. These NOE cross-peaks were commonly observed for i-motif structure (3,29). Weak intra-residue H1′–H6 and strong intra-residue H3′–H6 NOE cross-peaks were observed for the C2, C3 and T4 residues, indicating that the glycosidic angles and sugar puckers for these residues involved in the i-motif structure are the anti conformation and the C3′-endo/O4′-exo, respectively. This intra-residue NOE pattern was also commonly observed for i-motif structures (3,29). However, some NOE cross-peaks that were unusual for the i-motif were detected for the T-form in the 50 ms NOESY spectrum between the C1 and C2 residues of the same strand, such as C1 H3′–C2 H5, accompanying NOE cross-peaks between C1 H3′/H5′/H5″ and T4 CH3 in the wide groove (data not shown). As reported for typical i-motif structures, no NOE was observed between C(i) H3′ and C(i + 1) H5 of the covalently bonded adjacent cytidine residues in the NOESY spectra with short mixing times (29). These NOEs indicate that the H3′ and H5′/H5″ protons of the C1 deoxyribose of the T-form are moved to the wide groove and the phosphate backbone dihedral angles of C1–C2 are largely deviated from typical values of i-motif.
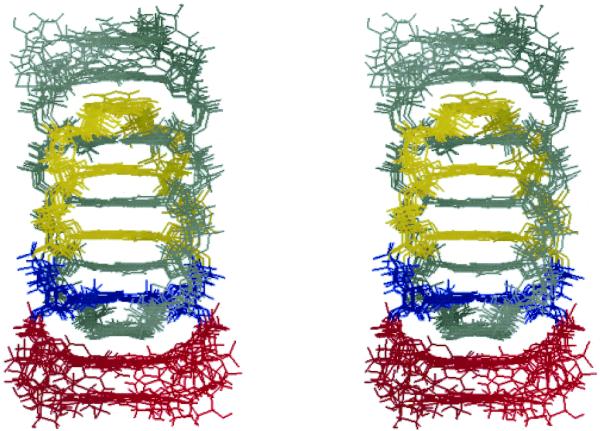
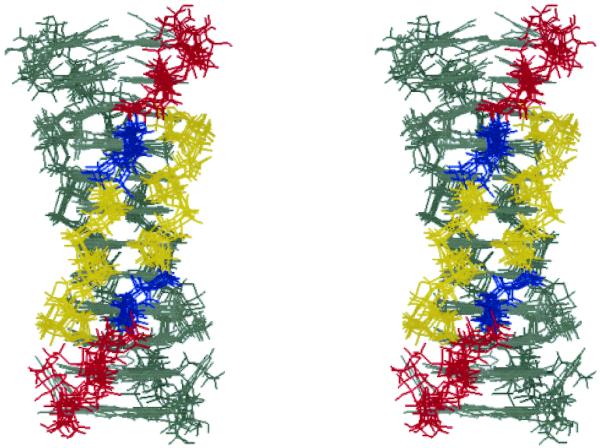
In order to investigate the helical geometry of the T-form, restrained molecular dynamics calculations were carried out and the helical parameters were obtained for the T-form. The molecular dynamics calculations were performed with 300 inter-proton distances (75 per strand). Seven lowest-energy structures out of 30 were chosen for the following structural analysis (Fig. S3). Energy terms of the lowest-energy structures, the largest violation and the number of the violations (>0.1 Å) are collected in Table S2. For all the chosen structures, there were no violations of the hydrogen bonds and chiral restraints, and the largest violation of the intra- and inter-residue NOEs were <0.2 Å. Root mean square deviations (r.m.s.d.) were calculated for each residue from the coordinates of the lowest-energy structures superimposed on the mean coordinate. The r.m.s.d. values per residue were 0.60–0.84 Å for C1–T4, indicating good convergence for these residues, while those for A5 and A6 residues were 1.24 and 2.37 Å, respectively (Table S2). Figures 4 and 5 show a superimposition of the seven lowest-energy structures aligned on the C1′ atoms of C1–A5 residues whose helical parameters were obtained by Curves 5.3 (30). The average ± standard deviation of r.m.s.d. values between each coordinate and the mean coordinate was 1.40 ± 0.13 Å.
Figure 4.
Stereoview of superposition of the seven lowest-energy structures of the T-form viewed from a wide groove. C1′ atoms of C1–A5 residues were superimposed. For one parallel duplex, cytidine residues are in yellow, thymidine in blue and adenosines in red. The other parallel duplex is in gray.
Figure 5.
Stereoview of superposition of the seven lowest-energy structures of the T-form viewed from a narrow groove. C1′ atoms of C1–A5 residues were superimposed. For one anti-parallel duplex, the bases are in gray, and the sugar–phosphate backbones are colored depending on the residue: yellow for cytidine, blue for thymidine and red for adenosine. The other anti-parallel duplex is in gray.
Investigation of the helical parameters provides us with detailed information about the local conformation of the T-form. Glycosidic torsion angles (χ), pseudo rotation angles (P) and selected helical parameters (buckle, propeller twist, rise and helical twist) are given in Table 1. Remarkable effects of the thymidine-intercalation on the geometrical parameters were detected for a large buckle angle for the C1·C1+ pair and a large rise parameter for the C1pC2 step. This result indicates that the exterior C1·C1+ base pair is significantly affected by the thymidine-intercalation and deviated from the ideal geometry of the i-motif. The non-planar C1·C1+ pair suggests that the hydrogen bonds of C1·C1+ are weaker than those of the ordinary C·C+ pairs. It was reported that an average distance of 3.1 Å between the adjacent stacked C·C+ pairs along the axis is shorter than that of DNA duplexes (3.4 Å) and that the cytosine rings never stack on top of other cytosine rings in the i-motif structure (31). The obtained rise parameter of 6.9 Å at the C1pC2 step means that the thymine ring of T4 is partly overlapped with the cytosine rings of C1 and C2. Sugar puckering of C1 was not converged, which is also different from an ideal i-motif puckering of C3′-endo/C4′-exo observed for C2, C3 and T4. Helical twist values at the C1pC2 and C3pT4 steps were slightly larger, as compared to reported typical twist values of 16–20° (32).
Table 1. Geometrical parameters of the T-form of d(CCCTAA)a.
| Base |
χ (°) |
P (°) |
Buckle (°) |
Propeller twist (°) |
| C1 | –146 ± 10 | 131 ± 50 | 29 ± 8 | –175 ± 7 |
| C2 | –125 ± 8 | 58 ± 6 | 3 ± 3 | –168 ± 3 |
| C3 | –123 ± 4 | 53 ± 12 | 4 ± 3 | –177 ± 3 |
| T4 | –128 ± 11 | 31 ± 14 | –2 ± 5 | –180 ± 4 |
| A5 |
–105 ± 40 |
100 ± 45 |
–8 ± 12 |
136 ± 58 |
| Step |
Rise (Å) |
Helical twist (°) |
|
|
| C1–C2 | 6.9 ± 0.4 | 25 ± 4 | ||
| C2–C3 | 6.1 ± 0.2 | 19 ± 5 | ||
| C3–T4 | 6.4 ± 0.3 | 27 ± 3 | ||
| T4–A5 | 6.1 ± 0.2 | –8 ± 10 |
aThe parameters were computed with the Curves 5.3 program (30), using the definitions therein. The values are the average ± standard deviation obtained for the seven lowest-energy structures: The χ and P parameters were obtained from 28 deoxyriboses, and the other helical parameters were from 14 parallel duplexes.
Denaturation profile and thermodynamic parameters of the three tetramers
In order to estimate the i-motif stability of [d(CCCTAA)]4, the denaturation profiles of the three i-motif structures were obtained by plotting the change of the peak areas in the 1D NMR spectra against various temperatures (Fig. S4). The peak areas of aromatic signals were used as markers for estimating the populations of the three forms. At low temperatures, the three conformations, R-, S- and T-form showed almost equal signal intensity although we could not precisely determine the intensity because of signal broadening. With an increase in temperature, the intensity of the R-form increased up to 25°C and then decreased, while the intensity of the S- and T-forms showed a constant decrease. In particular, the population of the T-form decreased more rapidly than that of the S-form and, above 40°C, the signals of the T-form were diminished. Apparent melting temperature (Tm) of [d(CCCTAA)]4 was determined by summing the three tetrad components and comparing the sum with the fraction of the single-strand. The Tm value obtained was 36°C at the strand concentration of 10 mM. This value was slightly higher than that of the previously reported value of 32°C for [d(CCCT)]4 (16).
For [d(CCCTAA)]4, the R-form was found to be the most stable of the three conformations in the temperature range of 15–50°C, the S-form the second and the T-form the least. With decreasing temperature, however, the population of the T-form conformation was increased. The denaturation profiles of these tetramers were studied thermodynamically in detail. To obtain the thermodynamic parameters (ΔH° and ΔS°), the equilibrium constants (K) between each i-motif conformation and four single-strands were calculated, and ln(K) was plotted against 1/T for each tetramer. The plots yielded straight lines for all the three tetramers, although a substantial ΔCp could not be ruled out due to the narrow temperature range, indicating the two-state transition model between each tetramer and the single-strands. Note that previously, the two-state transition model has been adopted successfully to explain the denaturation profile of the i-motif (5–7,16). According to the method described in Materials and Methods, ΔH° and ΔS° values of the formation of each i-motif structure from four single-strands were determined for all the topologies on the basis of the two-state transition model (summarized in Table 2). Both the ΔH° and ΔS° values of the R-form were found to be the least negative of all the three forms, while those values of the T-form were the most negative, and the S-form showed ΔH° and ΔS° values in between the R- and T-forms. The standard deviation of ΔG° values was larger than differences of the ΔG° values (Table 2). However, the population differences between the three conformations were reproducible in the repetitive measurements, and the differences of the ΔG° values (ΔΔG°) was significant (±0.2 kJ/mol). The large standard deviation of ΔG° was caused by the integration of the single-stranded species. The largest ΔΔG° value between the three forms was 2.4 kJ/mol at 20°C and <0.5 kJ/mol at 10°C. These values were consistent with the NMR observation that the three i-motif structures comparably exist with decreasing temperature. Thus it is deduced that the small negative TΔS° contributes to the stabilization of the R-form. On the other hand, the enthalpic term contributes to the stabilization of the T-form at low temperatures and, with increasing temperature, the entropic term takes part in the destabilization of the T-form.
Table 2. Thermodynamic parameters of the three i-motif structures of d(CCCTAA)a.
| |
ΔH° (kJ/mol–1)a |
ΔS° (J/K/mol)a |
ΔG° (kJ/mol)b |
|
R-form |
–285 |
–808 |
–48.6 |
|
S-form |
–313 |
–904 |
–47.9 |
| T-form | –343 | –1013 | –46.2 |
aCalculated by the NMR denaturation profile. Standard deviations of ΔH° and ΔS° values are ±13 kJ/mol and ±34 J/mol/K, respectively.
bCalculated at 20°C. The standard deviations of ΔG° and ΔΔG° (see text) are ±1.3 and 0.2 kJ/mol, respectively.
DISCUSSION
The present NMR study on d(CCCTAA) has shown that the three i-motif structures, R-, S- and T-forms (Fig. 1B), exist at equilibrium in solution. The intercalation topologies of the R- and S-forms were previously observed for d(CCCT) (16), while the topology of the T-form conformation has only been observed for d(CCCTAA). Since the T-form was not observed for d(TCm) and d(CnT) (m = 2–5, n = 3–4) (3,4,16,29), the internal stacking of thymidine residues was not considered to occur at either the 3′- or 5′-end of the i-motif structure. However, the present observation of the T-form indicates that the addition of the adenosine residue to the 3′-end of d(CCCT) largely stabilizes the T-form conformation as compared to the other two conformations and decreases the free energy differences between the conformations. Energy differences as small as 2.4 kJ/mol at 20°C enable us to observe the T-form by NMR spectroscopy. The denaturation profiles of the three d(CCCTAA) tetramers have shown that the T-form is less stable in the observed temperature range than the other two conformations (Figs 2 and S3). However, with decreasing temperature, the population of the T-form is increased by the contribution of the enthalpic energy term because the ΔH° value of the T-form is more negative than that of the R- and S-forms. What contributes to the large negative ΔH° value of the T-form? Generally, the i-motif can be stabilized by hydrogen bonding between cytosine and protonated cytosine, by base stacking between C·C+ pairs or between C·C+ and T·T/A·A pairs and by inter-molecular van der Waals contacts between the deoxyribose moieties of the anti-parallel strands in the narrow groove of i-motif. As compared to the R- and S-form, the topology of the T-form is characteristic in terms of the greater occurrence of base stacking, which results in many van der Waals contacts between the deoxyriboses in the narrow groove. The H1′–H1′ NOE cross-peaks of C1–T4, C2–T4 and C1–A5 (Fig. 3d) indicate that the interaction between the sugar–phosphate backbones in the narrow groove takes place in the wide range. Since these inter-molecular van der Waals contacts are considered to make the i-motif structure highly stable (3,16,33), the large negative ΔH° value of the T-form probably arises from the van der Waals contacts of the sugar–phosphate backbones in the narrow groove, leading to the stabilization of the T-form topology. Thus far, formation of the i-motif has also been confirmed by NMR and X-ray crystallography for many short C-rich oligodeoxynucleotides (3,4,12–14,29,31,34). In a number of these structural studies, most of the determined i-motif structures were found to form a single structure of a fully intercalated i-motif. Our previous study on d(CnT) demonstrated the equilibrium between the two different i-motif topologies of the R- and S-form in solution, indicating that the position of the thymidine residues is highly responsible for the occurrence of the two topologies (16). The current observation of the equilibrium between the three topologies clearly indicates that the number and position of T and A residues change the stability and structures of i-motif, and suggests that multiple i-motif structures including an unusual topology could occur for assembly of longer C-rich oligomers.
Another characteristic feature of [d(CCCTAA)]4 is that its S-form is less stable than the R-form in the temperature range of 15–50°C (Fig. S4). For the two [d(CCCT)]4 (R- and S-form), it was reported that the S-form is more stable than the R-form in the same temperature range (16). The differences in ΔH° and ΔS° between the R- and S-form [ΔΔH°S–R = ΔH° (S-form) – ΔH° (R-form) and ΔΔS°S–R = ΔS° (S-form) – ΔS° (R-form)] are negative for both the d(CCCT) and d(CCCTAA). These S-forms possess the same number of hydrogen bonds and base stacking between C·C+ pairs as the R-forms and, in addition, the T4·T4 base pair stacking on the C1·C1+ pair of the other duplex. The stacking of the T4·T4 pair and the presence of inter-strand van der Waals contacts between C1 and T4 are probably reflected by the negative ΔΔH°S–R values. The negative ΔΔS°S–R values for both d(CCCTAA) and d(CCCT) indicate that a ratio of the S-form to the R-form is increased with decreasing temperature. Although the ΔΔH°S–R and ΔΔS°S–R values have the same sign for both the sequences, ΔΔG°S–R [ΔG(S-form) – ΔG° (R-form)] values are positive and negative for d(CCCTAA) and d(CCCT) in the observed temperature range, respectively. In other words, the relative stability between the R- and S-form is changed by the addition of the adenosine residues at the 3′-terminus. A possible mechanism for the reversed stability is due to the van der Waals repulsion between the adenosine rings of the A5 residues, which are located outside of the T4·T4 pair of the same parallel duplex. Since the size of an adenosine ring is larger than that of a thymine ring, the stacking of A5 on T4 may weaken the enthalpic stabilizing factors of the S-form mentioned above, such as stacking of the T·T pair on the C1·C1 pair and the inter-strand van der Waals contacts between C1 and T4. The fact that addition of the adenosine residues to the 3′-end of d(CCCT) changes the relative stability between the R- and S-form indicates that inter- and intra-molecular interactions between the adenosine and thymidine residues influence i-motif folding topology and stability, and suggests that most common i-motif conformation in solution can be altered easily by external factors such as temperature, pH, salt concentration and, possibly, existence of proteins. The influence of the interactions between adenosine and thymidine residues on the i-motif folding may be more prominent for intra-molecular i-motif folding where the mutual position of adenosine and thymidine residues are fixed in the loops (14).
In the present study, we have demonstrated the conformational isomerization of the human telomere repeating unit and the small energetic difference between the isomeric structures, which is changed by temperature. A fragment of the C-rich strand of human telomeres containing four oligodeoxycytidine stretches, d[(CCCTAA)3CCC], has been reported to form an i-motif by intra-molecular folding at neutral pH and to have at least two intra-molecularly-folded isomers (6). Thus, the isomerization of i-motif structures probably occurs in C-repeats in telomeres if the C-rich 3′ termini take an i-motif structure during recombination, meiotic chromosome pairing or other genomic processes. The i-motif isomerization is of interest because it is feasible that the i-motif structure may not only work as a rigid structural element but may also regulate biological function together with the G-tetrad formation in the counter strand and related proteins. In addition, since the exchange between the isomers is very slow with increasing length of C-rich oligonucleotides (16), a minor conformation may occur not only thermodynamically but also kinetically. Since nuclear proteins that recognize the d(CCCTAA)n telomeric motif have been identified (9–11), the biological reason for the i-motif may also be proved in the near future. Recently, it was reported that the rapid shortening of human telomeres, which limits the proliferative capacity of most human cells, may not be caused by the incomplete lagging-strand replication but by the degradation of a 5′ C-rich strand (35). Also, lengths of G and C strand during synthesis of new telomere are coordinately regulated and a major regulatory step occurs in the C strand synthesis (36). These biological results about regulation of the C strand in telomeres suggest that the formation and isomerization of i-motif might play a relevant biological role in the regulatory processes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. Kazuyuki Akasaka for the use of the NMR facility. This work was supported by a Grant-in-Aid for Encouragement of Young Scientists from Japan Society for the Promotion of Science, to K.K. (11780426), a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, to K.M. (No. 11101001) and by a grant from the ‘Research for the Future’ program of the Japan Society for the Promotion of Science, to K.M. (JSPS-RFTF97I00301).
References
- 1.Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 2.Harley C.B. and Villeponteau,B. (1995) Telomeres and telomerase in aging and cancer. Curr. Opin. Genet. Dev., 5, 249–255. [DOI] [PubMed] [Google Scholar]
- 3.Gehring K., Leroy,J.L. and Guéron,M. (1993) A tetrameric DNA structure with protonated cytosine·cytosine base pairs. Nature, 363, 561–565. [DOI] [PubMed] [Google Scholar]
- 4.Leroy J.L., Gehring,K., Kettani,A. and Guéron,M. (1993) Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry, 32, 6019–6031. [DOI] [PubMed] [Google Scholar]
- 5.Manzini G., Yathindra,N. and Xodo,L.E. (1994) Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res., 22, 4634–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leroy J.L., Guéron,M., Mergny,J.L. and Hélène,C. (1994) Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res., 22, 1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mergny J.L., Lacroix,L., Han,X., Leroy,J.L. and Hélène,C. (1995) Intramolecular folding of pyrimidine oligodeoxynucleotides into an i-DNA motif. J. Am. Chem. Soc., 117, 8887–8898. [Google Scholar]
- 8.Ahmed S., Kintanar,A. and Henderson,E. (1994) Human telomeric C-strand tetraplexes. Nat. Struct. Biol., 1, 83–88. [DOI] [PubMed] [Google Scholar]
- 9.Marsich E., Piccini,A., Xodo,L.E. and Manzini,G. (1996) Evidence for a HeLa nuclear protein that binds specifically to the single-stranded d(CCCTAA)n telomeric motif. Nucleic Acids Res., 24, 4029–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsich E., Xodo,L.E. and Manzini,G. (1998) Widespread presence in mammals and high binding specificity of a nuclear protein that recognises the single-stranded telomeric motif (CCCTAA)n. Eur. J. Biochem., 258, 93–99. [DOI] [PubMed] [Google Scholar]
- 11.Lacroix L., Liénard,H., Labourier,E., Djavaheri-Mergny,M., Lacoste,J., Leffers,H., Tazi,J., Hélène,C. and Mergny,J.-L. (2000) Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res., 28, 1564–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang C.H., Berger,I., Lockshin,C., Ratliff,R., Moyzis,R. and Rich,A. (1995) Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere. Proc. Natl Acad. Sci. USA, 92, 3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger I., Kang,C., Fredian,A., Ratliff,R., Moyzis,R. and Rich,A. (1995) Extension of the four-stranded intercalated cytosine motif by adenine·adenine base pairing in the crystal structure of d(CCCAAT). Nat. Struct. Biol., 2, 416–425. [DOI] [PubMed] [Google Scholar]
- 14.Han X., Leroy,J.L. and Guéron,M. (1998) An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCT3CCT3ACCT3CC). J. Mol. Biol., 278, 949–965. [DOI] [PubMed] [Google Scholar]
- 15.Nonin S., Phan,A.T. and Leroy,J.L. (1997) Solution structure and base pair opening kinetics of the i-motif dimer of d(5mCCTTTACC): a noncanonical structure with possible roles in chromosome stability. Structure, 5, 1231–1246. [DOI] [PubMed] [Google Scholar]
- 16.Kanaori K., Maeda,A., Kanehara,H., Tajima,K. and Makino,K. (1998) 1H nuclear magnetic resonance study on equilibrium between two four-stranded solution conformations of short d(CnT). Biochemistry, 37, 12979–12986. [DOI] [PubMed] [Google Scholar]
- 17.Kang C.H., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Crystal structure of four-stranded Oxytricha telomeric DNA. Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- 18.Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 19.Kanehara H., Mizuguchi,M., Tajima,K., Kanaori,K. and Makino,K. (1997) Spectroscopic evidence for the formation of four-stranded solution structure of oligodeoxycytidine phosphorothioate. Biochemistry, 36, 1790–1797. [DOI] [PubMed] [Google Scholar]
- 20.Fasman G.D. (1975) Handbook of Biochemistry and Molecular Biology, 3rd Edn. CRC Press, Cleveland, OH.
- 21.Piantini U., Sørensen,O.W. and Ernst,R.R. (1982) Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc., 104, 6800–6801. [Google Scholar]
- 22.Rance M., Sørensen,O.W., Bodenhausen,G., Wagner,G., Ernst,R.R. and Wüthrich,K. (1983) Improved spectral resolution in COSY proton NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun., 117, 479–485. [DOI] [PubMed] [Google Scholar]
- 23.Braunschweiler L. and Ernst,R.R. (1983) Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson., 53, 521–528. [Google Scholar]
- 24.Davis D.G. and Bax,A. (1985) Assignment of complex proton NMR spectra via two-dimensional homonuclear Hartmann-Hahn spectroscopy. J. Am. Chem. Soc., 107, 2820–2821. [Google Scholar]
- 25.Jeener J., Meier,B.H., Bachmann,P. and Ernst,R.R. (1979) Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys., 71, 4546–4553. [Google Scholar]
- 26.Macura S., Huang,Y., Suter,D. and Ernst,R.R. (1981) Two-dimensional chemical exchange and cross-relaxation spectroscopy of coupled nuclear spins. J. Magn. Reson., 43, 259–281. [Google Scholar]
- 27.Blommers M.J.J., van de Ven,F.J.M., van der Marel,G.A., van Boom,J.H. and Hilbers,C.W. (1991) The three-dimensional structure of a DNA hairpin in solution two-dimensional NMR studies and structural analysis of d(ATCCTATTTATAGGAT). Eur. J. Biochem., 201, 33–51. [DOI] [PubMed] [Google Scholar]
- 28.Plateau P. and Guéron,M. (1982) Exchangeable proton NMR without base-line distortion, using strong pulse sequences. J. Am. Chem. Soc., 104, 7310–7311. [Google Scholar]
- 29.Leroy J.L. and Guéron,M. (1995) Solution structures of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connections between amino protons and sugar protons. Structure, 3, 101–120. [DOI] [PubMed] [Google Scholar]
- 30.Lavery R. and Sklenar,H. (1989) Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 31.Kang C.H., Berger,I., Lockshin,C., Ratliff,R., Moyzis,R. and Rich,A. (1994) Crystal structure of intercalated four-stranded d(C3T) at 1.4 Å resolution. Proc. Natl Acad. Sci. USA, 91, 11636–11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger I., Cai,L., Chen,L. and Rich,A. (1997) Energetics of the lattice: packing elements in crystals of four-stranded intercalated cytosine-rich DNA molecules. Biopolymers, 44, 257–267. [DOI] [PubMed] [Google Scholar]
- 33.Berger I., Egli,M. and Rich,A. (1996) Inter-strand C-HºO hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl Acad. Sci. USA, 93, 12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Cai,L., Zhang,X. and Rich,A. (1994) Crystal structure of a four-stranded intercalated DNA: d(C4). Biochemistry, 33, 13540–13546. [DOI] [PubMed] [Google Scholar]
- 35.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 36.Fan X. and Price,C.M. (1997) Coordinate regulation of G- and C strand length during new telomere synthesis. Mol. Biol. Cell, 8, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.