Abstract
Atg9 is a transmembrane protein essential for autophagy which cycles between the Golgi network, late endosomes and LC3-positive autophagosomes in mammalian cells during starvation through a mechanism that is dependent on ULK1 and requires the activity of the class III phosphatidylinositol-3-kinase (PI3KC3). In this study, we demonstrate that the N-BAR-containing protein, Bif-1, is required for Atg9 trafficking and the fission of Golgi membranes during the induction of autophagy. Upon starvation, Atg9-positive membranes undergo continuous tubulation and fragmentation to produce cytoplasmic punctate structures that are positive for Rab5, Atg16L and LC3. Loss of Bif-1 or inhibition of the PI3KC3 complex II suppresses starvation-induced fission of Golgi membranes and peripheral cytoplasmic redistribution of Atg9. Moreover, Bif-1 mutants, which lack the functional regions of the N-BAR domain that are responsible for membrane binding and/or bending activity, fail to restore the fission of Golgi membranes as well as the formation of Atg9 foci and autophagosomes in Bif-1-deficient cells starved of nutrients. Taken together, these findings suggest that Bif-1 acts as a critical regulator of Atg9 puncta formation presumably by mediating Golgi fission for autophagosome biogenesis during starvation.
Key words: Bif-1, Atg9 trafficking, Golgi fragmentation, membrane curvature, Beclin 1, UVRAG, class III PI3-kinase, autophagosome, starvation
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionally conserved bulk degradation process that is activated in response to a variety of intracellular and extracellular stresses including nutrient starvation.1,2 This self-digestive machinery is required for maintaining cellular homeostasis and is involved in a variety of physiological and pathophysiological processes including development, aging, neurodegenerative disorders, cancer and infectious diseases.3,4 The process of autophagic degradation is initiated by the sequestration of cytosolic components into crescent-shaped double membrane structures known as isolation membranes or phagophores.5,6 The isolation membranes are extended and sealed to form double-membrane vesicles termed autophagosomes, which then fuse with endosomes and/or lysosomes for hydrolytic degradation of the sequestered materials. The resulting macromolecules are then transported back into the cytosol for reuse.
The origin of autophagosomal membranes is still a topic under debate, although several candidates have been proposed as a membrane source for autophagosome formation. A previous study has shown that phosphatidylinositol 3-phosphate (PI3P)-enriched membrane structures called omegasomes are generated in the proximity of the endoplasmic reticulum (ER) and colocalize with autophagosome markers during starvation.7 Moreover, recent electron tomographic analyses revealed that the ER associates with isolation membranes and forms a cradle for the elongation of isolation membranes.8,9 While these reports highlight the ER as the primary origin of autophagosomal membranes, recent studies have demonstrated the involvement of alternative membrane sources in autophagosome biogenesis including the Golgi complex,10–12 mitochondria13 and the plasma membrane.14
Atg9 is a member of the Atg protein family that contains six transmembrane domains and plays an essential role in autophagosome formation.6,15–17 In yeast, Atg9 cycles between a perivacuolar site, known as the PAS (for phagophore assembly site or pre-autophagosomal structure), and peripheral sites including mitochondria, suggesting that Atg9-containing compartments are a source of membranes for the formation and/or expansion of autophagosomes.18,19 In contrast, mammalian Atg9 has been shown to localize to the trans-Golgi network (TGN) and late endosomes, but not to mitochondria.20,21 Starvation promotes the redistribution of Atg9 from the Golgi complex to the peripheral cytoplasm where it colocalizes with the autophagosome marker, LC3.20,21 Interestingly, the redistribution of Atg9 upon starvation requires the activation of class III phosphatidylinositol-3-kinase (PI3KC3) and Unc-51 like kinase (Ulk1).20
Here, we demonstrate that nutrient starvation induces the tubulation and fragmentation of Atg9-positive Golgi membranes in a manner that is dependent on the membrane curvature-driving protein Bif-1/Endophilin B1 and the PI3KC3 complex II. Starvation-induced Atg9 foci colocalize not only with Bif-1, but also with an early endosome marker, Rab5, and an autophagosome precursor marker, Atg16L. Knockout or knockdown of Bif-1 as well as inhibition of the PI3KC3 complex II, by PI3K inhibitor or knockdown of Beclin 1 or UVRAG impairs Golgi fission, Atg9 trafficking and LC3 foci formation. Moreover, starvation-induced Golgi fragmentation and Atg9 redistribution from the Golgi complex to peripheral sites require a functional N-BAR domain of Bif-1. Taken together, we propose that Bif-1-mediated fragmentation of the Golgi complex during nutrient starvation plays a crucial role in Atg9 trafficking and autophagosome biogenesis.
Results
Loss of Bif-1 suppresses the redistribution of Atg9 from the golgi complex to the peripheral cytoplasm during starvation.
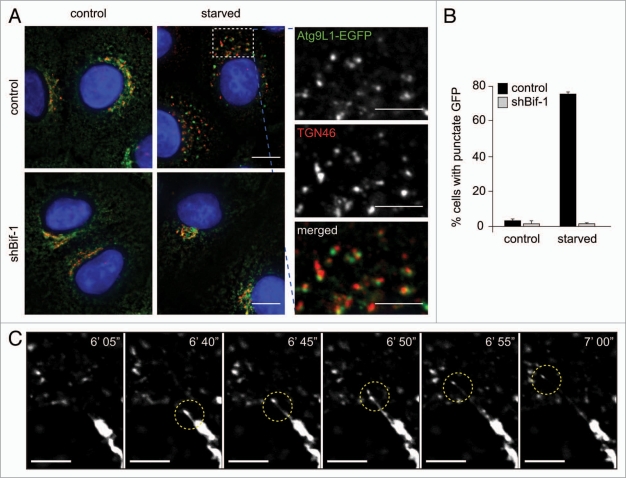
We have previously demonstrated that Bif-1 interacts with Beclin 1 through UVRAG and that loss of Bif-1 suppresses the formation of autophagosomes.22 Bif-1, also known as Endophilin B1 and SH3GLB1, is a member of the N-BAR and SH3 domain-containing endophilin protein family, which has been shown to tubulate liposomes in vitro23 and regulate the membrane dynamics of a variety of intracellular organelles.24,25 In response to starvation, Bif-1 forms foci in the cytoplasm where it colocalizes not only with LC3, but also with Atg5 and Atg9,22,26 suggesting that Bif-1 acts at the early stages of autophagosome formation. To determine whether Bif-1 is involved in Atg9 trafficking, we compared the localization of Atg9 in Bif-1 knockdown and control HeLa cells before and after starvation. In agreement with a previous study,20 Atg9 signals were detected in the cytoplasm including the juxtanuclear region where they partially colocalized with a trans-Golgi marker, TGN46, under normal culture conditions regardless of the presence or absence of Bif-1 expression (Fig. 1A). However, upon starvation, the juxtanuclear Atg9 signals were significantly reduced and numerous Atg9-positive punctate structures appeared in the peripheral cytoplasm of control HeLa cells (Fig. 1A and B). These starvation-induced Atg9-positive structures were mostly adjacent to or colocalized with TGN46-positive membranes. Interestingly, time-lapse microscopic analyses revealed that Atg9-positive membranes undergo continuous tubulation and fragmentation during nutrient starvation, resulting in the formation of cytoplasmic foci (Fig. 1C and Suppl. Video 1). In contrast, knockdown of Bif-1 abrogated starvation-induced tubulation and fission of Atg9-positive Golgi membranes (Fig. 1A and B and Suppl. Video 2). Similar results were obtained using Bif-1+/+ and −/− MEFs that were transfected with Atg9-EGFP and subjected to starvation or rapamycin treatment (Suppl. Fig. S1A). Importantly, we observed that endogenous Atg9 protein also requires Bif-1 to form foci in the peripheral cytoplasm during nutrient starvation (Suppl. Fig. S1B).
Figure 1.
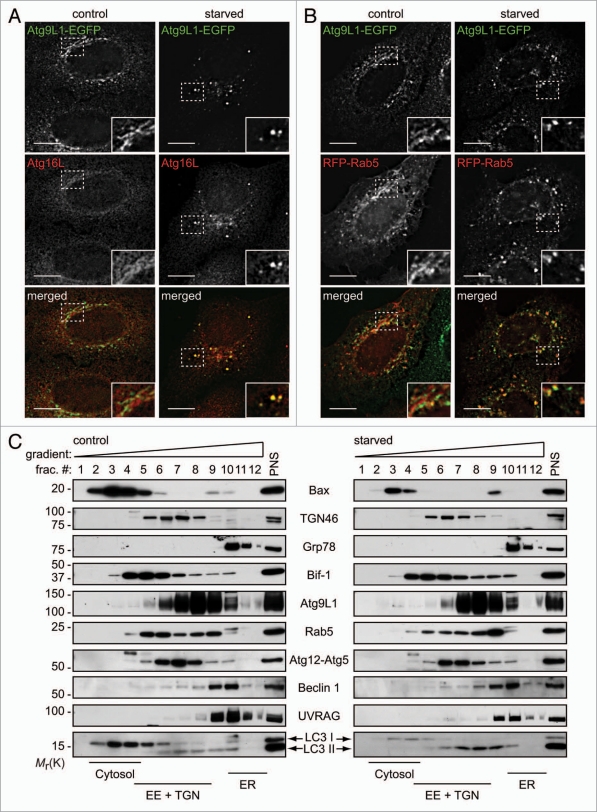
Bif-1 regulates the tubulation and fission of Atg9-positive membranes during starvation. (A) Control or Bif-1 knockdown HeLa cells stably expressing Atg9L1-EGFP were incubated in complete or starvation medium for 1.5 h. The cells were then immunostained for TGN46. The images were obtained using a fluorescence deconvolution microscope. Magnified images are shown in the right part. (B) The percentage of cells with GFP foci was calculated (mean ± SD.; n = 3 × 215). (C) HeLa cells stably expressing Atg9L1-EGFP were starved for 45 min and then analyzed by time-lapse fluorescent microscopy at 5-sec intervals. The scale bars represent 10 µm in (A), 5 µm in magnified images in (A and C).
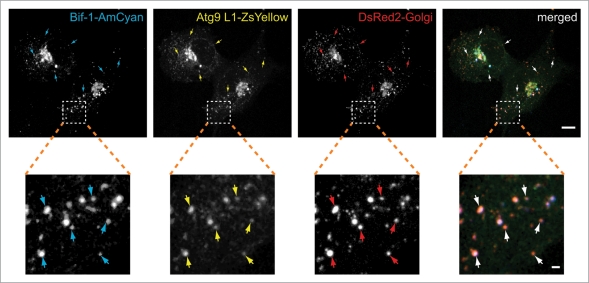
We have reported that in response to nutrient starvation, Bif-1 colocalizes with Atg9 and forms crescent-shaped small structures that expand by recruiting and fusing with other Atg9-positive foci to form vacuoles.26 Consistently, we detected endogenous Bif-1 on Atg9-positive foci induced by starvation (Suppl. Fig. 1C). As starvation-induced Atg9-positive compartments were closely associated with TGN46-positive membranes (Fig. 1A), we next examined whether Bif-1 and Atg9-positive punctate structures contain membranes derived from the Golgi complex. Consistent with our previous results,26 Bif-1 and Atg9 accumulated in cytoplasmic foci after starvation (Fig. 2). Notably, these punctate structures colocalized with, or were adjacent to, signals from the Golgi marker, indicating that these structures that are positive for both Bif-1 and Atg9 are, at least in part, derived from Golgi membranes.
Figure 2.
Starvation-induced Bif-1 and Atg9-positive foci are derived from Golgi membranes. COS7 cells co-transfected with Bif-1-AmCyan, Atg9L1-ZsYellow and DsRed-Monomer-Golgi were cultured in starvation medium for 1.5 h. The fluorescent images were obtained using a confocal microscope. Magnified images are shown in the bottom part. Arrows indicate representative colocalization of Bif-1, Atg9 and the Golgi marker. The scale bars represent 10 µm and 1 µm in the upper and bottom parts, respectively.
Bif-1 regulates the fragmentation of Golgi membranes during starvation.
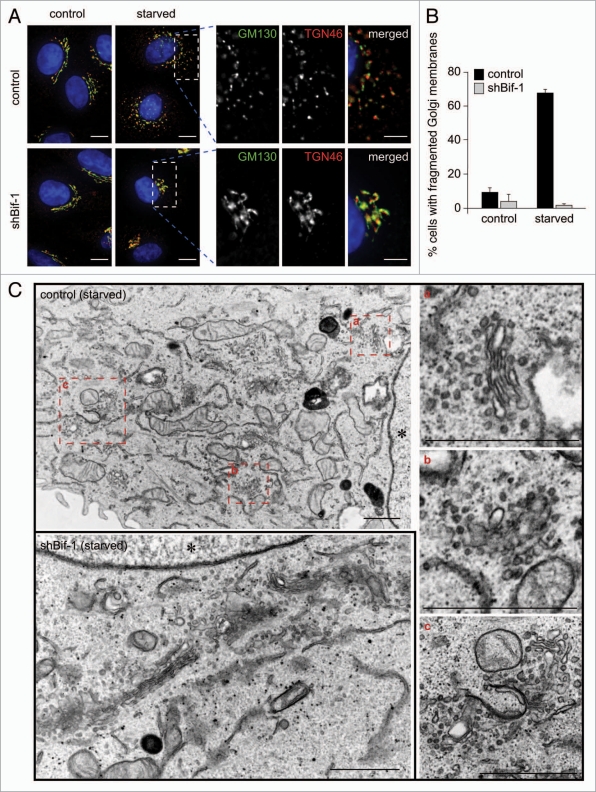
To investigate whether starvation-induced redistribution of Atg9 involves alteration of Golgi membrane structure, we first observed the morphology of the Golgi complex in Bif-1 knockdown and control HeLa cells under normal culture and starvation conditions. We found that starvation resulted in the fragmentation of both cis- and trans-Golgi membranes in control HeLa cells as determined by staining with anti-GM130 and anti-TGN46 antibodies, respectively (Fig. 3A and B). Similar results were obtained using the AcGFP-Golgi marker for fluorescent labeling of the Golgi complex (Suppl. Fig. S2A). The specificity of the AcGFP-tagged Golgi maker for Golgi membranes was confirmed by staining endogenous GM130 or TGN46 (Suppl. Fig. S2B). Electron microscopic analysis confirmed that fragmented Golgi membranes are dispersed throughout the cytoplasm of control HeLa cells that have been nutrient-starved (Fig. 3C). In contrast, the Golgi complex remained normal at the juxtanuclear region in Bif-1 knockdown cells after starvation (Fig. 3 and Suppl. Fig. S2A). Notably, the disassembly of the Golgi complex during mitosis was observed regardless of Bif-1 expression (Suppl. Fig. S2C). Furthermore, the morphology of the Golgi complex observed in Bif-1 knockdown cells was similar to that of control cells under normal culture conditions. These results indicate that Bif-1 plays a key role in starvation-induced fragmentation of Golgi membranes, but is not essential for the maintenance of normal Golgi complex morphology or the disassembly of the Golgi complex during mitosis.
Figure 3.
Bif-1 regulates the fragmentation of Golgi membranes during starvation. (A and B) Control or Bif-1 shRNA-expressing HeLa cells were incubated in complete or starvation medium for 1.5 h. The cells were then immunostained for GM130 (green) and TGN46 (red). The images were obtained using a fluorescence deconvolution microscope. Magnified images are shown in the right part. (B) The percentage of cells with fragmented TGN46-positive Golgi membranes was quantified (mean ± SD; n = 3 × 107). (C) HeLa cells incubated in complete or starvation medium for 1.5 h were analyzed by transmission electron microscopy. Magnified images are shown in the right. Asterisk indicates nucleus. The scale bars represent 10 µm in (A) and 1 µm in (C) and 5 µm in magnified images in (A).
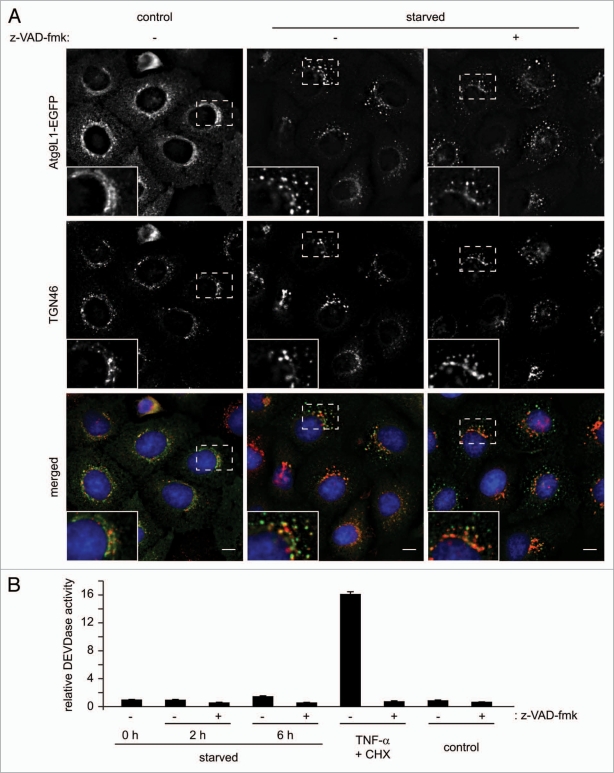
During the induction of apoptosis, it has been reported that the Golgi complex is fragmented in a caspase-dependent manner.27,28 To determine whether caspase activation is involved in the fragmentation of Golgi membranes during starvation, we starved cells in the presence or absence of a pan-caspase inhibitor, z-VAD-fmk. As shown in Figure 4A, inhibition of caspases failed to block the fragmentation of Golgi membranes. Importantly, the concentration of z-VAD-fmk used for these experiments was sufficient to suppress caspase-3/7 activation (Fig. 4B). Moreover, while the fragmentation of Golgi membranes occurred 1.5 h after starvation (Fig. 3), we did not observe elevated activity of caspase-3/7 until 6 h after starvation (Fig. 4B). Taken together, these results indicate that fragmentation of Golgi membranes during starvation occurs independently of caspase activation.
Figure 4.
Starvation-induced dispersal of Atg9 and TGN46-positive membranes does not require caspase activation. (A) HeLa cells stably expressing Atg9L1-EGFP were starved in the presence or absence of a pan-caspase inhibitor, z-VAD-fmk (20 µM) for 2 h and subjected to immunofluorescence microscopy using anti-TGN46 sheep polyclonal antibodies. Magnified images are shown in the insets. The scale bars represent 10 µm. (B) HeLa cells were incubated in starvation medium for 0, 2 and 6 h or treated with 1 ng/ml TNFα plus 10 µg/ml cycloheximide or control PBS for 6 h. Each treatment was performed in the presence or absence of 20 µM z-VAD-fmk. The caspase-3/7-like DEVDase activity was measured as previously described (n = 3).42
Dispersed Atg9-positive membranes induced by starvation contribute to autophagosome formation.
We next examined whether fragmented Atg9-containing Golgi membranes are involved in the formation of autophagosomes. Consistent with previous studies,20,21 a portion of Atg9 foci overlapped with GFP-LC3 signals in the cytoplasm of starved cells (Suppl. Fig. S3A), supporting a notion that Atg9-positive membranes derived from the Golgi complex may contribute to the formation of autophagosomes. As starvation promotes Atg9 localization not only to LC3-positive foci, but also to late endosomal membranes,20 and autophagosomes can fuse with endosomes during the maturation process,29 Atg9-positive membranes that exit from the Golgi complex may contribute to the biogenesis and/or maturation of autophagosomes. To determine the step at which Atg9-containing membranes contribute to autophagosome formation, we first examined whether starvation-induced Atg9 foci colocalize with an autophagosome precursor marker, Atg16L. As shown in Figure 5A, Atg16L signals were detected diffusely throughout the cytoplasm under normal culture conditions, as previously reported,30 with weak juxtanuclear signals. After starvation, we found that dispersed Atg9-containing compartments colocalized with Atg16L-positive foci. Similarly, Bif-1 colocalized with Atg16L in foci after starvation (Suppl. Fig. S3B). Interestingly, while less colocalization of TGN46-positive membranes with GFP-LC3 foci were detected after starvation, many of the Atg16L-positive foci were located adjacent to fragmented TGN46-positive membranes (Suppl. Fig. S4A). Taken together, these results indicate that Atg9-containing membranes derived from the Golgi complex are involved in the formation of early autophagosome precursors.
Figure 5.
Starvation promotes colocalization of Atg9 with Rab5 and Atg16L. (A) HeLa cells stably expressing Atg9L1-EGFP were starved for 1.5 h and subjected to immunofluorescent staining for Atg16L. Magnified images are shown as insets. (B) HeLa cells stably expressing Atg9L1-EGFP were transiently transfected with RFP-Rab5 for 24 h, incubated in complete or starvation medium for 1.5 h and analyzed by deconvolution fluorescent microscopy. (C) HeLa cells cultured in complete or starvation medium for 1.5 h were subjected to subcellular fractionation and immunoblot analyses using the indicated antibodies. The scale bars in (A and B) represent 10 µm.
We next examined whether starvation-induced Atg9-positive membranes translocate to endosomes for the expansion of autophagosomal membranes. We found that many of the Atg9-positive foci also colocalized with Rab5, an early endosome marker, after 2 h of starvation (Fig. 5B), while little colocalization was observed with Rab9, a late endosomes marker (Suppl. Fig. S3C). Subcellular fractionation analyses further confirmed that Rab5 signals in Atg9-rich fractions increased after starvation (Fig. 5C). Moreover, Rab5 was detected on Atg16L-positive foci after starvation (Suppl. Fig. S3D). Notably, while Bif-1 partially co-fractionated with TGN46, Rab5 and Atg9 under normal culture conditions, starvation promoted Bif-1 translocation into fractions enriched in Atg9, Rab5 and LC3 II (Fig. 5C). We noticed a decrease in the amount of the Atg12-Atg5 complex that co-fractionated with cytosolic Bax as well as LC3 I in both control and starved HeLa cells, while the majority of Atg12-Atg5 was found in the cytosolic fraction prepared from embryonic stem cells.31 In addition, unlike data previously reported using HEK293 cells,20 we did not observe clear colocalization of Atg9 with Rab9 in our system. Thus, the regulation of Atg protein localization in HeLa cells may differ from that of noncancerous cells.
The Bif-1 N-BAR domain plays a key role in Golgi fission, Atg9 trafficking and autophagosome formation.
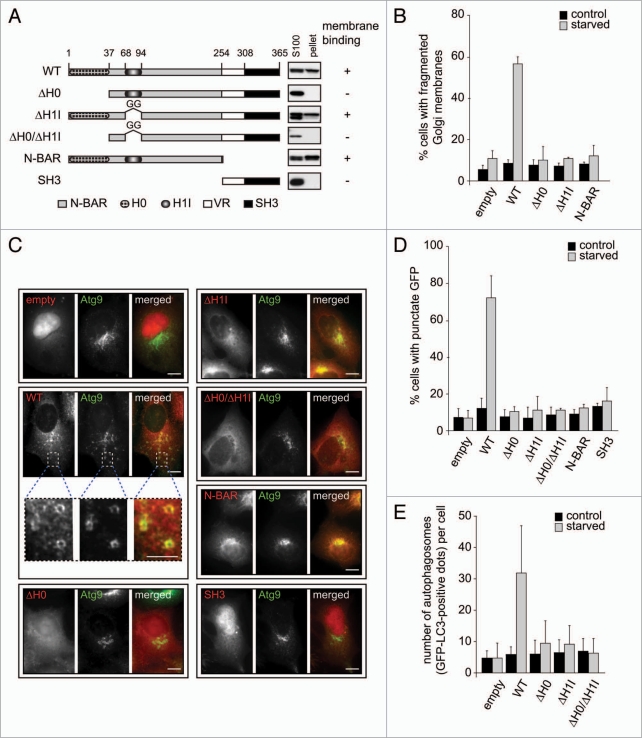
Crystallographic studies of the N-BAR domain of Endophilin A1, a close homologue of Bif-1, identified two distinct amphipathic segments referred to as helix 0 (H0) and helix 1 insert (H1I).32,33 The H0 region is responsible for the binding of the N-BAR domain to membranes, whereas the H1I ridge penetrates into the external leaflet of the bilayer, stabilizes the crescent-shaped N-BAR dimer, and promotes membrane curvature.32,33 Structure-based sequence alignment reveals that these two regions are conserved between Endophilin A1 and Bif-1.25,33 Moreover, it has been shown that the H0 region of Bif-1 indeed forms a helical structure and is capable of substantially penetrating into large unilamellar vesicles.34 While the SH3 domain is necessary for Bif-1 to form a protein complex with Beclin 1 through UVRAG, the N-BAR domain is required for Bif-1 to induce autophagosome formation.22 These findings suggest that the intrinsic membrane curvature-inducing activity of Bif-1 may contribute to the biogenesis of autophagosomal membranes. To examine the role of the Bif-1 N-BAR domain in autophagy, we generated a series of Bif-1 mutants in which the H0 (ΔH0), H1I (ΔH1I), H0 and H1I (ΔH0/ΔH1I), SH3 and variable region (N-BAR) or N-BAR (SH3) domain was deleted (Fig. 6A). In agreement with previous in vitro studies,32–34 a significant portion of purified recombinant Bif-1 and N-BAR proteins, but not the SH3 domain, co-fractionated with the Golgi marker, TGN38, as well as Atg9, in the pellet fraction when these recombinant proteins were incubated with membranes prepared from Bif-1-deficient MEFs (Fig. 6A and Suppl. Fig. S5A). These data indicate that Bif-1 indeed interacts with intracellular membranes through its N-BAR domain. The membrane-binding activity of Bif-1 is completely abolished by loss of the H0 region, whereas less effect was observed upon deletion of the H1I region, indicating that the N-terminal amphipathic helix is responsible for Bif-1 binding to membranes. Importantly, we found that deletion of the helix 0 in the Bif-1 N-BAR domain failed to restore the fission of Golgi membranes (Fig. 6B and Suppl. Fig. S5B) as well as Atg9 foci formation (Fig. 6C and D) in Bif-1-deficient cells after starvation, indicating that localization of Bif-1 to membranes is required for starvation-induced Golgi fragmentation and Atg9 trafficking. Moreover, expression of Bif-1 ΔH1I also failed to restore Golgi fission and Atg9 redistribution during starvation, suggesting that induction of membrane curvature through the H1I ridge is required for Bif-1 to induce the fission of Atg9-positive Golgi membranes.
Figure 6.
A functional N-BAR domain is required for Bif-1 to regulate Golgi fission, Atg9 trafficking and autophagosome formation. (A) Schematic of the domain structure of wild-type (WT) Bif-1 and various mutants. Bif-1 contains an N-terminal N-BAR domain, a variable region (VR) and a C-terminal SH 3 domain. The Bif-1 N-BAR domain is composed of two functional regions, helix 0 (H0) and helix 1 insertion (H1I). The membrane binding activity of Bif-1 and various mutants is shown on the right (see Suppl. Fig. S5A for further details). (B) Bif-1 knockdown HeLa cells were transfected with Bif-1 shRNA-resistant WT or mutant Bif-1-HcRed or empty HcRed vector for 24 h. After incubation in complete or starvation medium for 1.5 h, the cells were stained with anti-TGN46 sheep polyclonal antibody followed by secondary antibody conjugated with Alexa Fluor 468. The percentage of cells with fragmented TGN46-positive Golgi membranes was quantified (mean ± SD; n = 3 × 150). (C) Bif-1 knockdown HeLa cells stably expressing Atg9L1-EGFP were transfected with Bif-1 shRNA-resistant WT or mutant Bif-1-DsRed or empty DsRed vector for 24 h, then cultured in complete or starvation medium for 1.5 h. The images were obtained using a fluorescent microscope. (D) The percentage of DsRed-positive cells with GFP foci in (C) was counted using a fluorescent microscope (mean ± SD; n = 3 × 151). (E) HeLa cells stably expressing Bif-1 shRNA were co-transfected with GFP-LC3 and Bif-1 shRNA-resistant WT or mutant Bif-1-HcRed or empty HcRed1 vector for 24 h, then cultured in complete or starvation medium for 2 h. The images were obtained using a fluorescent microscope and the number of GFP-LC3 dots per HcRed-positive cell was quantified (mean ± SD.; n = 27). The scale bars represent 10 µm (5 µm in magnified images).
We next investigated whether the Bif-1 N-BAR domain plays a key role in autophagosome regulation. Consistent with our previous results,22 loss of Bif-1 suppressed GFP-LC3 foci formation induced by nutrient starvation (Fig. 6E and Suppl. Fig. S5C). Importantly, knockout of Bif-1 resulted in the inhibition of starvation-induced conversion of LC3-I to the lipidated form LC3-II in the presence of a lysosomal inhibitor, bafilomycin A1 (Suppl. Fig. S6), suggesting that loss of Bif-1 suppresses autophagy induction rather than promotes autophagosome maturation. Moreover, while expression of wild-type Bif-1 restored GFP-LC3 foci formation in Bif-1-deficient cells, this effect was not observed by expression of any of the N-BAR mutants (Fig. 6E and Suppl. Fig. S5C). Notably, deletion of H0, H1I or both domains had no effect on the interaction of Bif-1 with UVRAG (Suppl. Fig. S5D), indicating that the N-BAR mutants of Bif-1 are properly folded. Taken together, these results suggest that Bif-1 regulates the trafficking of Atg9 from the Golgi complex to the peripheral cytoplasm for the formation of autophagosomes by inducing Golgi membrane tubulation and fragmentation through its N-BAR domain.
The PI3KC3 complex plays a key role in Atg9 redistribution during nutrient starvation.
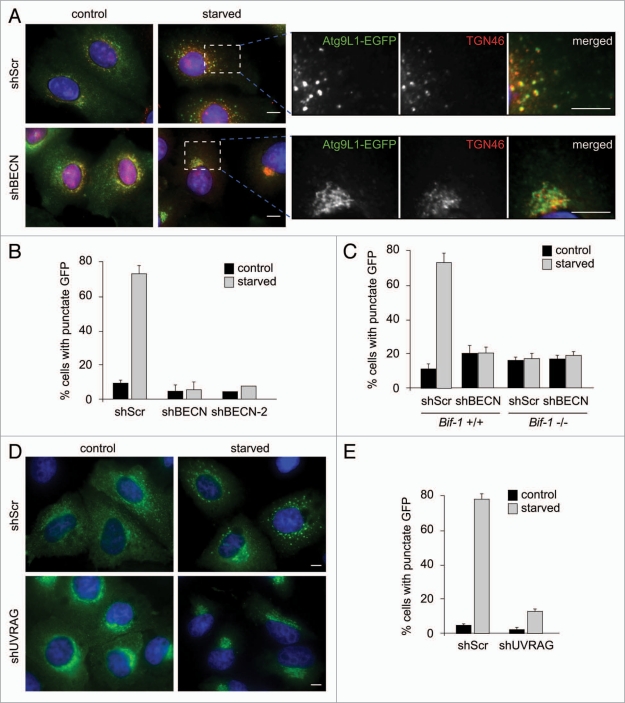
We have previously shown that the SH3 domain is required for Bif-1 to interact with the UVRAG-Beclin 1 complex.22 As deletion of not only the N-BAR domain, but also the SH3 domain, of Bif-1 failed to restore starvation-induced Atg9 foci formation in Bif-1-deficient cells, we next examined the role of the UVRAG-Beclin 1-PI3KC3 complex, also known as the PI3KC3 complex II, in Atg9 translocation during the induction of autophagy. Consistent with a previous study,20 treatment with a PI3-kinase inhibitor, wortmannin, suppressed starvation-induced redistribution of Atg9 in both HeLa cells and MEFs (Suppl. Fig. S7A–C). Similarly, suppression of Beclin 1 expression resulted in the inhibition of Atg9 foci formation during nutrient starvation (Fig. 7A–C and Suppl. Fig. S7D and E). Importantly, knockdown of Beclin 1 also suppressed starvation-induced Golgi fragmentation in HeLa cells (Fig. 7A). In contrast, essentially no effect of wortmannin treatment or Beclin 1 knockdown was observed in starved Bif-1-deficient cells, suggesting that Bif-1 and the PI3KC3 complex II act in a linear pathway to regulate Golgi fission and Atg9 redistribution during starvation. Indeed, the Bif-1 mutant that lacks the UVRAG-binding SH3 domain (Bif-1 N-BAR) failed to rescue defective Atg9 trafficking during autophagy induction in Bif-1-deficient cells (Fig. 6C and D) and knockdown of UVRAG greatly suppressed starvation-induced Atg9 foci formation (Fig. 7D and E and Suppl. Fig. S7F). Taken together, these results indicate that Bif-1, together with the PI3KC3 complex II, regulates Golgi fission and Atg9 trafficking during the induction of autophagy. However, while a small portion of UVRAG was detected at the TGN under normal culture conditions, neither Atg9-positive foci nor fragmented Golgi membranes colocalized with UVRAG after starvation (Suppl. Fig. S8).
Figure 7.
Bif-1 together with the PI3KC3 complex is required for Atg9 redistribution during starvation. (A) HeLa cells stably expressing Atg9L1-EGFP were infected with lentivirus expressing Beclin 1 shRNA (shBEC N or shBEC N-2) or scrambled shRNA (shScr) and subjected to selection with 1 µg/ml puromycin for 5 days. The cells were then incubated in complete or starvation medium for 1.5 h and subjected to immunofluorescent staining with an anti-TGN46 sheep polyclonal antibody followed by a secondary antibody conjugated with Alexa Fluor 568. Magnified images are shown in the right part. (B) The percentage of cells with cytoplasmic Atg9 foci in (A) was quantified (mean ± SD; n = 4 × 211 for shScr and shBEC N1, n = 219 for shBEC N-2). (C) Bif-1+/+ and Bif-1−/− MEFs stably expressing shBEC N or control shScr were transfected with Atg9L1-EGFP. Twenty-four hours after transfection, the cells were cultured in complete or starvation medium and the percentage of cells with GFP foci was quantified (mean ± SD; n = 3 × 67). (D) HeLa cells stably expressing Atg9L1-EGFP were infected with lentivirus expressing UVRAG shRNA (shUVRAG) or control shScr. After selection with puromycin for 5 days, the cells were incubated in complete or starvation medium for 1.5 h and subjected to fluorescent microscopic analysis. (E) The percentage of cells with cytoplasmic Atg9 foci in (D) was calculated (mean ± SD; n = 3 × 256). The scale bars represent 10 µm.
Discussion
In the present study, we have demonstrated that Bif-1 regulates the fission of Golgi membranes and the trafficking of Atg9 from the Golgi complex to autophagosomes during starvation. In response to nutrient starvation, Atg9-positive Golgi membranes were dispersed throughout the cytoplasm where they colocalized not only with Bif-1, but also with LC3 and Atg16L. Moreover, loss of Bif-1 resulted in the suppression of Golgi fission, Atg9 redistribution and autophagosome formation during starvation. These observations suggest that Atg9-positive membranes derived from the Golgi complex may contribute to the formation and/or expansion of autophagosome precursors. Indeed, our previous time-lapse studies show that an Atg9-Bif-1 positive punctum fuses with another Atg9-Bif-1 small membrane during the formation of a ring-shaped structure.26 It has been shown that Atg9 self-interaction is required for expansion of autophagosome precursors in yeast35 and that loss of Atg9 suppresses the lipidation of LC3 I and/or LC3 foci formation.15,20,21 Moreover, recent studies in yeast have shown that proteins involved in the secretary pathway at the Golgi complex play a key role in autophagosome formation.11,12 In addition, it has been reported that autophagosomes are formed through the fusion of isolation membranes with vesicles derived from the Golgi complex and late endosomes independent of Atg5/Atg7 in mammalian cells.10 These studies, taken together with our results, strongly support the notion that Golgi-derived membranes contribute to the formation of autophagosomes. However, the phenomenon of Golgi fragmentation after starvation has not been reported in previous studies.13,20 This discrepancy could be due to the difference in cell lines and/or starvation media used to induce autophagy. Interestingly, we also found that a portion of Atg9 colocalizes with a marker of early endosomes, Rab5. As plasma membrane that is internalized through clathrin-mediated endocytosis has been shown to regulate the early stages of autophagosome formation,14 we do not exclude the possibility that Atg9-positive structures used for the formation of pre-autophagosomal structures are derived from early endosomes.
The precise molecular machinery through which Bif-1 regulates the fission of Atg9-positive Golgi membranes during the induction of autophagy remains to be determined. However, our results demonstrate that loss of either the H0 or H1I region of the Bif-1 N-BAR domain suppresses Bif-1-mediated Atg9 puncta formation. As these regions are involved in membrane binding and tubulation,32,33 we propose a model in which Bif-1 regulates the redistribution of Atg9 from the Golgi complex to peripheral sites by deforming Golgi membranes into narrow tubules to facilitate membrane fission. Although the fission of Atg9-positive Golgi membranes occurs in a Bif-1-dependent manner during starvation, the morphology of the Golgi complex under normal culture conditions and the disassembly of the Golgi complex during mitosis are not affected by loss of Bif-1. Moreover, while loss of COPI causes fragmentation of the Golgi complex, it does not promote the formation of autophagosomes.36 Thus, the Golgi fission machinery, which generates Atg9-containing membranes for autophagosome formation, is uniquely controlled by Bif-1 during nutrient starvation.
Activation of PI3KC3 is essential for the trafficking of Atg9 and the formation of autophagosomes in both yeast and mammalian systems.20,37 Consistently, our results indicate that inhibition of the Beclin 1-PI3KC3 complex prevents Atg9 foci formation after nutrient starvation. Although the precise mechanism by which the activation of PI3KC3 regulates Atg9-positive foci formation is not fully understood, PI3P produced by this lipid kinase plays an important role in the recruitment of proteins required for Atg9 trafficking.6,38 In fact, it has been reported that production of PI3P is required for the autophagosomal localization of mAtg18/WIPI-1,39 the mammalian orthologue of yeast Atg18, which interacts with Atg9 and is required for Atg9 trafficking to the pre-autophagosomal structure.37 As starvation promotes Bif-1 translocation to Atg9-enriched fractions, it will be interesting to determine whether production of PI3P facilitates Bif-1 localization to fission sites for the generation of Atg9 vesicles. Our results also show that knockdown of UVRAG suppresses starvation-induced redistribution of Atg9 during starvation. Thus, UVRAG may be involved in the regulation of Atg9 trafficking during the induction of autophagy. However, we did not observe any significant colocalization of UVRAG with Atg9 foci after starvation, suggesting that UVRAG dissociates from Atg9-positive punctate structures after the fission event occurs. Clearly, further studies are required in order to determine the role of the PI3KC3 complex II in Atg9 trafficking.
Despite significant inhibition of autophagosome formation by loss of Bif-1 in our cell culture systems, the phenotypes of Bif-1-deficient mice are quite different from the embryonic lethal phenotype observed in mice lacking Beclin 1.22,40,41 This suggests that other proteins, such as its family member Endophilin B2, may functionally compensate for the loss of Bif-1 during embryonic development. Indeed, our unpublished data indicate that Endophilin B2 also localizes to the Golgi complex and interacts with UVRAG. Further studies on the characterization of Bif-1 and Endophilin B2 double knockout mice will allow insight into how these proteins work together in the regulation of Golgi remodeling and autophagosome biogenesis during development and tissue homeostasis. Nevertheless, this study has shown for the first time that the N-BAR domain-containing protein, Bif-1 plays an essential role in the fission of the Golgi complex during nutrient starvation presumably to deliver Atg9-positive membranes for autophagosome formation.
Materials and Methods
Plasmids and reagents.
The Bif-1 WT, ΔH0, ΔH1I, ΔH0/ΔH1I, N-BAR and SH3 cDNAs were amplified by PCR and subcloned into the EcoR I-Xba I sites of pEF6/myc-His A (Invitrogen, V962-20), the EcoR I-BamH I sites of pHcRed1-N1 (Clontech, 632424), pDsRed monomer-N1 (Clontech, 632465), pEGFP-N1 (Clontech, 6085-1) and pAmCyan1-N1 (Clontech, 632442) or the Nde I-Sap I sites of pTYB1 (New England Biolabs, N6701S) vectors. The Atg9L1 cDNA was generously provided by Dr. Stephen W. Scherer (The Hospital for Sick Children, Canada) and subcloned into the Nhe I-Hind III sites of the pZsYellow1-N1 (Clontech, 632445) and pEGFP-N1 vectors. The pDsRed-monomer-Golgi (632480) and pAcGFP-Golgi (632464) were purchased from Clontech. Lentiviral shRNA vectors targeting human or mouse Beclin 1 (RHS4533-NM_003766) and human UVRAG (RHS4533-NM_003369) were obtained from Open Biosystems. Control scrambled shRNA (1864) was obtained from Addgene. Antibodies were obtained from the following sources: mouse monoclonal anti-GM130 (BD Transduction Laboratories, 610822), sheep polyclonal anti-TGN46 (Novus Biologicals, NB110-60520), mouse monoclonal anti-α-tubulin (Sigma-Aldrich, T5168), rabbit polyclonal anti-Atg9A (Novus Biologicals, NB110-56893), rabbit polyclonal anti-LC3 (Novus Biologicals, NB100-2220 for immunoblot analyses; MBL, PM046 for immunofluorescent analyses), mouse monoclonal anti-Bif-1 (Imgenex, IMG-265A), goat polyclonal anti-Bif-1 (GeneTex, GTX21343), goat polyclonal anti-Beclin 1 (Santa Cruz, sc-10086), rabbit polyclonal anti-UVRAG (Bethyl Laboratories, A301-995A), mouse monoclonal anti-β-actin (Sigma-Aldrich, A5441), mouse monoclonal anti-Atg5 (MBL International, M153-3), rabbit polyclonal anti-Atg16L (MBL International, PM040), rabbit polyclonal anti-Rab5 (Abcam, ab18211), rabbit polyclonal anti-Grp78/BiP (StressGen, SPA-826), rabbit polyclonal anti-Bax (Santa Cruz, sc-493), goat polyclonal anti-c-Myc (Santa Cruz, sc-789-G), mouse monoclonal anti-Flag (Sigma-Aldrich, F1804), and rabbit polyclonal anti-Flag (Sigma-Aldrich, F7425). HeLa cells stably expressing Bif-1 shRNA and spontaneously immortalized Bif-1+/+ (WT) and −/− (KO) MEFs were previously described in reference 42.
Cell culture, transfection and shRNA-mediated gene silencing.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 100 µg/ml streptomycin and 100 U/ml penicillin. MEFs and COS7 cells were cultured in DMEM supplemented with 10% FBS, 100 µg/ml streptomycin and 100 U/ml penicillin. For transient transfection, FuGene HD reagent (Roche, 04709705001) or the Nucleofector system (Lonza, VCA-1001 for HeLa cells; VPD-1001 for MEFs) were used according to the manufacturer's protocols. To obtain stable transfectants, control or Bif-1 shRNA-expressing HeLa cells were transfected with pAtg9L1-EGFP or pAcGFP-Golgi using FuGene HD. Twenty-four hours after transfection, the cells were subjected to selection with 500 µg/ml Geneticin (Invitrogen, 10131-027) for 14 days. Lentivirus-mediated gene silencing was performed as previously described in reference 22. The sequences for shRNAs are: control scrambled shRNA (shScr, 5′-CCT AAG GTT AAG TCG CCC TCG-3′); mouse Beclin 1 (shBECN, 5′-GCG GGA GTA TAG TGA GTT TAA-3′), human Beclin 1 (shBECN, 5′-CCG ACT TGT TCC TTA CGG AAA-3′; shBECN-2, 5′-CTC AAG TTC ATG CTG ACG AAT-3′) and human UVRAG (shUVRAG, 5′-GCC CTT GGT TAT ACT GCA CAT-3′).
Autophagy induction and fluorescent microscopy.
To induce autophagy, cells grown on 0.1% gelatin-coated coverslips or NUNC Lab-Tek II chamber slides (Fisher Scientific, 12-565-2) were washed twice with phosphate-buffered saline (PBS), pH 7.4, once with starvation medium (DMEM without amino acids and glucose) (Invitrogen, REF. RR080014L1) and then incubated in starvation medium or control complete medium (DMEM supplemented with 10% FBS) for the indicated period of time. Cells were then fixed in 3.7% formaldehyde-PBS and mounted with DAPI. For immunofluorescent staining, cells were fixed in 4% paraformaldehyde-PBS, blocked in 3% bovine serum albumin (BSA)-PBS, stained with the indicated primary antibodies overnight at 4°C followed by incubation with fluorescent conjugated secondary antibodies and then mounted with DAPI-containing medium (Invitrogen, P36935). For intracellular localization analyses, cells were grown on gelatinized coverslips or NUNC Lab-Tek II chamber slides and transfected with the indicated plasmids for 24 h. The cells were then washed twice with PBS, once with starvation medium, incubated in starvation or complete medium for the indicated times, fixed in 3.7% formaldehyde/PBS and mounted with DAPI-containing medium. Fluorescent images were obtained using a Leica DMI6000 laser scanning confocal microscope or an OLYMPUS IX81 deconvolution microscope and analyzed using SlideBook 5.0 software (Intelligent Imaging Innovations).
Electron microscopy.
HeLa cells seeded on Thermanox plastic coverslips (Electron Microscopy Sciences, 72280) were incubated in complete or starvation medium for 1.5 h and then fixed in 2% paraformaldehyde-2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.3, for 1.5 h at room temperature. The cells were then incubated in post-fixation buffer (1% osmium tetroxide/1.5% potassium ferrocyanide-0.1 M sodium cacodylate, pH 7.3) overnight, dehydrated in a graded series of ethanol, embedded in EMbed 812 resin (Electron Microscopy Sciences, 14120), and sectioned at a thickness of 70–90 nm. The sections were mounted on mesh copper grids, stained with aqueous uranyl acetate and lead citrate and analyzed using a JEOL JEM 1400 transmission electron microscope.
Subcellular fractionation.
HeLa cells were incubated in complete or starvation medium for 1.5 h, washed twice with ice-cold PBS and harvested in homogenization buffer (0.25 M sucrose, 140 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0). After centrifugation at 250x g at 4°C, the resultant cell pellets were resuspended in 0.9 ml of homogenization buffer containing protease and phosphatase inhibitor cocktails, passed through a 27-guage syringe needle and then centrifuged at 500x g at 4°C in order to obtain post-nuclear supernatants (PNSs). The PNSs were loaded on 7.2 ml of a 5–40% continuous OptiPrep (SIGMA, D1556) gradient prepared in homogenization buffer and centrifuged at 36,000 rpm for 20 h at 4°C using a SW-41 swing rotor. Twelve fractions were collected from the top of the gradient and analyzed by immunoblotting with the indicated antibodies.
Recombinant proteins and membrane binding assays.
Recombinant human Bif-1 WT (1–365), ΔH0 (37–365), ΔH1I (1–67 + GG + 95–365), ΔH0/ΔH1I (37–67 + GG + 95–365), N-BAR (1–255) and SH3 (M + 256–365) were purified using the IMPACT system (New England Biolabs, E6900S) as previously described in reference 43. For the intracellular membrane sedimentation assay, Bif-1−/− MEFs were resuspended in PBS containing 1 mM PMSF, then sonicated for 5 seconds 4 times on ice. After centrifugation at 2,000x g for 5 min at 4°C, the resultant supernatant (500 µg) was incubated with 500 ng of purified recombinant Bif-1 or Bif-1 mutant proteins for 30 min at room temperature and then centrifuged at 100,000x g for 30 min at 4°C to obtain the cytosolic fraction (S100). The pellet was washed once with PBS and then lysed in 1x SDS-PAGE loading buffer in the same volume as the S100 fraction to obtain the membrane fraction. The resultant S100 and membrane fractions were subjected to western blot analysis with the indicated antibodies.
Acknowledgements
We thank N. Nakamura for technical advice on immunofluorescent analyses and K. El-Bayoumy for critical reading of the manuscript. This work is supported by grants from the James & Esther King Biomedical Research Program (08KN-15-17228) to Y.T., National Institutes of Health (CA82197 and CA129682), American Cancer Society (RSG-05-244-01-CCG) and Flight Attendant Medical Research Institute (FAMRI062463-CIA) to H.G.W.
Abbreviations
- Atg
autophagy-related
- BECN
Beclin 1
- Bif-1
bax-interacting factor-1
- COPI
coat protein complex I
- H0
helix 0
- H1I
helix 1 insert
- LC3
microtubule-associated protein 1 light chain 3
- N-BAR
N-terminal amphipathic helix and Bin/amphiphysin/Rvs
- PI3KC3
class III phosphatidylinositol-3-kinase
- SH3
src homology 3
- UVRAG
ultraviolet radiation resistance-associated gene
- ULK1
Unc51-like kinase 1
- TGN
trans-Golgi network
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/14015
Supplementary Material
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimori T, Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr Opin Cell Biol. 2008;20:401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 7.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 9.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 10.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 11.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec Proteins Are Required for Autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi Is Required for the Expansion of the Autophagosomal Phagophore in Yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 17.Webber JL, Young AR, Tooze SA. Atg9 trafficking in Mammalian cells. Autophagy. 2007;3:54–56. doi: 10.4161/auto.3419. [DOI] [PubMed] [Google Scholar]
- 18.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 21.Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–18290. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huttner WB, Schmidt A. Lipids, lipid modification and lipid-protein interaction in membrane budding and fission— insights from the roles of endophilin A1 and synaptophysin in synaptic vesicle endocytosis. Curr Opin Neurobiol. 2000;10:543–551. doi: 10.1016/s0959-4388(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi Y, Meyerkord CL, Wang HG. BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1. Autophagy. 2008;4:121–124. doi: 10.4161/auto.5265. [DOI] [PubMed] [Google Scholar]
- 27.Chiu R, Novikov L, Mukherjee S, Shields D. A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J Cell Biol. 2002;159:637–648. doi: 10.1083/jcb.200208013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane JD, Lucocq J, Pryde J, Barr FA, Woodman PG, Allan VJ, Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J Cell Biol. 2002;156:495–509. doi: 10.1083/jcb.200110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 30.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Etxebarria A, Terrones O, Yamaguchi H, Landajuela A, Landeta O, Antonsson B, et al. Endophilin B1/Bif-1 stimulates BAX activation independently from its capacity to produce large scale membrane morphological rearrangements. J Biol Chem. 2009;284:4200–4212. doi: 10.1074/jbc.M808050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 38.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 40.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi Y, Karbowski M, Yamaguchi H, Kazi A, Wu J, Sebti SM, et al. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol Cell Biol. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi H, Woods NT, Dorsey JF, Takahashi Y, Gjertsen NR, Yeatman T, et al. SRC directly phosphorylates Bif-1 and prevents its interaction with Bax and the initiation of anoikis. J Biol Chem. 2008;283:19112–19118. doi: 10.1074/jbc.M709882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.