Abstract
Background
Nicotine induces the proliferation of non–small cell lung cancer (NSCLC) cells via nicotinic acetylcholine receptors and the arrestin, β1 (ARRB1) protein. However, whether ARRB1 translocates to the nucleus upon nicotinic acetylcholine receptor activation and how it regulates growth of human NSCLCs are not known.
Methods
We investigated nuclear localization of ARRB1 in human NSCLC cell lines (A549 and H1650), normal lung cell lines (NHBE and SAEC), and lung cancer tissue microarray. A549 cells were transfected with ARRB1-specific short hairpin RNA (A549-sh) to knockdown ARRB1 expression, or with empty vector (A549-EV), to examine the role of ARRB1 in the mitogenic and antiapoptotic effects of nicotine, binding of ARRB1 to E2F transcription factors, and the role of ARRB1 in nicotine-induced expression of E2F-regulated survival and proliferative genes cell division cycle 6 homolog (CDC6), thymidylate synthetase (TYMS), and baculoviral IAP repeat–containing 5 (BIRC5). Real-time polymerase chain reaction was performed for quantitative analysis of mRNA expression. Chromatin immunoprecipitation assays were performed on A549 cells and fresh-frozen human NSCLC tumors (n = 8) to examine the binding of ARRB1, E1A binding protein (EP300), and acetylated histone 3 (Ac-H3) on the E2F-regulated genes. All statistical tests were two-sided.
Results
Nicotine induced the nuclear translocation of ARRB1 in NSCLC and normal lung cells, and lung tumor tissues from smokers showed an increased nuclear localization. The mitogenic and antiapoptotic effects of nicotine were reduced in A549-sh cells. Nuclear ARRB1 bound to E2F transcription factors in normal lung cells, NSCLC cells, and tumors. Nicotine treatment induced a statistically significant increased expression of E2F-regulated genes in A549-EV but not in A549-sh cells; the maximum difference being observed in BIRC5 (A549-EV vs A549-sh, mean fold-increase in mRNA level upon nicotine treatment = 20.7-fold, 95% confidence interval = 19.2- to 22.2-fold, vs mean = 0.8-fold, 95% confidence interval= 0.78- to 0.82-fold, P < .001). Furthermore, nicotine induced the binding of ARRB1, EP300, and Ac-H3 on E2F-regulated genes.
Conclusion
Nicotine induced the nuclear translocation of ARRB1 and showed increased expression of proliferative and survival genes, thereby contributing to the growth and progression of NSCLCs.
CONTEXT AND CAVEATS
Prior knowledge
ARRB1 has been shown to have a role in invasion and proliferation of many cancers, including nicotine-induced proliferation of human non–small cell lung cancers (NSCLCs). Whether ARRB1 translocates to the nucleus and the mechanism of regulation of cell proliferation are not known.
Study design
Expression and nuclear localization of ARRB1 in NSCLC cell lines, normal lung cells, microarrays, and human NSCLC tumors were investigated. Knockdown of ARRB1 expression was performed to study its role in nicotine-induced cell proliferation and protective effect against apoptosis. Genes involved in ARRB1-mediated regulation of these functions were identified via DNA-protein binding experiments.
Contribution
ARRB1 translocated to the nucleus on induction with nicotine and regulated genes involved in cell survival and proliferation.
Implications
Nicotine-induced proliferation of human NSCLCs is regulated by ARRB1 and may be involved in growth and metastasis of NSCLCs, particularly in tobacco smokers.
Limitations
There could be other mechanisms involved in nicotine-induced survival and proliferation of NSCLCs. Also, other genes that may be regulated by ARRB1 are not shown in this study.
From the Editors
Non–small cell lung cancer (NSCLC) accounts for 80% of all lung cancer cases and demonstrates a strong association with tobacco use (1,2). Nicotine, the psychoactive and addictive component of tobacco, has been shown to induce cell proliferation, angiogenesis, epithelial to mesenchymal transition, and metastasis of NSCLCs through nicotinic acetylcholine receptors (nAChRs) (3–6). Furthermore, nicotine demonstrates antiapoptotic properties in NSCLC cells in vitro (5,7,8). Tobacco smoke is associated with 60% of all reported NSCLCs (1), suggesting that tobacco components like nicotine and its derivatives contribute to signaling pathways involved in the growth and progression of human NSCLCs.
Several convergent studies have shown that the alpha (α) and beta (β) subunits of nAChR have potential tyrosine phosphorylation sites (9–11), and cellular v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) (SRC) may have a role in the tyrosine phosphorylation of nAChR subunits in chicken myoblasts (8). Nicotinic receptors are ion-channel receptors with no inherent tyrosine kinase activity in their transmembrane domains (12–14). Therefore, an important question that emerged was how the binding of nicotine to nAChRs caused the activation of SRC. We recently found that the binding of nicotine to nAChRs leads to the formation of an oligomeric complex between nAChR, SRC, and arrestin, β1 (ARRB1), which was vital for nicotine-induced proliferation of human NSCLCs (15).
In mammals, the arrestin family has four members (16,17)—ARRB1 (also known as arrestin-2), ARRB2 (also known as arrestin, β2, or arrestin-3), ARRB3 (also known as retinal X-arrestin or arrestin-4), and SAG (S-antigen; also known as arrestin-1). ARRB1 and ARRB2 are ubiquitous, multifunctional, scaffolding proteins that are involved in the termination or desensitization of signals arising from activated G-protein-coupled receptors (GPCRs) (18). Besides being scaffolding proteins for GPCRs, ARRB1 and ARRB2 regulate structurally diverse receptors like Notch, endothelin A receptor, frizzled, smoothened, and the nicotinic cholinergic receptors (15,19–23). ARRB1 also regulates multiple intracellular signaling proteins involved in cell proliferation and differentiation, such as SRC, mitogen-activated protein kinases, alpha regulatory subunit A of protein phosphatase 2 (PP2R1A) (protein phosphatase 2, regulatory subunit A, alpha), and components of the wingless-type MMTV integration site family member (WNT) signaling pathway (21,24,25). ARRB1 and ARRB2 also facilitate receptor ubiquitination and regulate chemotaxis mediated by the chemokine (C-X-C motif) receptor 4 (CXCR4) (20,26–29).
Emerging evidence suggests that ARRB1 and ARRB2 can translocate to the nucleus in response to opioid peptides (30,31). The activation of GPCR-delta (δ) and kappa (κ) opioid receptors by enkephalin-derived peptides like the delta peptide [D-Ala2,D-Leu5]Enkephalin has been shown to induce translocation of ARRB1 to the nucleus where it bound to specific promoters of genes like cyclin-dependent kinase inhibitor 1B (CDKN1B) and FBJ murine osteosarcoma viral oncogene homolog (FOS) in human embryonic kidney (HEK) 293 cells (30,31).
Furthermore, nuclear ARRB1 facilitates the recruitment of the E1A binding protein EP300 (also known as histone acetyltransferase p300) on the promoters of CDKN1B and FOS, and results in an increased acetylation of histone H4, reorganization of chromatin, and increased transcription of CDKN1B and FOS (30,31). Nuclear ARRB1 suppresses the activation of nuclear factor kappa B and functions as corepressors of the androgen receptor in prostate cancer (32). In addition, ARRB1 has been shown to promote multiple steps of cancer progression including invasion and metastasis in breast, ovarian, and colon cancers, and skin papillomas (20,26,27,29,33). Because ARRB1 does not contain a discernible DNA-binding domain (16,34), it is probable that it binds to the CDKN1B and FOS promoters in an indirect manner via binding to transcription factors that directly bind to CDKN1B and FOS promoters.
Our previously published data showed that proliferative signaling via the nAChRs induced the dissociation of E2F transcription factor 1 (E2F1) from retinoblastoma 1 (RB1) protein (15). The E2F family of transcription factors plays a central role in regulating cell cycle progression, and the E2F-RB1 pathway is dysregulated in about 90% of lung cancers (35,36). The inactivation of RB1 by mutation of the gene or phosphorylation causes it to dissociate from E2F1. The free E2F1 then binds to specific E2F-binding sites on the promoters of proliferative genes like cell division cycle 6 homolog (CDC6), cell division cycle 25 homolog A (CDC25A), thymidylate synthetase (TYMS), and cyclin E1 (CCNE1) and stimulates the transcription of genes, leading to S-phase entry (35–37). We further observed that nicotine induced the binding of E2F1, E2F2, and E2F3 transcription factors to the promoters of proliferative genes like CDC6 and CDC25A, whereas dissociation of RB1 from the E2F transcription factors, as well as the above-mentioned promoters, resulted in promoter activation (15).
It is not known whether the binding of nicotine to nAChRs cause nuclear localization of ARRB1 in human NSCLC cells. Because the E2F family of transcription factors plays a vital role in nAChR signaling, it is probable that ARRB1 interacts with the E2F transcription factors to regulate nicotine-induced cell proliferation and cell survival in human NSCLCs. In this study, we investigated the role of nuclear ARRB1 in mitogenic and antiapoptotic functions of nicotine in survival and proliferation of human NSCLC cells.
Patients, Materials, and Methods
Cell lines, Cell Culture, and Reagents
Human lung adenocarcinoma epithelial cell lines A549 and H1650 were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Adenocarcinomas are a subset of NSCLC cells. The HEK293 cell line was purchased from the ATCC and maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 2 mM glutamine, 100 U/mL penicillin, 50 μg/mL streptomycin, and 10% fetal bovine serum (FBS). DMEM and all supplements were purchased from Mediatech, Inc (Manassas, VA), except for FBS, which was purchased from Hyclone Laboratories (Rockford, IL). A549 cells were maintained in Ham's F-12K medium (Mediatech, Inc) supplemented with 2 mM glutamine, 100 U/mL penicillin, 50 μg/mL streptomycin, and 10% FBS. The phenotypes of the cells were verified by ATCC by immunostaining for specific markers. The protocols for verifying the cell lines are available on the ATCC Web site (http://www.atcc.org/Portals/1/Pdf/CellBiologyStandards.pdf). H1650 cells were grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Mediatech, Inc) supplemented with 2 mM glutamine, 100 U/mL penicillin, 50 μg/mL streptomycin, and 10% FBS. Primary normal human bronchial epithelial cells (NHBEs) and primary small airway epithelial cells (SAECs) were purchased from Lonza (Basel, Switzerland). NHBEs were cultured in bronchial epithelial basal medium (BEBM) supplemented with 0.1% human epidermal growth factor, 0.4% bovine pituitary extract, 0.1% insulin, 0.1% gentamycin, 0.1% retinoic acid, 0.1% transferrin, 0.1% triiodothyronine, and 0.1% epinephrine. SAECs were maintained in small airway basal medium (SABM) supplemented with 5% fat-free bovine serum albumin, and other supplements were the same as that of BEBM, described above. BEBM and SABM and all supplements were purchased from Lonza. All cells were cultured at 37°C in a humidified 5% CO2 incubator. The phenotypes and genotypes of the NHBEs were verified by Lonza by immunostaining for specific markers. A Certificate of Analysis confirming the authentication of cells was obtained from Lonza.
Nicotine, α-bungarotoxin (α7-nAChR subunit inhibitor), and dihydro-beta-erythroidine (DhβE; α3/β2-nAChR antagonist) were purchased from Sigma-Aldrich (St Louis, MO). The phosphatidylinositol 3-kinase, catalytic, delta polypeptide (PIK3CD) inhibitor LY294002 (2-morpholin-4-yl-8-phenylchromen-4-one) was purchased from Cell Signaling Technology (Danvers, MA). The inhibitor of α7-nAChR subunit methyllycaconitine (MAA) and α-lobeline (α4/β2–nAChR inhibitor) were purchased from Tocris Biosciences (Ellisville, MO). The SRC inhibitor PP2 (4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyramidine) was purchased from Enzo Life Sciences (Plymouth Meeting, PA) and delta peptide [D-Ala2,D-Leu5]Enkephalin was purchased from Bachem California (Torrance, CA).
Human Lung Tumor Samples
Fresh-frozen human lung adenocarcinoma tissues (n = 8) and matched distant normal lung tissues (n = 8) from patients who were active smokers or had a smoking history were retrieved from the Moffitt Cancer Center Tumor Bank. Eight frozen lung adenocarcinoma tissues were paired with matched distant normal lung tissues and were used for preparing lysates for immunoprecipitation, described later. Two pairs of frozen lung adenocarcinoma and matched distant normal lung tissues were used to prepare lysates for the chromatin immunoprecipitation (ChIP) assay, also described later. The use of all NSCLC tumor tissues and normal tissues was approved by the patients and the Institutional Review Board of the University of South Florida.
Monoclonal and Polyclonal Antibodies
Rabbit anti-human E2F1, rabbit anti-human baculoviral IAP repeat–containing 5 (BIRC5), rabbit anti-human EP300, rabbit anti-human-α7-nAChR, mouse anti-human E2F1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-human RB1 was purchased from BD Biosciences (San Jose, CA). Rabbit anti-human phosphorylated SRC (Tyr416), rabbit anti-human extracellular regulated kinase (ERK) 1/2 (Thr202/Tyr204), rabbit anti-human poly (ADP-ribose) polymerase (PARP-1), rabbit anti-human ERK1/2, rabbit anti-human v-akt murine thymoma viral oncogene homolog 1 (AKT1), and rabbit anti-human phospho-AKT1 (Thr308) antibodies were purchased from Cell Signaling Technology. Mouse anti-human ARRB1 was purchased from BD Biosciences, rabbit anti-human ARRB1 was purchased from EMD Biosciences (Darmstadt, Germany), and rabbit anti-human ARRB1 was purchased from Novus Biologicals (Littleton, CO). Mouse anti-human SRC, mouse anti-human pan β-nAChR, and rabbit anti-human acetylated histone H3 antibodies were purchased from Millipore (Billerica, MA). Mouse anti-FLAG, mouse anti-human tubulin, alpha 4a (TUBA4A), and mouse anti-human actin, beta (ACTB) were purchased from Sigma-Aldrich. Mouse anti-HA monoclonal antibody was purchased from Covance (San Diego, CA). Mouse anti-human X-linked inhibitor of apoptosis (XIAP) was purchased from Enzo Life Sciences. Secondary antibodies rabbit anti-mouse IgG and donkey anti-rabbit IgG were purchased from Pierce Biotechnologies (Rockford, IL). Secondary antibodies rabbit anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 555 were purchased from Invitrogen (Carlsbad, CA).
Stable Transfection With Short Hairpin RNA
The knockdown of ARRB1 expression in A549 cells was performed by stable transfection of a microRNA-adapted short hairpin RNA (shRNA) construct in retroviral pSM2c vector to specifically target ARRB1 mRNA in A549 human NSCLC cells. The shRNA construct, empty vector pSM2c, and the shRNAmir library were purchased from Open Biosystems (Huntsville, AL). The ARRB1 target sequence was 5′-TGCTGTTGACAGTGAGCGCCGACATTGTATTTGAGGACTTTAGTGAAGCCACAGATGTAAAGTCCTCAAATACAATGTCGTTGCCTACTGCCTCGGA-3′.
A549 cells were transfected with the ARRB1-specific shRNA construct using Lipofectamine 2000 transfection reagent (Invitrogen) following the manufacturer’s instructions. The transfected cells were selected using Ham’s F-12K medium containing 10% FBS and 1 μg/mL puromycin (Open Biosystems). A549 cells transfected with the empty vector pSM2c (A549-EV) were used as the control for all experiments. A549-ARRB1-shRNA (A549-sh) cells stably expressing ARRB1 and A549-EV cells were maintained in Ham’s F-12K medium containing 10% FBS and 1 μg/mL puromycin. A549-RB1-shRNA (sh6 cells) cells stably expressing RB1-specific shRNA and A549-control nontargeting-shRNA (NH9 cells) have been previously (38) described and were maintained in Ham’s F-12K medium containing 10% FBS and 1 μg/mL puromycin.
Cell Proliferation Assay
A549 cells were plated in poly-D-lysine–coated eight-well chamber slides at a density of 10 000 cells per well and rendered quiescent by serum starvation in serum-free Ham’s F-12K medium for 36 hours (15,39). Cells were treated with 1 μM nicotine or medium containing 10% FBS for 18 hours, and a proliferation assay was performed using the 5-bromo-2-deoxy-uridine (BrdU) Labeling and Detection kit II (Roche Applied Science, Indianapolis, IN). BrdU-positive cells in the S-phase were visualized by microscopy, and a quantitative analysis of proliferating cells was done by counting three high-power fields of 100 cells in quadruplicate at ×200 magnification on a Leica DMILB inverted phase contrast microscope (Leica Microsystems, Wetzlar, Germany). The magnitude of BrdU incorporation in control cells was considered to be equal to 1, and the BrdU incorporation observed in nicotine-treated and medium containing 10% FBS-treated cells was calculated as fold-increase relative to the control cells. The experiment was performed two independent times with two replicates in each experiment.
Transient Transfection
HEK293 cells were cultured to 70% confluence in antibiotic-free DMEM containing 10% FBS and transfected with the following constructs using Lipofectamine 2000 transfection reagent (Invitrogen). The gene constructs used for transfection were pcDNA3-HA-E2F1 and pcDNA3-FLAG-rat ARRB1 (wild type) (Supplementary Methods, available online). Two micrograms each of pcDNA3-HA-E2F1, pcDNA3-FLAG-rat ARRB1 (wild type), or a combination of both were transfected in HEK293 cells using Lipofectamine 2000 following the manufacturer’s instructions. Cells transfected with the empty vector pCDNA3 were used as control (mock transfected). Twenty-four hours after transfection, lysates were prepared for immunoprecipitation and immunoblotting, as described below.
Immunoprecipitation and Immunoblotting
NHBE and SAEC cells were rendered quiescent by incubating in BEBM and SABM, respectively, without any supplements, at 37°C for 24 hours (15). A549 cells were rendered quiescent by serum starvation in serum-free Ham’s F-12K medium for 36 hours. All quiescent cells were treated with 1 μM nicotine at 37°C for varying time periods ranging from 5 minutes to 2 hours (40). Subsequently, the cells were harvested and washed three times with ice-cold phosphate-buffered saline (PBS; 138 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 0.06 mM Na2HPO4·7H2O). Cell lysates made from exponentially growing cultures of A549 cells in Ham’s F-12K medium containing 10% FBS were referred to as asynchronous A549 lysates.
Cell lysates were prepared from NHBE, SAEC, and HEK293 transfected cells by adding 70 μL of lysis buffer (20 mM Tris–HCl [pH 7.6], 0.5% Nonidet-40 [new name IGEPAL-CA-630], 250 mM NaCl, 3 mM EGTA, 3 mM EDTA, 4 μM dithiothreitol [DTT], 5 mM phenylmethylsulfonylfluoride [PMSF], 1 mM sodium fluoride, 1 mM sodium orthovanadate, 25 μg/mL leupeptin, 5 μg/mL pepstatin, 5 μg/mL aprotinin, 25 μg/mL trypsin–chymotrypsin inhibitor) to 20 μL of packed cell volume (40). The lysate was rotated at 4°C for 30 minutes and subsequently centrifuged at 15 000g at 4°C for 15 minutes. The protein concentration of the clarified lysate was measured using a Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA).
Fresh-frozen human NSCLC tumor tissues (n = 8) from patients and matched normal lung tissues (n = 8) were teased with a sterile surgical forceps on frosted slides (Fisher Scientific, Pittsburgh, PA) to homogenize the tissues. The tissues were resuspended in 1 mL of ice-cold red blood cell lysis buffer (0.14 M NH4Cl and 0.017 M Tris–HCl [pH 7.2]) and incubated on ice for 5 minutes. The cells were washed in red blood cell lysis buffer three times until red blood cells were no longer visible. Subsequently, one packed cell volume of cell lysis buffer (20 mM Tris–HCl [pH 7.6], 0.5% IGEPAL-CA-630, 3 mg/ml bovine serum albumin, 250 mM NaCl, 3 mM EGTA, 3 mM EDTA, 4 μM DTT, 5 mM PMSF, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 25 μg/mL leupeptin, 5 μg/mL pepstatin, 5 μg/mL aprotinin, 25 μg/mL trypsin–chymotrypsin inhibitor) was added to the tissues, and lysates were rotated at 4°C for 45 minutes. The rest of the process was the same as described in the above paragraph.
For co-immunoprecipitation, the cell lysates containing 250 μg of total proteins were incubated with 1 μg of the following antibodies: Mouse anti-human ARRB1, mouse anti-human pan-β-nAChR, mouse anti-influenza HA antibody, rabbit anti-human E2F1, mouse anti-human RB1, and rabbit anti-human EP300. The total reaction volume was adjusted to 100 μL with immunoprecipitation buffer (2 mM HEPES [pH 7.9], 40 mM KCl, 0.001 mM MgCl2, 25 mM EGTA, 1 mM EDTA, 1% IGEPAL-CA-630, 1 mM DTT, 5 mM PMSF, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 25 μg/mL leupeptin, 5 μg/mL pepstatin, 5 μg/mL aprotinin, 25 μg/mL trypsin–chymotrypsin inhibitor) and rotated on a nutator at 4°C for 1 hour. After 1 hour, 50 μL of 1:1 protein G-Sepharose slurry was added (GE Healthcare, Piscataway, NJ) and the mixture was rotated at 4°C for another 2 hours. The beads were washed four times in immunoprecipitation buffer. Bound proteins were eluted in sodium dodecyl sulfate (SDS) sample buffer (0.06 M Tris–HCl [pH 6.8], 10% glycerol, 2% SDS, and 100 mM DTT, 0.2% w/v bromophenol blue) and resolved on a 10% SDS–polyacrylamide gel. The proteins on the SDS–polyacrylamide gel were transferred to 0.45 μm nitrocellulose membranes (Bio-Rad Laboratories), and the interacting proteins were detected by immunoblotting.
Briefly, nitrocellulose membranes were incubated in a blocking solution containing 5% nonfat dry milk in PBS containing 0.1% Tween-20 (Fisher Scientific) at room temperature for 1 hour. Membranes were incubated at 4°C overnight with the primary antibodies—mouse anti-human ARRB1 (1:500 dilution), mouse anti-human TUBA4A (1:2000 dilution), mouse anti-human ACTB (1:50 000 dilution), rabbit anti-human E2F1 (1:300 dilution), rabbit anti-human p-AKT1 (1:300 dilution), rabbit anti-human AKT1 (1:1000 dilution), rabbit anti-human p-SRC (1:300 dilution), mouse anti-human SRC (1:1000 dilution), rabbit anti-human ERK1/2 (1:1000 dilution), mouse anti-HA (1:1000 dilution), mouse anti-FLAG (1:1000 dilution), mouse anti-human XIAP (1:500 dilution), rabbit anti-human BIRC5 (1:300 dilution), rabbit anti-human α7-nAChR (1:300 dilution), rabbit-anti-human PARP-1 (1:1000 dilution), and mouse anti-human RB1 (1:500 dilution) followed by incubation with horseradish peroxidase–conjugated anti-mouse IgG (1:3000 dilution) or anti-rabbit IgG (1:3000 dilution) at room temperature for 1 hour. Antibody–protein complexes were detected using enhanced chemiluminescence immunoblotting detection reagent (GE Healthcare). The immunoblot blot signals were quantified using Alpha Imager HP gel documentation system (Cell Biosciences, Santa Clara, CA).
As a control for the experiment, one-third of the amount of protein was analyzed on the SDS–polyacrylamide gel for each immunoprecipitation reaction (39,40). The experiment was performed two independent times with two independent sets of cell lysates and tumor lysates.
Immunofluorescence and Confocal Microscopy
For immunostaining, A549, NHBE, and H1650 cells were plated onto poly-D-lysine–coated eight-well glass chamber slides (7000 cells per well) and rendered quiescent (serum starved for 36 hours) as described earlier. A549 and H1650 cells were treated with 1 μM nicotine at 37°C for 15 minutes, and NHBE cells were treated with the same concentration of nicotine for 15 minutes, 30 minutes or 1 hour. The cells were fixed with 10% formalin and permeabilized with 0.2% Triton-X-100 in PBS at room temperature for 5 minutes. Subsequently, cells were washed three times for 5 minutes each in PBS and treated with blocking buffer (5% normal goat serum [Thermo Scientific, Rockford, IN] in PBS) at room temperature for 1 hour. The cells were incubated with primary antibody rabbit anti-human ARRB1 (1:200 dilution), overnight at 4°C, as described previously (41). In case of double immunofluoresence experiments, the primary antibodies were rabbit anti-human ARRB1 (1:200 dilution) and mouse anti-human E2F1 (1:200 dilution) (41). The cells were washed three times for 15 minutes each in PBS. The cells were then incubated with secondary antibody goat anti-rabbit Alexa Fluor 555 (1:1000 dilution) for single immunofluorescence experiments and with goat anti-rabbit Alexa Fluor 555 (1:1000 dilution) and goat anti-mouse-Alexa Flour 488 (1:200 dilution) for double-immunofluorescence experiments, at room temperature for 45 minutes. The cells were washed three times for 15 minutes each in PBS and mounted with VECTASHIELD mounting medium with 4′6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Cells were observed using a Leica TCS SP5 confocal microscope (Leica Microsystems) at ×630 magnification. Interference between fluorescence signals was avoided by capturing the images using multitracking mode (excitation 480 or 555 nm, emission 570 nm). All experiments were repeated three independent times.
Preparation of Nuclear and Cytosolic Extracts
A549 cells (1 × 107) were rendered quiescent in serum-free Ham’s F-12K medium for 36 hours, as before. Cells were then treated with 1 μM α-bungarotoxin, 1 μM MAA, 1 μM DhβE, 10 μM PP2, and 10 μM LY294002, in 10 mL serum-free Ham’s F12K medium. After 30 minutes, 1 μM nicotine was added to the mix and the cells were incubated at 37°C for 15 minutes. As a positive control, A549 cells were treated with 10 μM delta peptide ([D-Ala2,D-Leu5]Enkephalin) for 15 minutes. A549 cells were washed twice in ice-cold PBS and then scraped from the dishes in 1 mL PBS and transferred to microtubes (42,43). Cells were centrifuged at 2000g for 5 minutes, the supernatant was discarded, and the cell pellet was allowed to swell and lyse after the addition of 100 μL hypotonic buffer (10 mM [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] or HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.5% IGEPAL-CA-630, 1 mM DTT, and 0.5 mM PMSF). The lysates were incubated on ice for 10 minutes and then centrifuged at 7200g at 4°C for 5 minutes. Pellets containing crude nuclei were resuspended in 50 μL of extraction buffer (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA, 1 mM DTT, 5 μg/mL leupeptin, 5 μg/mL pepstatin, 5 μg/mL trypsin–chymotrypsin inhibitor, 5 μg/mL aprotinin, and 1 mM PMSF) and then incubated on ice for 30 minutes. The samples were then centrifuged at 16 000g at 4°C for 10 minutes to obtain supernatants containing nuclear extracts. Nuclear extracts were stored at −70°C, until use (42,43).
To prepare the cytosolic extracts, A549 cells (1 × 107) were treated as described above and subsequently washed twice in PBS, scraped from the dishes in 1 mL PBS, and sonicated using Branson Sonifer 450 (Branson Ultrasonics, Danbury, CT) at power 4 for 30 seconds to disrupt the membranes. Cytosolic extracts were prepared (44) by differential centrifugation at 15 000g for 15 minutes and stored at −70°C, until use. Protein content of nuclear and cytosolic extracts was determined as before using Bio-Rad Protein Assay Kit.
The purity of the nuclear extract was assessed by performing an immunoblot analysis for PARP-1 using rabbit-anti-human PARP-1 antibody (1:1000 dilution). Similarly, the integrity of the cytosolic fraction was examined by using mouse anti-human TUBA4A antibody (1:2000 dilution).
ChIP Assay
Quiescent (serum starved) A549 cells (2.5 × 107) were incubated with 1 μM nicotine in 100 mm tissue culture dishes (BD Biosciences, Franklin Lakes, NJ) at 37°C for 24 hours. Cells were treated with 1% formaldehyde for 10 minutes at room temperature for cross-linking the DNA to the proteins. The cross-link reaction was terminated by addition of 0.125 M glycine (Fisher Bioreagents, Fisher Scientific, Pittsburgh, PA). The cells were scraped, washed in ice-cold PBS, and centrifuged at 800g at 4°C for 5 minutes. Subsequently, the pellet was resuspended in cell lysis buffer (44 mM Tris–HCl [pH 8.1], 1% SDS, 1 mM EDTA [pH 8.0]). The cells were sonicated twice for 15 seconds each. Subsequently, the cell lysates were centrifuged at 10 000g at 4°C for 15 minutes. The soluble chromatin was precleared with 100 μL of protein G-Sepharose. A 20% aliquot of the precleared chromatin was used as the input for the ChIP assay. The remainder of the precleared chromatin was diluted with ChIP dilution buffer (16 mM Tris–HCl [pH 8.1], 250 mM NaCl, 0.1% SDS, 1% Triton-X-100, 1.2 mM EDTA) and rotated overnight with primary antibody. The following primary antibodies were used at 10 μg concentration for each ChIP reaction: rabbit anti-human E2F1, rabbit anti-human EP300, rabbit anti-human acetylated histone H3, mouse anti-human-ARRB1, rabbit anti-human ARRB1 (EMD Biosciences), or rabbit anti-human-ARRB1 (Novus Biologicals). Rabbit anti-mouse IgG antibody was used as the irrelevant antibody for all ChIP reactions. The next day, 60 μL of 1:1 protein G-Sepharose was added to the immune complexes, and the mixture was rotated at 4°C for 2 hours. The beads were washed five times with ChIP dilution buffer and eluted with ChIP elution buffer (0.1 M sodium bicarbonate, 1% SDS, 5 mM NaCl). The cross-links were reversed by incubation at 65°C for 4 hours. DNA was isolated by ethanol precipitation. The associated proteins with the DNA were digested with 50 μg Proteinase K at 37°C for 30 minutes. DNA was purified by phenol to chloroform ratio extraction method followed by ethanol precipitation. Purified DNA was resuspended in 50 μL water. The differential binding between proteins and BIRC5, CDC6, CDC25A, TYMS, and FOS promoter DNA was examined by polymerase chain reaction (PCR). The primers are listed in the Supplementary Methods (available online).
The ChIP assay on human NSCLC tumors (n = 2) and matched distant normal lung tissue (n = 2) was performed using 30 mg of each tumor tissue or normal lung tissue sample per immunoprecipitation reaction. Each tissue sample was transferred into a tube containing 10 mL PBS (supplemented with 10 μg/mL leupeptin, 10 μg/mL aprotinin, 1 mM PMSF) per gram of tissue. Protein–DNA cross-linking reaction was done using 1% formaldehyde at room temperature with gentle agitation. After the cross-linking reaction, tissue samples were disaggregated in ice-cold PBS using a Dounce homogenizer with type B pestle (Kontes Glass Company, Vineland, NJ). The rest of the procedure was similar to that of the cultured cells, as described above. Each ChIP assay was performed two independent times with independent sets of cell lysates and tumor lysates.
Transfection With Small Interfering RNA and Assays With Transfected Cells
Double-stranded small interfering RNA (siRNA) for ARRB1 (sequence: 5′-AAAGCCUUCUGCGCGGAGAAU-3′) was purchased from Qiagen (Valencia, CA) for one of our previous studies (18). Transient transfection of A549 cells with 50 pmol siRNA was performed using the Oligofectamine Transfection Reagent (Invitrogen) according to the manufacturer’s instructions. Eighteen hours after transfection, the cells were rendered quiescent (serum starved) and then treated with 1 μM nicotine for 24 hours. Lysates were prepared for immunoblotting or ChIP assays. The primary and secondary antibodies for immunoblot and ChIP assays have been described before.
RNA Isolation and Real-Time PCR
A549-EV and A549-sh cells (1 × 107 cells) were rendered quiescent (serum starved) and subsequently treated with 1 μM nicotine or Ham’s F-12K medium containing 10% FBS for 24 hours, and total RNA was isolated using RNeasy Mini Kit (Qiagen). First-strand cDNA was synthesized in a 20 μL reaction volume using the Bio-Rad iScript system (Bio-Rad Laboratories). Quantitative real-time PCR (qPCR) was performed with SYBR Green Supermix Taq Kit (Bio-Rad Laboratories) and analyzed on iCycler, MyiQ Single Color Real-Time PCR Detection System (Bio-Rad Laboratories), equipped with Optical System Software version 1.0. The PCR conditions were 10 minutes at 95°C, 1 minute at 55°C, 40 cycles of 15 seconds at 95°C, followed by 30 seconds at 55°C. The real-time PCR primers sequences for BIRC5, TYMS, CDC6, and 18S RNA were published previously (38,45) and are listed in the Supplementary Methods (available online).
The mRNA expression data were normalized using 18S RNA as internal control, and the fold-change in the expression levels was determined using the quiescent cells as control. Each qPCR analysis was performed two independent times.
Apoptosis Assay
A549-EV and A549-sh cells were plated on poly-D-lysine–coated eight-well chamber slides at a density of 10 000 cells per well and were rendered quiescent (serum starved as before). Subsequently, these cells were incubated with 1 μM nicotine at 37°C for 30 minutes, then 20 μM cisplatin was added, and the cells were incubated at 37°C for 24 hours (7). Apoptosis was measured by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay using a DeadEnd TUNEL Colorimetric Kit (Promega, Madison, WI) according to the manufacturer’s instructions. The TUNEL assays were repeated twice with two replicates in each of the experiments. TUNEL-positive apoptotic cells were visualized by microscopy, and a quantitative analysis of apoptosis was done by counting three high-power fields of 50 TUNEL-positive cells in quadruplicate at ×200 magnification on a Leica DMILB inverted phase contrast microscope (Leica Microsystems). The percentage of TUNEL-positive cells was calculated. The data were graphically represented by calculating the means and the 95% confidence intervals (CIs) for two independent experiments.
Immunohistochemical Staining of ARRB1 in Lung Cancer Tissue Microarray Slide
A human lung cancer tissue microarray slide (catalog number IMH-358; Imgenex, San Diego, CA) was immunostained for ARRB1. The slide contained 59 tissue sections (referred to as cores) that included normal lung tissues, different carcinomas of the lung, and metastatic carcinomas. The 59 cores consisted of nine normal lung tissues adjacent to the tumors, 40 primary tumors of the lung, and 10 metastatic carcinomas (46). The normal lung tissues, primary and metastatic carcinomas were histologically characterized by Imgenex and have been described elsewhere (http://www.imgenex.com/tissue_array_tds.php?id=924). The tissue microarray slide was deparaffinized by baking at 62°C for 1 hour and then immersed twice in 100% xylene at room temperature for 10 minutes each, followed by incubating in 100% ethyl alcohol for 10 minutes, and rehydrated with decreasing concentrations (90%, 80%, 70% and 50%; vol/vol in water) of ethyl alcohol for 5 minutes each. At the final step the slide was rinsed in cold water and subjected to microwave antigen retrieval for 20 minutes on 70% power, with a 1 minute cooling period after every 5 minutes in 0.1 M sodium citrate buffer (pH 6.0). Sections were cooled for 20 minutes, rinsed three times in distilled water, twice in PBS, and the rest of the staining was done following the manufacturer’s protocol (VECTASTAIN Elite ABC Kit [Universal], Vector Laboratories). The kit contained blocking serum, secondary antibody and avidin–biotin–horseradish peroxidase complex. The slide was blocked with blocking serum for 30 minutes at room temperature followed by incubation with primary antibody rabbit anti-human-ARRB1 (1:500 dilution; Novus Biologicals) at 4°C, overnight (39). As negative staining controls, separate slides were stained wherein the primary antibody was replaced with either 1% bovine serum albumin in PBS to rule out the effect of endogenous peroxidase, or isotype-matched irrelevant rabbit IgG to test for antibody specificity. The slide was rinsed three times in PBS for 10 minutes each and incubated with secondary antibody for 30 minutes at room temperature. Following three rinses in PBS again for 10 minutes each, the slide was incubated with avidin–biotin–horseradish peroxidase complex for 30 minutes at room temperature. To detect the bound antibody, the slide was treated with peroxidase substrate kit (Vector Laboratories), wherein 3,3′-diaminobenzidine (DAB) was the chromogen, and color developed within 2–3 minutes of adding the DAB solution. After a final rinse in distilled water, sections were lightly counterstained in hematoxylin, dehydrated by immersing in increasing concentrations of ethyl alcohol (50%, 70%, 80%, 90%, and 100%; vol/vol in water) for 3 minutes each and finally immersed in 100% xylene twice for 2 minutes each. The slides were mounted in Clarion mounting medium (Santa Cruz Biotechnology). Immunostained slides were scanned using the Aperio Automatic Scanning System from Applied Imaging (San Jose, CA) and scored by two pathologists (Alexis Lopez and D. Coppola). The semiquantitative score was reached by taking into consideration both cellularity and intensity of expression (semiquantitative score = cellularity × intensity). Cellularity was scored as follows: a score of 3 equals to greater than 66% cellularity, a score of 2 equals to 34%–65% cellularity, and a score of 1 equals to less than 33% cellularity. Intensity was scored as follows: a score of 3 equals to strong intensity, a score of 2 equals to moderate intensity, and a score of 1 equals to weak intensity (47). The semiquantitative score of the normal lung tissue and primary tumors or metastatic tumors was calculated using two-sided Student t test for statistical significance. The data were considered statistically significant when the P value was less than .05. Total and nuclear expression of ARRB1 were analyzed and plotted as fold-change. The images were captured at ×200 magnification. The experiment was repeated twice with two independent arrays.
Statistical Analysis
All data have been graphically represented and statistically analyzed using Microsoft Office Excel 2003 (Microsoft Corporation, Redmond, WA). In all analyses, means and 95% confidence intervals were estimated. Statistical analysis of mRNA levels, BrdU incorporation assays, TUNEL assays, and protein densitometric analysis were performed using a two-sided Student t test. P values less than .05 were considered statistically significant. All statistical tests were two-sided.
Results
Effect of ARRB1 on Nicotine-Induced Activation of nAChRs and Cell Proliferation
To investigate the role of ARRB1 in nicotine-induced and nAChR-mediated NSCLC cell proliferation, we generated a human NSCLC cell line (A549) stably expressing ARRB1-specific shRNA (A549-sh). A549 cells stably transfected with the empty vector (A549-EV) were used as control. Cell lysates made from exponentially growing cultures of A549 cells (referred to as asynchronous A549) in Ham’s F-12K containing 10% FBS were used as the positive control for the immunoblotting experiments (Figure 1, A). We first confirmed the knockdown of ARRB1 expression (Figure 1, A). Next, we examined if reduced ARRB1 expression showed an effect on nicotine-induced proliferation of A549 cells by performing BrdU incorporation assays (Figure 1, B). We found that nicotine induced a statistically significant increase of BrdU incorporation in A549-EV cells (untreated vs nicotine-treated A549-EV cells; mean fold-increase in BrdU incorporation = 1, 95% CI = 1- to 1-fold, vs 2.5-fold, 95% CI = 2.4- to 2.6-fold, P < .001). In contrast, the increase in BrdU incorporation was statistically significantly reduced in nicotine-treated A549-sh cells, relative to A549-EV cells (nicotine treated A549-EV vs nicotine-treated A549-sh cells, mean fold-decrease in BrdU incorporation = 2.5- vs 1.1-fold; difference of mean fold-decrease = 1.4-fold, 95% CI = 0.1- to 0.1-fold, P < .001). The treatment of A549-EV and A549-sh cells with medium containing 10% FBS induced similar magnitude of BrdU incorporation (untreated vs 10% FBS-treated A549-EV cells, mean fold-increase in BrdU incorporation = 1-fold, 95% CI = 1- to 1-fold, vs 3.0-fold, 95% CI = 2.9- to 3.1-fold, P < .001; untreated vs 10% FBS-treated A549-sh cells, mean fold-increase in BrdU incorporation = 1-fold, 95% confidence interval = 1- to 1-fold, vs 2.7-fold, 95% confidence interval = 2.6- to 2.8-fold, P < .001).
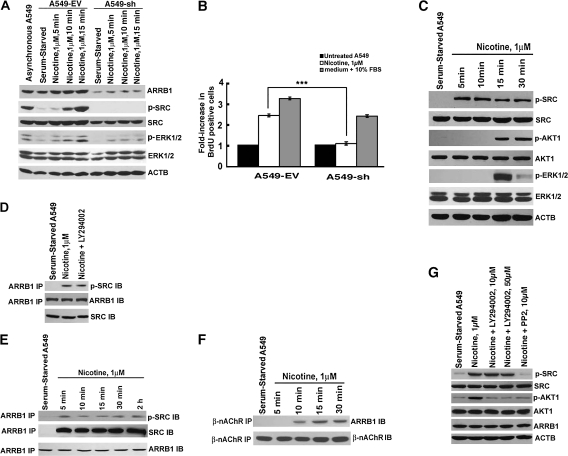
Figure 1.
Effect of reduced ARRB1 expression on nicotinic acetylcholine receptor (nAChR) signaling in a human non–small cell lung cancer (NSCLC) cell line. A) ARRB1 expression and phosphorylated levels of nAChR signaling pathway proteins. Human NSCLC cell line, A549, was stably transfected with an empty vector (A549-EV) or ARRB1-specific shRNA (A549-sh). Cells were serum starved for 36 hours and then induced with nicotine for 5, 10, and 15 minutes. Cell lysates were analyzed by immunoblot assay, and representative immunoblot of two independent experiments shows the levels of ARRB1, p-SRC, and p-ERK1/2. Total SRC and total ERK1/2 levels are also shown. Cell lysates from exponentially growing cultures of A549 cells (asynchronous) were used as the positive control, and mouse anti-human actin, beta (ACTB) was used as a loading control. B) Effect of nicotine on the proliferation of A549-EV and A549-sh cells. Serum-starved A549-EV and A549-sh cells were treated with 1 μM nicotine for 18 hours, and cell proliferation was measured by 5-bromo-2-deoxy-uridine (BrdU) incorporation assay. The means and 95% confidence intervals of two independent experiments are shown. ***P < .001. C) Real-time kinetics of nAChR signaling. Serum-starved A549 cells were treated with 1 μM nicotine for 5, 10, 15, and 30 minutes. Cell lysates were analyzed by immunoblot assay and representative immunoblot of two independent experiments show the levels of p-SRC, p-AKT1, and p-ERK1/2. Total SRC, AKT1, and ERK1/2 levels are also shown. ACTB was used as the loading control for the experiment. D) Effect of PIK3CD/AKT pathway inhibitor, LY294002, on the binding of SRC and ARRB1. Serum-starved A549 cells were treated with 1 μM nicotine in the presence or absence of 10 μM of LY294002. Cell lysates were immunoprecipitated with rabbit anti-human ARRB1 antibody, and immunoblot analysis was done with mouse anti-human SRC antibody. E) Nicotine-induced time-dependent binding of ARRB1 and SRC. Serum-starved A549 cells were treated with 1 μM nicotine for 5, 10, 15, 30 minutes, and 2 hours. Cell lysates were immunoprecipitated with rabbit anti-human ARRB1 antibody, and immunoblot analysis was done with mouse anti-human SRC antibody. F) Sequential order of binding of ARRB1 and SRC, and ARRB1 and nAChRs. Serum-starved A549 cells were treated with 1 μM nicotine for 5, 10, 15, and 30 minutes. Cell lysates were immunoprecipitated with mouse anti-human-pan-β-nAChR antibody, and immunoblot analysis was done with rabbit anti-human ARRB1 antibody. G) Effect of PP2 on nicotine-induced phosphorylation of AKT1. Serum-starved A549 cells were treated with 1 μM nicotine for 15 minutes in the presence of 10 μM or 50 μM LY294002. The expression of p-SRC and p-AKT1 was analyzed by immunoblotting. In addition, the levels of total SRC, AKT1, and ARRB1 were also assessed by immunoblotting. ACTB was used as a loading control. All immunoblotting results are representative of two independent experiments. IB = immunoblot; IP = immunoprecipitation; p = phosphorylated. All P values were calculated using a two-sided Student t test.
Our published data and other studies have shown that ARRB1 plays a vital role in nicotine-induced proliferation of human NSCLC and the mechanism involves SRC, AKT1, and ERK1/2 signaling pathways (4,5,15,48). Therefore, to investigate the effect of reduced ARRB1 expression on nicotine-induced signaling pathways, we examined the phosphorylated levels of SRC and ERK1/2 in A549-EV and A549-sh cells. We observed that A549-sh cells showed reduced levels of phosphorylated SRC and ERK1/2 proteins when induced with nicotine (Figure 1, A).
In our previous studies, we have shown that the binding of nicotine to nAChRs induced the formation of an oligomeric complex of β-nAChR, ARRB1, and SRC (15,49). These signaling events led to downstream phosphorylation of SRC, AKT1, and ERK1/2; however, the sequential order of these signaling events is not known. Therefore, we conducted a series of experiments to gain insight into the real-time kinetics of signaling events upon nicotine treatment of A549 cells. Immunoblot analysis of serum-starved A549 cells treated with nicotine showed an increased level of phosphorylated SRC within 5 minutes of nicotine treatment (Figure 1, C). Previous studies have shown that the PIK3CD/AKT pathway plays a vital role in the antiapoptotic effects of nicotine (5,7,50). The treatment of NSCLC cell lines as well as normal lung cells with nicotine is reported to induce phosphorylation of AKT1 at residues threonine-308 and serine-473, which protects NSCLC cells from apoptosis induced by DNA damaging agents (5,50,51). Therefore, we investigated the role of ARRB1 in nicotine-induced phosphorylation of AKT1 in A549 cells. When A549-EV and A549-sh cells were treated with nicotine, we observed an increased level of phosphorylated AKT1 and ERK1/2 in A549-sh cells after 15 minutes, suggesting that AKT1 and ERK1/2 may be phosphorylated subsequent to increased phosphorylation of SRC (Figure 1, C). The pro-metastatic effects of ARRB1 in colon cancer were found to involve the sequential activation of SRC and the PIK3CD/AKT pathways (27), so we investigated whether nicotine treatment induced a similar sequential order of signaling events in NSCLC cells. A549 cells were treated with nicotine in the presence or absence of the PIK3CD/AKT pathway inhibitor, LY294002, and immunoprecipitation and immunoblot analysis of the cell lysates showed that inhibiting the PIK3CD/AKT pathway had no effect on interaction between ARRB1 and SRC (Figure 1, D). Our previous studies demonstrated that nicotine induced the formation of an oligomeric complex comprising of β-nAChR, SRC, and ARRB1 (15). However, the real-time kinetics of association among β-nAChR, SRC, and ARRB1 were not known. To investigate the sequential order of these events in real time, we performed immunoprecipitation and immunoblot analysis of binding between ARRB1 and SRC at different time points after nicotine treatment of A549 cells. We observed that the interaction between ARRB1 and SRC occurred within 5 minutes of nicotine treatment (Figure 1, E) and interaction between β-nAChR and ARRB1 occurred later—within 10 minutes of nicotine treatment (Figure 1, F). Nicotine-induced increased level of phosphorylated SRC was unaffected by treatment with LY294002 in A549 cells (Figure 1, G), suggesting that activation of the PIK3CD/AKT pathway is not necessary for the increase in phosphorylated SRC. In contrast, the SRC inhibitor PP2 showed a substantial reduction in the level of phosphorylated AKT1, suggesting that the phosphorylation of AKT1 upon nicotine treatment was probably dependent on SRC.
Effect of Nicotine on Subcellular Localization of ARRB1
Previous studies have shown that ARRB1 can translocate to the nucleus in response to the binding of delta peptide to the δ- and κ-opioid receptors (30,34). The nuclear ARRB1 was found to participate in chromatin-remodeling pathways involving CDKN1B and FOS promoters (30,34). Therefore, we investigated whether nicotine was able to induce nuclear localization of ARRB1 in normal lung cells namely NHBEs and SAECs and A549 and H1650 NSCLC cell lines. Subcellular location of ARRB1 was studied by immunofluorescence experiments. We observed that ARRB1 was ubiquitously distributed in quiescent NHBE cells and treatment with 1 μM nicotine induced the translocation of a subset of ARRB1 to the nucleus within 15 minutes of treatment (Figure 2, A). A substantial amount of the ARRB1 remained in the nucleus even after 1 hour of nicotine treatment.
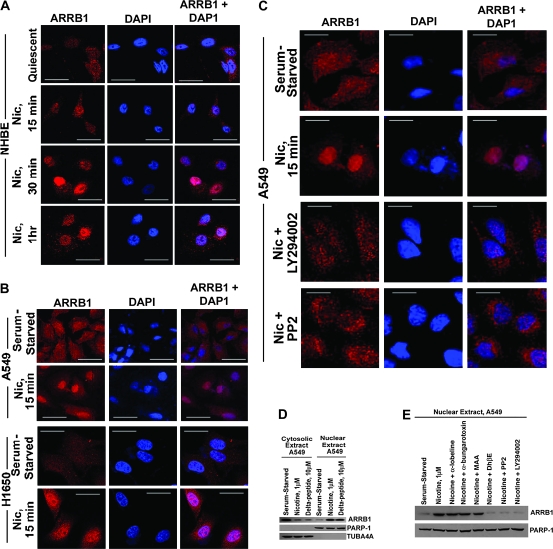
Figure 2.
Effect of nicotine on subcellular localization of ARRB1 in multiple types of human lung cells. A) Effect of nicotine on the localization of ARRB1 in NHBE normal lung cells. Quiescent NHBEs were treated with 1 μM nicotine for varying time points, and the localization of ARRB1 was analyzed by immunofluorescence (×630 magnification, scale bar = 10 μm). The cells were counterstained with the nuclear marker, 4′6-diamidino-2-phenylindole (DAPI). B) Effect of nicotine on translocation of ARRB1 in non–small cell lung cancers. Serum-starved A549 and H1650s were treated with 1 μM nicotine (denoted as Nic) for 15 minutes, and the localization of ARRB1 was analyzed by immunofluorescence (×630 magnification, scale bar = 10 μm). The cells were counterstained with the nuclear marker, DAPI. C) Effect of PIK3CD/AKT and SRC pathway inhibitors on nicotine-induced nuclear localization of ARRB1. Serum-starved A549 and H1650s were treated with 1 μM nicotine in the presence or absence of LY294002 (PIK3CD/AKT pathway inhibitor) or PP2 (SRC pathway inhibitor) for 15 minutes, and the localization of ARRB1 was visualized by confocal microscopy (×630 magnification, scale bar = 10 μm). The cells were counterstained with the nuclear marker, DAPI. D) Effect of nicotine on localization of ARRB1 in subcellular fractions of A549 cells. Nuclear and cytosolic extracts were prepared from serum-starved and nicotine-treated A549 cells. Immunoblotting analysis was performed to examine the nuclear localization of ARRB1. As a positive control, immunoblotting was performed to assess the localization of ARRB1 in nuclear and cytosolic extracts prepared from A549 cells treated with 10 μM delta peptide. E) Effect of DhβE (α3/β2-nicotinic acetylcholine receptor [nAChR] antagonist), LY294002, or PP2 on nicotine-induced nuclear translocation of ARRB1 in A549 cells. Serum-starved A549 cells were treated with 1 μM nicotine in the presence or absence of different nAChR antagonists α-lobeline (α4/β2-nAChR antagonist), α-bungarotoxin (α7-nAChR subunit antagonist), MAA (α7-nAChR subunit antagonist), and DhβE. LY294002 or PP2 for 15 minutes and nuclear and cytosolic extracts were prepared. The subcellular localization of ARRB1 was examined by immunoblotting. The integrity of the nuclear extracts was confirmed by immunoblotting for PARP-1. Similarly, the purity of cytoplasmic extracts was assessed by analyzing the level of TUBA4A. All the immunoprecipitation-immunoblotting and immunofluorescence assays are representative of two independent experiments. Nic = nicotine.
Treatment of serum-starved A549 and H1650 cells with 1 μM nicotine for 15 minutes caused a robust nuclear localization of ARRB1 (Figure 2, B). Next, we investigated if LY294002 and PP2 showed an effect on nicotine-induced nuclear translocation of ARRB1 at 15 minutes in A549 cells and observed that ARRB1 translocation was substantially blocked by these inhibitors (Figure 2, C).
To confirm these findings, we performed a subcellular fractionation of nicotine-treated A549 cells and assessed the level of ARRB1 in the cytosolic and nuclear extracts by immunoblot analysis (Figure 2, D). We detected ARRB1 predominantly in the cytosolic extract in serum-starved A549 cells, and treatment with 1 μM nicotine showed an enrichment of ARRB1 protein in the nuclear extract. The delta peptide was used as the positive control because it is known to induce nuclear translocation of ARRB1 protein in HEK293 cells (30,34). Furthermore, nuclear translocation of ARRB1 was inhibited by DhβE (α3/β2-nAChR antagonist), but unaffected by α-lobeline (α4/β2-nAChR antagonist), α-bungarotoxin (α7-nAChR antagonist), or MAA (α7-nAChR antagonist) (Figure 2, E). Immunoblotting for PARP-1 and TUBA4A confirmed the integrity of nuclear and cytosolic extracts, respectively. These results indicated that the translocation of ARRB1 to the nucleus was nAChR-dependent.
Effect of Nicotine on Interaction Between ARRB1 Protein and E2F Pathway
We have previously shown that A549 cells treated with 1 μM nicotine recruit E2F1 transcription factor on proliferative promoters of CDC6, CDC25A, and BIRC5 genes (10,44). Because ARRB1 has no discernible DNA-binding domain (16,52), we hypothesized that it may affect the expression of proliferative genes via interaction with transcription factors like E2F1. Therefore, we investigated whether ARRB1 could bind to E2F1 in the nucleus. The ability of endogenous ARRB1 to bind to E2F1 in a signal-dependent process was examined by double-immunofluorescence experiments. Nicotine-treated and serum-starved (quiescent) A549 cells were immunostained for ARRB1 and E2F1 (Figure 3, A). Although we detected only negligible association between ARRB1 and E2F1 proteins in quiescent A549 cells, we found that nicotine treatment induced the translocation of a subset of ARRB1 to the nucleus, where it colocalized with E2F1 transcription factor.
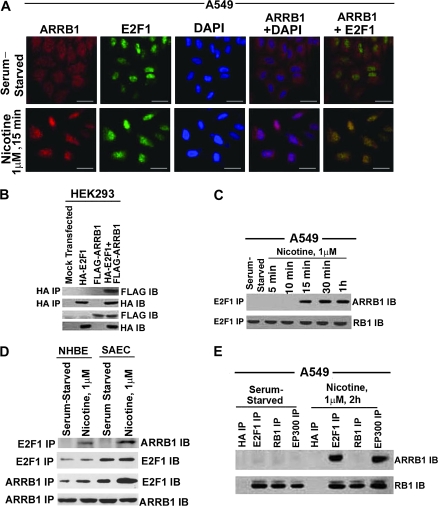
Figure 3.
Effect of nicotine on association between E2F1 and ARRB1 in human non–small cell lung cancer cells. A) Serum-starved A549 were treated with 1 μM nicotine for 15 minutes, and localization of ARRB1 and E2F1 was analyzed by double-immunoflourescence staining followed by confocal microscopy (×630 magnification, scale bar = 5 μm). The cells were counterstained with the nuclear marker, 4′6-diamidino-2-phenylindole. Overlay of the images show yellow spots indicating colocalization in the bottom right panel. B) Effect of overexpression of pcDNA3-FLAG-rat ARRB1 (wild type) and pcDNA3-HA-E2F1 in HEK293 cells. HEK293 cells were transfected with the above plasmids, and the physical interaction between ARRB1 and E2F1 was analyzed 24 hours after transfection by immunoprecipitation with mouse anti-HA antibody followed by immunoblotting using mouse anti-FLAG antibody. In addition, immunoblot analysis was done for FLAG and HA expression. C) Real-time kinetics of ARRB1–E2F1 interaction upon nicotine treatment of A549 cells. Serum-starved A549 cells were treated with 1 μM nicotine for 15 minutes, 30 minutes, and 1 hour, and the physical interaction between ARRB1 and E2F1 was analyzed by immunoprecipitation–immunoblot assay. Furthermore, the E2F1 immunoprecipitates were immunoblotted for RB1 to demonstrate equivalent amount of E2F1. D) Effect of nicotine on the association of ARRB1 and E2F1 in normal lung epithelial cells, NHBEs and SAECs. Quiescent NHBEs and SAECs were treated with 1 μM nicotine for 15 minutes, and the physical interaction between ARRB1 and E2F1 was analyzed by immunoprecipitation–immunoblot analysis. Furthermore, the E2F1 and ARRB1 immunoprecipitates were immunoblotted with E2F1 and ARRB1 to demonstrate equivalent amounts of protein. E) Effect of nicotine on binding of ARRB1 to E2F1, RB1 and EP300. Serum-starved A549 cells were treated with 1 μM nicotine for 2 hours. The binding of ARRB1 to E2F1, EP300, and RB1 was analyzed by immunoprecipitation–immunoblotting experiment. Immunoprecipitation with anti-mouse HA antibody was used as the negative control for the experiment. The above-mentioned immunoprecipitates were immunoblotted for RB1 protein. All the immunofluorescence and immunoprecipitation–immunoblotting experiments are representative of two independent experiments. IB = immunoblot; IP = immunoprecipitation.
To confirm the above results, we performed immunoprecipitation and immunoblot analysis on HEK293 cells transiently transfected with pcDNA3-HA-E2F1 pcDNA3-FLAG-rat ARRB1 or a combination of both. We observed that E2F1 and ARRB1 proteins interacted in these transfected cells (Figure 3, B). Also, association between endogenous E2F1 and endogenous ARRB1 was detected at 15 minutes, 30 minutes, and 1 hour of nicotine treatment of quiescent A549 cells (Figure 3, C). A direct association between E2F1 and ARRB1 proteins was also observed in human normal lung cells, NHBEs, and SAECs, treated with 1 μM of nicotine for 15 minutes (Figure 3, D). Because RB1 is a major regulator of E2F1 (37), we next examined whether ARRB1 could also interact with RB1. Previous studies have shown that nuclear localization of ARRB1 is accompanied by the recruitment of both ARRB1 and EP300 to the CDKN1B and FOS promoters facilitating histone acetylation and transcription of genes (30,31,34). We next assessed whether a similar event occurred in response to nicotine treatment of NSCLC cells, which led to enhanced levels of E2F-responsive proliferative genes. Both quiescent and nicotine-treated A549 cells showed that there was no association between ARRB1 and RB1, but in nicotine-treated cells, ARRB1 associated with E2F1 and EP300 (Figure 3, E).
Furthermore, to identify the domain of the E2F1 protein that binds with ARRB1 protein, we performed in vitro GST pull-down assays (Supplementary Methods, available online), with different constructs expressing different domains of E2F1 (39,53). The GST-RB1 construct was used as a positive control because RB1 is known to interact with E2F1 protein. We tested the binding of GAL4-E2F1(amino acids 304–437) and E2F1(amino acids 89–304)-VP16 with ARRB1 (Supplementary Figure 1, A and B, available online). The binding between ARRB1 and E2F1 was mediated through amino acids 89–304 of E2F1, and its transactivation domain was not needed (Supplementary Figure 1, B, available online). Additional GST pull-down assays showed that amino acids 103–284 of E2F1 were required for the binding of E2F1 to ARRB1 (Supplementary Figure 1, B, available online). Next, we investigated the regions of ARRB1 protein required for its association with E2F1. We observed that amino acids 1–163 of ARRB1 were required for binding with E2F1 (Supplementary Figure 1, C, available online). In addition to E2F1, other members of the E2F pathway like E2F2 and E2F3 also showed binding with ARRB1 upon nicotine treatment (Supplementary Figure 2, A and B, available online).
Nuclear ARRB1 and Histone Acetylation at Proliferative Promoters
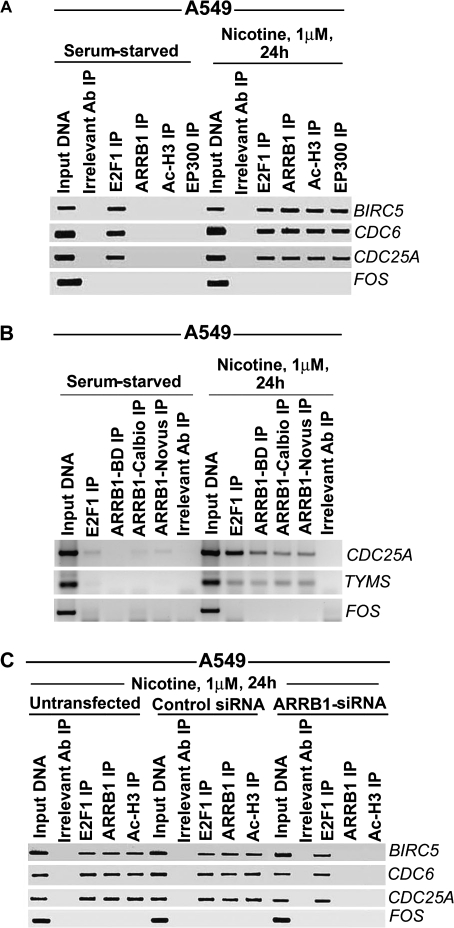
Next, we performed in vivo ChIP assays to investigate the effect of ARRB1 protein on histone acetylation at BIRC5, CDC6, and CDC25A promoters in response to nicotine treatment in NSCLC cells. Quiescent A549 cells were treated with 1 μM nicotine for 24 hours, and the binding between the E2F-responsive promoters (BIRC5, CDC6, and CDC25A) and E2F1, ARRB1, acetylated histone H3 (Ac-H3), and EP300 proteins was investigated (Figure 4, A). We detected binding between the promoters and E2F1 protein in quiescent cells, but not with ARRB1, Ac-H3, and EP300 proteins; however, induction with nicotine showed binding between the promoters and ARRB1, Ac-H3, and EP300 proteins. The FOS promoter was used as a negative control because it not regulated by E2F (39,40,53,54). Furthermore, the recruitment of ARRB1 on the E2F-responsive CDC25A and TYMS promoters (35,39,40,54–56) was confirmed by ChIP assays using three different commercial antibodies to ARRB1; all gave similar results (Figure 4, B). Knockdown of ARRB1 mRNA expression using ARRB1-specific siRNAs abolished the recruitment of EP300 and Ac-H3 proteins on BIRC5, CDC6, and CDC25A promoters in response to nicotine (Figure 4, C, and Supplementary Figure 3, available online). Collectively, these results showed that recruitment of ARRB1 to E2F-responsive promoters was a specific event induced by nicotine in cultured human NSCLC cells.
Figure 4.
Role of ARRB1 in the binding of E2F1, Ac-H3, and EP300 with E2F1-responsive promoters upon nicotine treatment of A549 cells. A) Effect of nicotine on the recruitment of E2F1, ARRB1, Ac-H3, and EP300 to E2F1-responsive promoters. Serum-starved A549 cells were treated with 1 μM nicotine for 24 hours, and the relative binding of E2F1, ARRB1, Ac-H3 and EP300 with E2F1-responsive promoters like BIRC5, CDC6 and CDC25A was measured by chromatin immunoprecipitation (ChIP) assay. FOS promoter was used as a negative control. B) ChIP assays were performed using three antibodies against ARRB1 from three different vendors. Serum-starved A549 cells were incubated with 1 μM nicotine for 24 hours, and the relative binding of ARRB1 with BIRC5, CDC6, and CDC25A promoters was examined. FOS promoter was used as a negative control. C) Effect of silencing of ARRB1 by siRNA on nicotine-induced binding of Ac-H3 and EP300 with the E2F1-responsive promoters. A549 cells were transfected with ARRB1 siRNA or control siRNA. The relative binding of E2F1, ARRB1, and Ac-H3 in untransfected and transfected cells was analyzed by ChIP assay. Rabbit anti-mouse IgG was used as the irrelevant antibody for all ChIP reactions. The input DNA lane represents one-fifth of the precleared chromatin used in each ChIP reaction. The results presented in this figure are representative of two independent transfection experiments and ChIP assays. Ab = antibody; IP = immunoprecipitation.
Effect of Nicotine and Serum on the Recruitment of ARRB1 and Ac-H3 on E2F-Responsive Proliferative Promoters
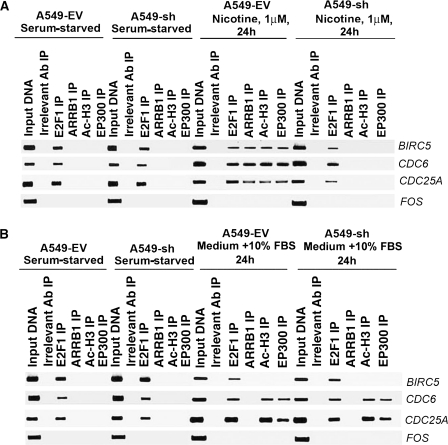
Taken together, the results of ChIP assays (Figure 4) showed that the treatment of human NSCLC cells with nicotine promoted histone acetylation on E2F-responsive promoters like CDC25A and TYMS. Next, ChIP assays were performed to analyze the binding of ARRB1 and Ac-H3 to E2F-responsive promoters in response to nicotine treatment vs 10% FBS treatment in A549-EV and A549-sh cells. Quiescent A549-EV and A549-sh cells showed binding of E2F1 alone on BIRC5, CDC6, and CDC25A promoters (Figure 5, A). Nicotine treatment induced the binding of E2F1, ARRB1, EP300, and acetylated H3 on these promoters in A549-EV cells and not in A549-sh cells (Figure 5, A). However, there was no difference in the recruitment of EP300 and Ac-H3 when both A549-EV and A549-sh cells were incubated in medium containing 10% FBS (Figure 5, B) showing that ARRB1 functions predominantly in response to nicotine-induced signaling but not in response to serum.
Figure 5.
Effect of suppression of ARRB1 on nicotine-induced vs serum-induced association of E2F1, ARRB1, Ac-H3, and EP300 with E2F-1 responsive promoters. A) Effect of nicotine on the binding of E2F1, ARRB1, Ac-H3, and EP300 with BIRC5, CDC6, and CDC25A promoters in A549-EV and A549-sh cells. Serum-starved A549-EV and A549-sh cells were treated with 1 μM nicotine for 24 hours, and the association of E2F1, ARRB1, Ac-H3, and EP300 with BIRC5, CDC6, and CDC25A promoters was assessed by chromatin immunoprecipitation (ChIP) assays. FOS promoter was used as a negative control. B) Effect of serum (medium containing 10% FBS) on the association of E2F1, ARRB1, Ac-H3, and EP300 with BIRC5, CDC6, and CDC25A promoters in A549-EV and A549-sh cells. Serum-starved A549-EV and A549-sh cells were incubated with medium containing 10% fetal bovine serum for 24 hours, and the association of E2F1, ARRB1, Ac-H3, and EP300 with BIRC5, CDC6, and CDC25A promoters was examined by ChIP assays. FOS promoter was used as a negative control. Rabbit anti-mouse IgG was used as the irrelevant antibody for all ChIP reactions. The input lane represents one-fifth of the precleared chromatin used in each ChIP reaction. The results presented in this figure are representative of two independent ChIP assays. Ab = antibody; IP = immunoprecipitation.
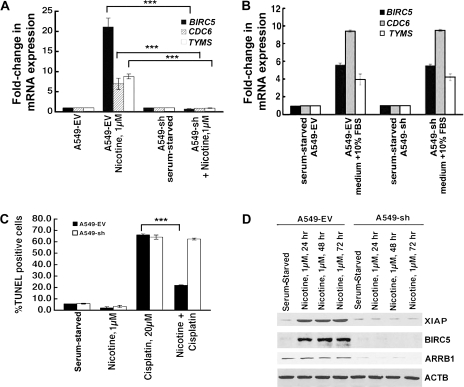
Effect of ARRB1 on the Expression of E2F-Regulated Proliferative Genes
Next, we conducted a series of experiments to examine whether ARRB1 was essential for nicotine-induced expression of E2F-regulated genes involved in cell proliferation. Real-time PCR analysis showed that nicotine statistically significantly induced the levels of BIRC5, CDC6, and TYMS genes in A549-EV cells but not in A549-sh cells (for BIRC5, A549-EV vs A549-sh, mean fold-increase in mRNA upon nicotine treatment = 20.7-fold, 95% CI = 19.2- to 22.2-fold, vs 0.8-fold, 95% CI = 0.78- to 0.82-fold, P < .001; for TYMS, A549-EV vs A549-sh, mean fold-increase in mRNA upon nicotine treatment = 7.5-fold, 95% CI = 6.0- to 8.0-fold, vs 1.0-fold, 95% CI = 0.99- to 1.001-fold, P < .001; for CDC6, A549-EV vs A549-sh, mean fold-increase in mRNA upon nicotine treatment = 9.1-fold, 95% CI = 8.6- to 9.7-fold, vs 1.0-fold, 95% CI = 0.99- to 1.001-fold, P < .001) (Figure 6, A). However, the treatment of cells with medium containing 10% FBS induced the above-mentioned promoters in both A549-EV and A549-sh cells to a comparable extent (Figure 6, B) (P >.05). We have previously shown that nicotine protected NSCLC cells from anticancer drug-induced apoptosis via an increased expression of XIAP and BIRC5 (7). We next examined whether ARRB1 plays a role in the antiapoptotic effects of nicotine. Analysis of apoptosis by TUNEL assays showed that nicotine-treated A549-sh cells regained their sensitivity to cisplatin-induced apoptosis compared with nicotine-treated A549-EV cells (nicotine and cisplatin-treated A549-EV vs nicotine and cisplatin-treated A549-sh cells, mean percentage-increase in TUNEL-positive cells = 25%, 95% CI = 24.9% to 25.1%, vs 64.6%, 95% CI = 64.1% to 65.1%, P < .001) (Figure 6, C).
Figure 6.
Effect of ARRB1-specific siRNA on nicotine-induced overexpression of E2F-responsive genes and on antiapoptotic effects of nicotine. A) Effect of silencing of ARRB1 on BIRC5, CDC6, and CDC25A mRNA levels in response to nicotine treatment in A549-EV and A549-sh cells. Serum-starved A549-EV and A549-sh cells were treated with 1 μM nicotine for 24 hours. Subsequently, the levels of BIRC5, CDC6, and TYMS mRNA were analyzed by real-time polymerase chain reaction (PCR). The means and 95% confidence intervals from two independent experiments are presented ***P < .001. B) Effect of suppression of ARRB1 levels on serum-induced overexpression of BIRC5, CDC6, and CDC25A mRNA. Serum-starved A549-EV and A549-sh cells were incubated with medium containing 10% fetal bovine serum for 24 hours, and the levels of BIRC5, CDC6, and TYMS mRNA were analyzed by real-time PCR. C) Role of ARRB1 in antiapoptotic effects of nicotine. Serum-starved A549-EV and A549-sh cells were treated with 1 μM nicotine for 30 minutes, then 20 μM cisplatin was added, and the cells were incubated for 24 hours. Apoptosis was measured by terminal deoxynucleotidyl transferase dUTP nick end labeling assay. The means and 95% confidence intervals from two independent experiments are shown ***P < .001. D) Effect of suppression of ARRB1 levels on the induction of XIAP and BIRC5 in response to nicotine. Serum-starved A549-EV and A549-sh cells were treated with 1 μM nicotine for 24, 48, or 72 hours. The expression levels of XIAP, BIRC5, and ARRB1 were analyzed by immunoblotting. ACTB was used as a loading control. All P values were calculated using a two-sided Student t test.
Additionally, nicotine treatment of A549-sh cells showed decreased expression of XIAP and BIRC5 compared with A549-EV cells (Figure 6, D). Taken together, ARRB1 appeared to play a critical role in the proliferative and antiapoptotic signaling by nAChRs (56–58) by facilitating the expression of E2F-regulated genes, BIRC5, TYMS, and CDC6, and thereby contributing to nicotine-mediated growth of NSCLC cells.
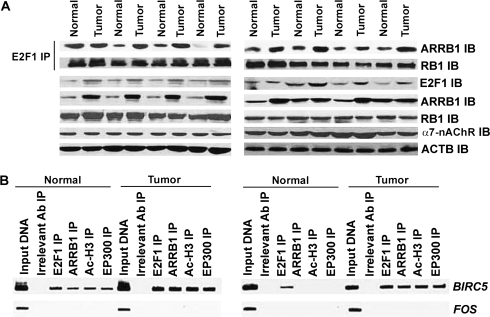
ARRB1 Expression and ARRB1-E2F1 Complexes in Human NSCLC Tumors From Smokers
Our next objective was to assess whether ARRB1 expression or the binding of ARRB1 to E2F1 (referred herewith as ARRB1–E2F1 complexes) were increased in human NSCLC tumors relative to normal lung tissues. Human NSCLC tumors (n = 8) were paired with matched distant normal lung tissue samples (n = 8) obtained from NSCLC patients who were active smokers or had a smoking history. Immunoprecipitation and immunoblot analysis of the lysates of these samples showed that the amount of ARRB1–E2F1 complex was increased in seven out of eight tumors (Figure 7, A) relative to matched distant normal lung tissue. Densitometric analysis demonstrated that the magnitude of ARRB1–E2F1 complex was statistically significantly higher (P < .05) in the NSCLC tumor tissue relative to matched distant normal lung tissue (Supplementary Figure 4, A, available online). Levels of E2F1 and ARRB1 were greater in six of eight NSCLC tumor samples compared with normal lung tissue, whereas the levels of α7-nAChR and RB1 were similar in tumor and normal tissues (Figure 7, A). Furthermore, ChIP assays on four human NSCLC tumor samples showed increased levels of E2F1, ARRB1, EP300, and Ac-H3 associated with BIRC5, CDC6, and CDC25A promoters in tumor tissues compared with matched distant normal lung tissue (Figure 7, B, and Supplementary Figure 4, B, available online). These results indicated that the nuclear ARRB1 probably played a role in the enhanced growth of NSCLCs.
Figure 7.
The levels of ARRB1–E2F1 complexes and association of E2F1, ARRB1, Ac-H3, and EP300 with BIRC5 promoter in human NSCLC tumors. A) The levels of ARRB1–E2F1 complexes in human NSCLC tumor tissues relative to matched distant normal lung tissues. Tissue lysates were prepared from human NSCLC tumors (n = 8) and matched distant normal lung tissue (n = 8). Immunoprecipitation–immunoblotting analysis was performed to compare the levels of ARRB1–E2F1 complexes in the above lysates. The E2F1 immunoprecipitates were analyzed for the level of RB1 protein by immunoblotting. The NSCLC tumor tissues were also analyzed for levels of E2F1, ARRB1, RB1, and α7-nAChR relative to matched distant normal lung tissue. ACTB was used as a loading control. B) Relative association of E2F1, ARRB1, Ac-H3, and EP300 with BIRC5 promoter in human NSCLC tumor tissue relative to matched distant normal lung tissue. Chromatin immunoprecipitation (ChIP) assays were performed to compare the levels of E2F1, ARRB1, Ac-H3, and EP300 bound to BIRC5 promoter in these tissues. FOS promoter was used as a negative control. Rabbit anti-mouse IgG was used as the irrelevant antibody for all ChIP reactions. The input DNA lane represents one-fifth of the precleared chromatin used in each ChIP reaction. The results presented in this figure are representative of two independent immunoprecipitation–immunoblotting assays. Ab = antibody; IB = immunoblot; IP = immunoprecipitation.
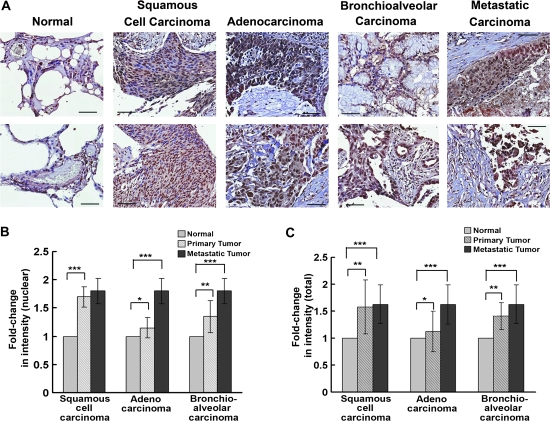
ARRB1 Levels in Human NSCLCs
Human lung cancer tissue microarrays were immunostained for ARRB1 using rabbit anti-human ARRB1 antibody to determine whether total ARRB1 and nuclear ARRB1 levels were increased in human NSCLC tissues relative to matched distant normal lung tissues. Primary squamous cell carcinomas and bronchioalveolar carcinomas showed elevated levels of ARRB1 relative to matched distant normal lung tissues (Figure 8, A). Furthermore, ARRB1 appeared to be more nuclear in the tumor sections (normal lung tissue vs primary squamous cell carcinoma, mean fold-increase in nuclear ARRB1 expression = 1-fold, 95% CI = 1- to 1-fold, vs 1.7-fold, 95% CI = 1.4- to 2-fold, P < .001; normal lung tissue vs primary bronchioalveolar cell carcinoma, mean fold-increase in nuclear ARRB1 expression = 1-fold, 95% CI = 1- to 1-fold, vs 1.4-fold, 95% CI = 1.1- to 1.7-fold, P < .01). Highest levels of nuclear ARRB1 were observed in metastatic cancers (normal lung tissue vs metastatic adenocarcinoma, mean fold-increase in nuclear ARRB1 expression = 1-fold, 95% CI = 1- to 1-fold, vs 1.8-fold, 95% CI = 1.5- to 2.1-fold, P<.001; normal lung tissue vs metastatic bronchioalveolar cell carcinoma, mean fold-increase in nuclear ARRB1 expression = 1-fold, 95% CI = 1- to 1-fold, vs 1.8-fold, 95% CI = 1.5- to 2.1-fold, P < .001) (Figure 8, B). Total ARRB1 levels were elevated in distant metastatic lung tumors (normal lung tissue vs metastatic carcinoma, mean fold-increase in total ARRB1 expression = 1-fold, 95% CI = 1- to 1-fold, vs 1.6-fold, 95% CI = 1.3- to 1.9-fold, P < .001), but the primary lung adenocarcinoma did not have substantially higher level of total ARRB1 than the normal tissue (Figure 8, C). Taken together, these results indicate that elevated levels of ARRB1 in squamous cell carcinoma, bronchioalveolar carcinoma, and metastatic adenocarcinoma may contribute at least, in part, to the growth and metastasis of NSCLC (56–58).
Figure 8.
Total ARRB1 expression and its subcellular localization in human lung cancer tissue microarray. A) Immunohistochemical staining of ARRB1 in human lung cancer tissue arrays. Immunostaining was performed using rabbit anti-human ARRB1 antibody and representative images of ARRB1 expression in normal lung tissue, squamous cell carcinomas, adenocarcinomas, bronchioalveolar carcinomas, and metastatic carcinomas are shown. Magnification = ×200. Scale bar = 10 μm. B) Quantitative analysis of nuclear ARRB1 in lung tissue microarray. The immunostaining of ARRB1 in primary squamous cell carcinomas (n = 13), adenocarcinomas (n = 4), bronchioalveolar carcinomas (n = 4), and metastatic carcinomas (n = 10) relative to normal tissue (n = 9) was quantified by using semiquantitative scoring method based on cellularity and intensity of expression. Normal lung tissue vs squamous cell carcinomas, ***P < .001; normal lung tissue vs adenocarcinomas, *P < .4; normal lung tissue vs bronchioalveolar carcinomas, **P < .1; and normal lung tissue vs metastatic carcinomas, ***P < .003. C) Quantitative analysis of total ARRB1 in lung tissue microarray. The immunostaining of total ARRB1 in primary squamous cell carcinomas, adenocarcinomas, bronchioalveolar carcinomas, and metastatic carcinomas, relative to normal tissue, was quantified by using semiquantitative scoring method based on cellularity and intensity of expression. Normal lung tissue vs squamous cell carcinomas, ***P < .01; normal lung tissue vs adenocarcinomas, *P < .5; normal lung tissue vs bronchioalveolar carcinomas, **P < .1; and normal lung tissue vs metastatic carcinomas, ***P < .004. The means and 95% confidence intervals of two independent arrays are shown. All P values were calculated using a two-sided Student t test.
Discussion
In this study, we showed for the first time that ARRB1 was vital to the proliferative and antiapoptotic activity of nicotine in human NSCLC cells. In addition, our data demonstrated that nicotine could induce the translocation of ARRB1 to the nucleus in normal lung cells and NSCLC cells. Nuclear ARRB1 directly bound to the E2F family of transcription factors in both NSCLC cell lines and in human NSCLC tumors. The levels of ARRB1–E2F complex were greater in NSCLC tumor tissues of smokers compared with matched distant normal lung tissue. ARRB1 was detected on the promoters of E2F-responsive proliferative and prosurvival genes where it facilitated histone acetylation. These results indicated that ARRB1 was involved in regulating the expression of E2F target proliferative genes like CDC6, TYMS, and prosurvival genes like BIRC5. Finally, analysis of ARRB1 levels in human NSCLC tumor microarray revealed enhanced nuclear and total ARRB1 levels in tumor tissues compared with normal lung tissue. Taken together, this study shows a novel function for ARRB1 in nicotine-induced survival of human NSCLC and the mitogenic pathways involved in the process.
ARRB1 is a scaffolding protein involved in GPCR desensitization (16,52) and can also facilitate the functioning of non-GPCRs like Frizzled, Notch, and the nAChRs (15,19–21). Here, we show for the first time that nAChR signaling in normal lung epithelial cells and NSCLCs is at least, in part, mediated by the nuclear functions of ARRB1. The nuclear localization of ARRB1 was found to be dependent on nAChR function SRC and PIK3CD/AKT pathway. Given the established role of SRC and AKT1 in tumor progression and metastasis (59,60), it is probable that nuclear ARRB1-induced gene regulation plays a role in facilitating the survival and growth of NSCLC tumors (20,26,27,29,33,52). The enrichment of ARRB1 was observed in proliferative promoters like CDC25A, CDC6, and prosurvival promoters like BIRC5, which are known to promote tumor growth (7).
This study also shows that nuclear ARRB1 can directly bind to the E2F family of transcription factors. The treatment of A549 NSCLC cells with nicotine led to the formation of an oligomeric complex comprising of ARRB1, E2F1, and EP300, which facilitated the acetylation of histones and E2F1, inducing the transcription of proliferative and prosurvival genes. Our findings are in agreement with previous studies, which showed that nuclear translocation of ARRB1 in response to delta-opioid receptor signaling increased the transcription of cell cycle regulatory genes like CDKN1B and FOS (30,31). Previous studies have shown that CDKN1B is an E2F-responsive gene (35,37,55) that is frequently dysregulated in NSCLCs (61). Therefore, the ARRB1-E2F1-signaling axis might represent a novel pathway of gene expression in NSCLC that regulates proliferation and survival.
Our data suggest that ARRB1 facilitates the acetylation of histones in NSCLCs, specifically in response to nicotine and not in response to serum. It is also possible that nicotine-induced nuclear ARRB1 enhances the acetylation of other cellular proteins, leading to transcriptional activation. For example, acetylation of E2F1 has been found to increase its affinity for E2F1–DNA-binding sites, leading to enhanced transactivation of E2F1-responsive promoters (62–64). Our previous studies have shown that vascular endothelial growth factor enhances the acetylation of E2F1 and promotes the recruitment of acetyl transferases like cAMP-response element-binding protein binding protein (CREBBP), EP300 and P300/CREBBP associated factor (KAT2B) on the pro-angiogenic fms-related tyrosine kinase-1 (FLT-1), and kinase insert domain receptor (KDR) promoters (65). It is known that the acquisition of angiogenic phenotype is vital for the sustained growth of NSCLCs. Therefore, it is possible that the ARRB1-E2F1 pathway contributes to the progression of NSCLCs, by facilitating the expression of genes like FLT-1 and KDR (66–68).
Although, we describe results that are novel and relevant to NSCLC, this study has a few limitations. For example, our results do not shed light on the specific functions of the binding of nuclear ARRB1 with individual E2F family members. It remains a possibility that the transcriptional regulatory functions of ARRB1 (in response to nicotine) are mediated at least, in part, by transcription factors other than E2F. The extent of such interactions and their relevance to nicotine-induced signaling in NSCLC is not known. Similarly, it is not clear from the above studies whether the only genes that are regulated are those involved in proliferation and survival. A broader analysis of gene expression profiles would be necessary to answer these questions.
In this study, we showed for the first time that the association of ARRB1, E2F1, and EP300 on proliferative promoters occurred in response to nicotine and not in response to serum. The above observation is associated with increased magnitude of ARRB1–E2F1 complex in NSCLC tumors from patients who were either current smokers or had a smoking history. The development of NSCLCs shows a strong epidemiological association with cigarette smoking (1). Our data raise the possibility that nicotine-induced nuclear translocation of ARRB1, and its subsequent binding to E2F1 may be an important event mediating the growth and progression of NSCLCs as a result of exposure to tobacco carcinogens. Considering that exposure to nicotine induces epithelial–mesenchymal transition (69) and metastasis of NSCLC (46,69–72), these observations seem to suggest that nuclear functions of ARRB1 might be contributing to the growth and metastasis of NSCLCs, especially in smokers.
Funding
National Cancer Institute, National Institutes of Health (CA127725 to S.P.C.); Young Clinical Scientist Award, Flight Attendant’s Medical Research Institute (82115 to P.D.).
Supplementary Material
Footnotes
The authors are solely responsible for the design of the study, the analysis and interpretation of the data, the writing of the article, and the decision to submit the article for publication.
We thank the Moffitt Cancer Tissue Bank for providing frozen human non–small cell lung cancer and normal lung tissues for the study. We thank the Histopathology and Microscopy Core Facility, Moffitt Cancer Center, for their technical assistance. We are grateful to Dr Alexis Lopez for scoring the lung tissue microarray.
References
- 1.Johnson BE. Tobacco and lung cancer. Prim Care. 1998;25(2):279–291. doi: 10.1016/s0095-4543(05)70064-6. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts and Figures. Atlanta, GA: ACS; 2002. [Google Scholar]
- 3.Schuller HM, Orloff M. Tobacco-specific carcinogenic nitrosamines. Ligands for nicotinic acetylcholine receptors in human lung cancer cells. Biochem Pharmacol. 1998;55(9):1377–1384. doi: 10.1016/s0006-2952(97)00651-5. [DOI] [PubMed] [Google Scholar]
- 4.Schuller HM, Jull BA, Sheppard BJ, Plummer HK. Interaction of tobacco-specific toxicants with the neuronal α7 nicotinic acetylcholine receptor and its associated mitogenic signal transduction pathway: potential role in lung carcinogenesis and pediatric lung disorders. Eur J Pharmacol. 2000;393(1–3):265–277. doi: 10.1016/s0014-2999(00)00094-7. [DOI] [PubMed] [Google Scholar]
- 5.West KA, Brognard J, Clark AS, et al. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111(1):81–90. doi: 10.1172/JCI16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Calaf GM, Hei TK. Malignant transformation of human bronchial epithelial cells with the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Int J Cancer. 2003;106(6):821–826. doi: 10.1002/ijc.11319. [DOI] [PubMed] [Google Scholar]
- 7.Dasgupta P, Kinkade R, Joshi B, Decook C, Haura E, Chellappan S. Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc Natl Acad Sci U S A. 2006;103(16):6332–6337. doi: 10.1073/pnas.0509313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manackjee R, Minna JD. Opioid and nicotine receptors affect growth regulation of human lung cancer cell lines. Proc Natl Acad Sci U S A. 1990;87(9):3294–3298. doi: 10.1073/pnas.87.9.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charpantier E, Wiesner A, Huh KH, et al. Alpha7 neuronal nicotinic acetylcholine receptors are negatively regulated by tyrosine phosphorylation and Src-family kinases. J Neurosci. 2005;25(43):9836–9849. doi: 10.1523/JNEUROSCI.3497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohamed AS, Swope SL. Phosphorylation and cytoskeletal anchoring of the acetylcholine receptor by Src class protein-tyrosine kinases. Activation by rapsyn. J Biol Chem. 1999;274(29):20529–20539. doi: 10.1074/jbc.274.29.20529. [DOI] [PubMed] [Google Scholar]
- 11.Kumar P, Meizel S. Nicotinic acetylcholine receptor subunits and associated proteins in human sperm. J Biol Chem. 2005;280(27):25928–25935. doi: 10.1074/jbc.M502435200. [DOI] [PubMed] [Google Scholar]
- 12.Gotti C, Clementi F. Neuronal nicotinic receptors: from structure to pathology. Prog Neurobiol. 2004;74(6):363–396. doi: 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Itier V, Bertrand D. Neuronal nicotinic receptors: from protein structure to function. FEBS Lett. 2001;504(3):118–125. doi: 10.1016/s0014-5793(01)02702-8. [DOI] [PubMed] [Google Scholar]
- 14.Lindstrom J. Ion Channels. New York, NY: Plenum Press; 1996. Neuronal nicotinic acetylcholine receptors; pp. 377–450. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta P, Rastogi S, Pillai S, et al. Nicotine induces cell proliferation by beta-arrestin-mediated activation of Src and Rb-Raf-1 pathways. J Clin Invest. 2006;116(8):2208–2217. doi: 10.1172/JCI28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308(5721):512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 17.Lefkowitz RJ, Whalen EJ. Beta-arrestins: traffic cops of cell signaling. Curr Opin Cell Biol. 2004;16(2):162–168. doi: 10.1016/j.ceb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Ahn S, Nelson CD, Garrison TR, Miller WE, Lefkowitz RJ. Desensitization, internalization, and signaling functions of beta-arrestins demonstrated by RNA interference. Proc Natl Acad Sci U S A. 2003;100(4):1740–1744. doi: 10.1073/pnas.262789099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 2005;7(12):1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 20.Rosano L, Cianfrocca R, Masi S, et al. Beta-arrestin links endothelin A receptor to beta-catenin signaling to induce ovarian cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2009;106(8):2806–2811. doi: 10.1073/pnas.0807158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, ten Berge D, Brown J, et al. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science. 2003;301(5638):1391–1394. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Ren XR, Nelson CD, et al. Activity-dependent internalization of smoothened mediated by beta-arrestin 2 and GRK2. Science. 2004;306(5705):2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 23.Wilbanks AM, Fralish GB, Kirby ML, Barak LS, Li YX, Caron MG. Beta-arrestin 2 regulates zebrafish development through the hedgehog signaling pathway. Science. 2004;306(5705):2264–2267. doi: 10.1126/science.1104193. [DOI] [PubMed] [Google Scholar]
- 24.Luttrell LM, Ferguson SS, Daaka Y, et al. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283(5402):655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 25.Shenoy SK, Drake MT, Nelson CD, et al. Beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281(2):1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 26.Li TT, Alemayehu M, Aziziyeh AI, et al. Beta-arrestin/Ral signaling regulates lysophosphatidic acid-mediated migration and invasion of human breast tumor cells. Mol Cancer Res. 2009;7(7):1064–1077. doi: 10.1158/1541-7786.MCR-08-0578. [DOI] [PubMed] [Google Scholar]
- 27.Buchanan FG, Gorden DL, Matta P, Shi Q, Matrisian LM, DuBois RN. Role of beta-arrestin 1 in the metastatic progression of colorectal cancer. Proc Natl Acad Sci U S A. 2006;103(5):1492–1497. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galliera E, Jala VR, Trent JO, et al. Beta-arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279(24):25590–25597. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- 29.Tilley DG, Kim IM, Patel PA, Violin JD, Rockman HA. Beta-arrestin mediates beta1-adrenergic receptor-epidermal growth factor receptor interaction and downstream signaling. J Biol Chem. 2009;284(30):20375–20386. doi: 10.1074/jbc.M109.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang J, Shi Y, Xiang B, et al. A nuclear function of beta-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell. 2005;123(5):833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Ma L, Pei G. Beta-arrestin signaling and regulation of transcription. J Cell Sci. 2007;120(Pt 2):213–218. doi: 10.1242/jcs.03338. [DOI] [PubMed] [Google Scholar]
- 32.Lakshmikanthan V, Zou L, Kim JI, et al. Identification of betaArrestin2 as a corepressor of androgen receptor signaling in prostate cancer. Proc Natl Acad Sci U S A. 2009;106(23):9379–9384. doi: 10.1073/pnas.0900258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun KS, Lao HC, Trempus CS, Okada M, Langenbach R. The prostaglandin receptor EP2 activates multiple signaling pathways and beta-arrestin1 complex formation during mouse skin papilloma development. Carcinogenesis. 2009;30(9):1620–1627. doi: 10.1093/carcin/bgp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaulieu JM, Caron MG. Beta-arrestin goes nuclear. Cell. 2005;123(5):755–757. doi: 10.1016/j.cell.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle. 2009;8(4):532–535. doi: 10.4161/cc.8.4.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem. 2007;102(4):1400–1404. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- 37.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9(9):713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 38.Kinkade R, Dasgupta P, Carie A, et al. A small molecule disruptor of Rb/Raf-1 interaction inhibits cell proliferation, angiogenesis, and growth of human tumor xenografts in nude mice. Cancer Res. 2008;68(10):3810–3818. doi: 10.1158/0008-5472.CAN-07-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dasgupta P, Sun J, Wang S, et al. Disruption of the Rb–Raf-1 interaction inhibits tumor growth and angiogenesis. Mol Cell Biol. 2004;24(21):9527–9541. doi: 10.1128/MCB.24.21.9527-9541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Ghosh RN, Chellappan SP. Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation. Mol Cell Biol. 1998;18(12):7487–7498. doi: 10.1128/mcb.18.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J Biol Chem. 2006;281(5):2951–2959. doi: 10.1074/jbc.M508669200. [DOI] [PubMed] [Google Scholar]
- 42.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucl Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dignam JD, Martin PL, Shastry BS, Roeder RG. Methods Enzymol. Maryland Heights, MO: Elsevier; 1983. Eukaryotic gene transcription with purified components; pp. 582–598. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez ER, Hu JL, Zhong S, Shen P, Greene MJ, Housley PR. Potentiation of glucocorticoid receptor-mediated gene expression by heat and chemical shock. Mol Endocrinol. 1994;8(4):408–421. doi: 10.1210/mend.8.4.8052262. [DOI] [PubMed] [Google Scholar]
- 45.Rastogi S, Joshi B, Dasgupta P, Morris M, Wright K, Chellappan S. Prohibitin facilitates cellular senescence by recruiting specific corepressors to inhibit E2F target genes. Mol Cell Biol. 2006;26(11):4161–4171. doi: 10.1128/MCB.02142-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis R, Rizwani W, Banerjee S, et al. Nicotine promotes tumor growth and metastasis in mouse models of lung cancer. PLoS One. 2009;4(10):e7524. doi: 10.1371/journal.pone.0007524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein M, Vignaud JM, Hennequin V, et al. Increased expression of the vascular endothelial growth factor is a pejorative prognosis marker in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2001;86(2):656–658. doi: 10.1210/jcem.86.2.7226. [DOI] [PubMed] [Google Scholar]
- 48.Heeschen C, Weis M, Aicher A, Dimmler S, Cooke JP. A novel angiogenic pathway mediated by non-neuronal nicotinic acetylcholine receptors. J Clin Invest. 2002;110(4):527–536. doi: 10.1172/JCI14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dasgupta P, Chellappan SP. Nicotine-mediated cell proliferation and angiogenesis: new twists to an old story. Cell Cycle. 2006;5(20):2324–2328. doi: 10.4161/cc.5.20.3366. [DOI] [PubMed] [Google Scholar]
- 50.West KA, Linnoila IR, Belinsky SA, Harris CC, Dennis PA. Tobacco carcinogen-induced cellular transformation increases activation of the phosphatidylinositol 3’-kinase/Akt pathway in vitro and in vivo. Cancer Res. 2004;64(2):446–451. doi: 10.1158/0008-5472.can-03-3241. [DOI] [PubMed] [Google Scholar]
- 51.Schuller HM, Plummer HK, III, Jull BA. Receptor-mediated effects of nicotine and its nitrosated derivative NNK on pulmonary neuroendocrine cells. Anat Rec. 2003;270A(1):51–58. doi: 10.1002/ar.a.10019. [DOI] [PubMed] [Google Scholar]
- 52.Lefkowitz RJ, Rajagopal K, Whalen EJ. New roles for beta-arrestins in cell signaling: not just for seven-transmembrane receptors. Mol Cell. 2006;24(5):643–652. doi: 10.1016/j.molcel.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Dasgupta P, Betts V, Rastogi S, et al. Direct binding of apoptosis signal-regulating kinase 1 to retinoblastoma protein: novel links between apoptotic signaling and cell cycle machinery. J Biol Chem. 2004;279(37):38762–38769. doi: 10.1074/jbc.M312273200. [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Nath N, Fusaro G, Chellappan S. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol Cell Biol. 1999;19(11):7447–7460. doi: 10.1128/mcb.19.11.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polager S, Ginsberg D. E2F—at the crossroads of life and death. Trends Cell Biol. 2008;18(11):528–535. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Egleton RD, Brown KC, Dasgupta P. Nicotinic acetylcholine receptors in cancer: multiple roles in proliferation and inhibition of apoptosis. Trends Pharmacol Sci. 2008;29(3):151–158. doi: 10.1016/j.tips.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Lindstrom JM. Vol. 998. New York: New York Academy of Sciences; 2003. Nicotinic acetylcholine receptors of muscles and nerves: comparison of their structures, functional roles, and vulnerability to pathology; pp. 41–52. [DOI] [PubMed] [Google Scholar]
- 58.Cook JP, Bitterman H. Nicotine and angiogenesis: a new paradigm for tobacco-related diseases. Ann Med. 2004;36(1):33–40. doi: 10.1080/07853890310017576. [DOI] [PubMed] [Google Scholar]
- 59.Brunton VG, Frame MC. Src and focal adhesion kinase as therapeutic targets in cancer. Curr Opin Pharmacol. 2008;8(4):427–432. doi: 10.1016/j.coph.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 60.Sarker D, Reid AH, Yap TA, de Bono JS. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res. 2009;15(15):4799–4805. doi: 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]
- 61.Neuhaus EM, Mashukova A, Barbour J, Wolters D, Hatt H. Novel function of beta-arrestin2 in the nucleus of mature spermatozoa. J Cell Sci. 2006;119(Pt 15):3047–3056. doi: 10.1242/jcs.03046. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19(4):662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozaki T, Okoshi R, Sang M, Kubo N, Nakagawara A. Acetylation status of E2F-1 has an important role in the regulation of E2F-1-mediated transactivation of tumor suppressor p73. Biochem Biophys Res Commun. 2009;386(1):207–211. doi: 10.1016/j.bbrc.2009.06.035. [DOI] [PubMed] [Google Scholar]
- 64.Daury L, Chailleux C, Bonvallet J, Trouche D. Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 2006;7(1):66–71. doi: 10.1038/sj.embor.7400561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pillai S, Kovacs M, Chellappan S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: role of acetylation. Cancer Res. 2010;70(12):4931–4940. doi: 10.1158/0008-5472.CAN-10-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trombino S, Bisio A, Catassi A, Cesario A, Falugi C, Russo P. Role of the non-neuronal human cholinergic system in lung cancer and mesothelioma: possibility of new therapeutic strategies. Curr Med Chem Anticancer Agents. 2004;4(6):535–542. doi: 10.2174/1568011043352687. [DOI] [PubMed] [Google Scholar]
- 67.Trombino S, Cesario A, Margaritora S, et al. Alpha7-nicotinic acetylcholine receptors affect growth regulation of human mesothelioma cells: role of mitogen-activated protein kinase pathway. Cancer Res. 2004;64(1):135–145. doi: 10.1158/0008-5472.can-03-1672. [DOI] [PubMed] [Google Scholar]
- 68.Catassi A, Servent D, Paleari L, Cesario A, Russo P. Multiple roles of nicotine on cell proliferation and inhibition of apoptosis: implications on lung carcinogenesis. Mutat Res. 2008;659(3):221–231. doi: 10.1016/j.mrrev.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 69.Dasgupta P, Rizwani W, Pillai S, et al. Nicotine induces cell proliferation, invasion and epithelial-mesenchymal transition in a variety of human cancer cell lines. Int J Cancer. 2009;124(1):36–45. doi: 10.1002/ijc.23894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paleari L, Catassi A, Ciarlo M, et al. Role of alpha7-nicotinic acetylcholine receptor in human non-small cell lung cancer proliferation. Cell Prolif. 2008;41(6):936–959. doi: 10.1111/j.1365-2184.2008.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Paleari L, Cesario A, Fini M, Russo P. Alpha7-nicotinic receptor antagonists at the beginning of a clinical era for NSCLC and Mesothelioma? Drug Discov Today. 2009;14(17–18):822–836. doi: 10.1016/j.drudis.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 72.Paleari L, Grozio A, Cesario A, Russo P. The cholinergic system and cancer. Semin Cancer Biol. 2008;18(3):211–217. doi: 10.1016/j.semcancer.2007.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.