SUMMARY
Current ongoing genome-wide association studies represent a powerful approach to uncover common unknown genetic variants causing common complex diseases. The discovery of these genetic variants offers an important opportunity for early disease prediction, prevention and individualized treatment. We describe here a method of combining multiple genetic variants for early disease prediction, based on the optimality theory of the likelihood ratio. Such theory simply shows that the receiver operating characteristic (ROC) curve based on the likelihood ratio (LR) has maximum performance at each cutoff point and that the area under the ROC curve (AUC) so obtained is highest among that of all approaches. Through simulations and a real data application, we compared it with the commonly used logistic regression and classification tree approaches. The three approaches show similar performance if we know the underlying disease model. However, for most common diseases we have little prior knowledge of the disease model and in this situation the new method has an advantage over logistic regression and classification tree approaches. We applied the new method to the Type 1 diabetes genome-wide association data from the Wellcome Trust Case Control Consortium. Based on five single nucleotide polymorphisms (SNPs), the test reaches medium level classification accuracy. With more genetic findings to be discovered in the future, we believe a predictive genetic test for Type 1 diabetes can be successfully constructed and eventually implemented for clinical use.
Keywords: Backward clustering, Classification tree, Cross validation, Logistic regression
1. Introduction
Early disease prediction and prevention is one of the most promising strategies in health care. It can not only prevent mortality, but also decrease morbidity and public health costs (Etzioni et al., 2003). Predictive genetic tests, which use genetic markers - e.g., single nucleotide polymorphisms (SNPs) - to predict an individual’s future risk of disease, form one of the most appealing early disease prediction methods. Such tests can be conducted early in life (e.g., at birth) and, by use of appropriate prevention strategies, prevent individuals from contracting a disease. With the current intensive research on common complex diseases, in particular with the completion of genome-wide association studies, developing predictive genetic tests for common complex diseases has been initiated (e.g. Type 2 Diabetes (Weedon et al., 2006)). These tests have the potential to be the cornerstone of future genomic medicine and are anticipated to have a large impact on health care (Epstein, 2006).
Most diseases are normally caused by more than one genetic variant. To improve disease prediction, we should combine the information from all available risk variants instead of using only one of them. One way to form a predictive genetic test from multiple risk factors is to use logistic regression (Weedon et al., 2006), the most commonly used method for classification/prediction. However, a recent study (Pepe, Cai, and Longton, 2006) showed that when applied for multiple predictors, logistic regression is not always optimal because it assumes that the underlying link function is logit, an assumption that may not hold. There is also an increasing use of classification trees for predictive purposes. Compared to logistic regression, a classification tree is said to have the advantage of identifying important interactions in the data; but, in a real data example, Austin (2007) found logistic regression performed better than the classification tree method. Theoretically, a decision rule based on the likelihood ratio is optimal (Egan, 1975; McIntosh and Pepe, 2002) and the receiver operating characteristic (ROC) curve based on likelihood ratios (LRs), the optimal ROC curve, attains the highest classification/diagnostic accuracy in terms of the area under the ROC curve (AUC) (Lu and Elston, 2008). Thus, compared to logistic regression, use of the optimal ROC curve appears to be a promising general strategy.
Baker (2000) noted the optimal properties of the likelihood ratio for combining multiple predictors and developed three nonparametric approaches to incorporate multiple biomarkers for cancer prediction. These approaches can be extended for constructing predictive genetic tests from multiple genetic markers. Unlike many cancer biomarkers, genetic markers are usually categorical. Moreover, with genetic markers we face issues such as marker selection (i.e., eliminating “noise” markers), and unknown disease model (e.g., unknown mode of inheritance and unknown interaction model). Simply applying Baker’s methods without considering these issues will lead to over-fitting. Concentrating on these issues, we propose here a robust and powerful method for developing predictive tests using genetic markers.
This article is organized as follows. In section 2, we briefly review the concept of the optimal ROC curve and its utility for combining multiple cancer biomarkers. Based on this concept, in section 3 we propose two forms of the robust optimal ROC curve-based method for building predictive genetic tests, one that requires knowledge of the disease model and one that requires no assumptions about the underlying disease model. In section 4, we evaluate the two forms of the method with simulation studies and compare their performance with logistic regression, classification trees, and two approaches proposed by Baker (2000). In section 5, we illustrate the method using the Wellcome Trust genome-wide association data for Type 1 diabetes. We conclude with a discussion in section 6.
2. The Optimal ROC Curve
True positive rates (TPRs) and false positive rates (FPRs) are the two basic measures of classification accuracy for a test. The TPR is defined as the probability that the test result (x) is positive given the patient develops the disease (S =1): TPR = P(x =1|S =1). Similarly, we define the FPR as the probability that the test result (x) is positive given the patient does not develop the disease (S =0): FPR =P(x =1|S =0). For most predictive tests, there are several pairs of TPRs and FPRs available, and we should use the entire spectrum of TPR and FPR pairs to evaluate the overall classification accuracy of the test (Zweig and Campbell, 1993). For that purpose, the ROC curve is a useful tool. The ROC curve plots a test’s TPR against its FPR for continually changing cutoff points over the whole range of possible test results, and has been recognized as a global measure of a test’s accuracy (McClish, 1989). Since the ROC curve is a two-dimensional plot, a one-dimensional summary index of the ROC curve will often be more convenient and useful, and the most popular one is the area under the ROC curve (AUC).
Like the TPR and FPR, the LR is also a popular measure of test’s classification accuracy, and is defined as the ratio of two density functions, evaluated at x, conditional on disease status S: LR (x) = P (x|S =1)/P (x|S =0). The LR is useful for generating the optimal ROC curve. By definition (Egan, 1975), the optimal ROC curve consists of the entire set of TPR and FPR pairs resulting from the continually changing LR from its largest value to its smallest value. The optimal ROC curve is the best for each point on the curve in terms of 1) maximizing the TPR for any fixed value of the FPR, 2) minimizing the FPR for any fixed value of the TPR, 3) minimizing the overall misclassification probability, and 4) minimizing the expected cost (Egan, 1975; McIntosh and Pepe, 2002). The optimal ROC curve achieves the highest classification accuracy in terms of the AUC (Lu and Elston, 2008).
Although the optimal ROC curve was introduced several decades ago, its usefulness for combining multiple tests has only recently been recognized (Baker, 2000; McIntosh and Pepe, 2002). Based on the concept of the optimal ROC curve, Baker (2000) proposed three nonparametric methods: the unordered optimal method, the jagged ordered, and the rectangular ordered optimal robust methods. Unlike the unordered optimal method that ranks the individuals simply according to their LRs, the two other methods rank the individuals based on both the LRs and certain assumptions (e.g., that higher biomarker values lead to greater probability of disease). In a real data application, Baker (2000) fitted the three nonparametric methods and logistic regression in a training sample and then compared them in a separate validation sample. The nonparametric methods showed better performance than did logistic regression in the validation sample. However, Baker (2000) found that the unordered optimal method led to overly optimistic ROC performance in the training sample.
3. Optimal robust ROC Curve Estimation
The optimal ROC curve can be used to construct a predictive genetic test from multiple genetic markers, but genetic marker data bring up special issues that must be addressed, such as marker selection, unknown mode of inheritance and unknown interaction model. As we show in the simulations presented below, directly applying the optimal ROC curve to the genetic markers without considering these issues leads to a serious overestimation of the test’s performance. To obtain a more robust estimate, we propose two ways of estimating the optimal ROC curve depending on whether or not the disease model is known.
3.1. Estimation When There Is Prior Knowledge of the Disease Mode of Inheritance
Ideally, if we know the causal loci, their mode of inheritance and the interaction model, we can incorporate this into the model and hence estimate the underlying true optimal ROC curve. Let yi (yi ∈ S) be the binary response measurement and xi =(xi1, xi2, …, xip) (xij ∈ (gj1, gj2, …, gjmj)) be the measurement of p disease loci for the i-th individual. The marginal distribution of genotypes given disease status (S = 0, 1) can be calculated as
| (1) |
where IA (i) is the indicator function defined as . Given the mode of inheritance, we can combine the genotypes that have the same relative risks. For instance, for a single SNP marker j that has three genotypes, A A, A Ā and Ā Ā, if A is dominant we cluster the genotypes A A and A Ā into one group and calculate the corresponding frequency as . Given the disease prevalence ρ, we calculate the population genotype frequencies: P (gjkj) =ρP (gjkj|S =1)+(1 − ρ)P(gjkj|S =0). If the variants at the p loci cause the disease independently (i.e., no interaction), then, based on a multiplicative model, the probabilities of the multi-locus genotype Gl =(g1k1, g2k2, …, gpkp) given disease status can be expressed as
| (2) |
where ρ and L respectively denote the disease prevalence and the total number of multi-locus genotypes possible for the p disease loci, and if all p loci are in linkage equilibrium. We then rank the multi-locus genotypes according to their LRs, defined by LRl =P(Gl|S =1)/P(Gl|S =0), and plot the ROC curve. This represents the empirical optimal ROC curve, consisting of the set of TPR and FPR pairs:
| (3) |
where G(ζ) is the ζ-th multi-locus genotype when ranked by its LR value. As we have shown previously, it has the highest AUC (Lu and Elston, 2008) given by:
| (4) |
where TPR(0) = FPR(0) = 0.
This approach provides a simple way to approximate the underlying optimal ROC curve by incorporating the correct disease model (i.e., mode of inheritance) and by making the multiplicative model assumption. Following the same strategy, as described in more detail in Web Appendix A, we can extend this approach to incorporate interacting loci. This method is ideal for diseases that have been well studied. However, in most cases the disease model is not well understood. The disease susceptibility loci that we have discovered may not even be causal loci. Therefore, we now propose an approach that does not require prior knowledge about the disease model.
3.2. Estimation When There Is No Prior Knowledge of the Disease Model
In this approach, we start with all the data, treating each multi-locus genotype as a separate cluster, and then implement a backward clustering algorithm to group the multi-locus genotypes and so reduce the model complexity. Based on the prediction AUC calculated from K-fold cross validation, we choose the most parsimonious model with the appropriate number of multi-locus genotype clusters.
Suppose we have L possible multi-locus genotypes ( ) created from p loci. The p loci are disease susceptibility loci detected from previous association studies. We first estimate from the data the distribution of all the p -locus genotypes, separately in the case and control samples,
| (5) |
The p -locus genotypes are then ranked by their likelihood ratios, , to estimate the optimal ROC curve. Since the ROC curve depends on only the ranks of the test results, we use these ordered p -locus genotypes, , to represent our full model when constructing the optimal ROC curve. Once we have formed this optimal ROC curve, we use it to estimate the area under the optimal ROC curve and denoted this estimate .
Since this full model G(0) has the largest number of multi-locus genotype clusters (i.e., each multi-locus genotype represents a separate cluster) and includes all p loci, it most likely over-fits the data, and is hence biased upward. To reduce the model’s complexity, we gradually combine the multi-locus genotypes together and search for the best model that has a more accurate AUC estimate. At each step of the backward clustering process, we consider all possible multi-locus genotype clusterings that can be formed when two one-locus genotypes are pooled together, and choose the clustering that leads to a minimum decrease in the AUC when applied to the data. Thus, suppose we have p′ disease susceptibility loci at backward clustering step t (i.e. p−p′ loci have been eliminated prior to step t). For a genotype pair at locus j, we combine the two corresponding multi-locus genotypes that contain or and rank all the multi-locus genotypes (some now being combined together) according to their LRs. We then form the optimal ROC curve, which is denoted , and calculate the corresponding AUC, . Step t consists of repeating this for all possible pairs of one-locus genotypes at all p′ loci, and the best candidate model at step t, denoted G(t), is chosen based on having the highest AUC.
We repeat the clustering process steps and so progressively combine the multi-locus genotypes until all multi-locus genotypes finally cluster into one group. As a result, we obtain a series of candidate models, G(0), G(1), …, G(T), where G(T) is the model with only one cluster of multi-locus genotypes, which has an AUC value of 0.5. Note that for p diallelic SNPs, the maximum number of backward clustering steps, T, is 2p and for a real data application it would be fewer if not all multi-locus genotypes are represented in the data. Clearly, nether G(0) nor G(T) is the appropriate model for building the test. G(0), the most complex model with the largest number of multi-locus genotype clusters and including all p loci, has a high but over-fitted AUC, while G(T), the simplest model with only one cluster of multi-locus genotypes and excluding all p loci, gives a useless AUC. Letting nc(0), nc(1), …, nc(T), the numbers of multi-locus genotype clusters for candidate models G(0), G(1), …, G(T), describe the model complexity, a parsimonious model with a modest model complexity nc(m) should exist between G(0) and G(T) that has a relatively high and accurate AUC. To find the most appropriate number of multi-locus genotype clusters, nc(m), we use K-fold cross-validation.
In K-fold cross validation, we randomly partition the data into K subsets. K−1 subsets are used for the purpose of training and the remaining subset is used for the purpose of validation. We first apply the above clustering algorithm to the training dataset to find the candidate models, , k =1, 2, ···, K, and then apply these candidate models (i.e., the order of the multi-locus genotypes from the training dataset) to the validation dataset to construct the ROC curve and calculate the AUC. We repeat the cross-validation process K times, with each of the K subsets used exactly once as the validation dataset. The K results are then averaged to provide an estimate of the prediction AUC, which is denoted , t =0,1, …, T. nc(m) is then chosen to be the value of nc(t) that maximizes the , and the corresponding model, G(m), is chosen as the most parsimonious model.
A practical issue in the cross-validation process is that by chance some of the multi-locus genotypes in the validation dataset may not be found in the training dataset, and hence we cannot order these multi-locus genotypes. Instead of treating them as missing, we put them into clusters whose order can be inferred from the multi-locus genotypes present in the training dataset. In other words, we follow the same strategy of gradually clustering the multi-locus genotypes in the training dataset until we arrive at clusters that are present in the validation dataset, in the sense that each cluster in the training dataset corresponds to a cluster in the validation dataset, though the latter may not include all the multi-locus genotypes present in the training dataset cluster. We then calculate the LR statistics for the clusters that are in the training dataset, and use those to infer the multi-locus genotypes’ order.
This backward clustering algorithm applies naturally to both the disease model and marker selection. Ideally, by clustering the multi-locus genotypes on a particular locus, the method automatically approaches the marker’s mode of inheritance or eliminates a “noise” marker. For instance, if at a particular step the multi-locus genotypes with either A A or A Ā genotype at SNP j are grouped together, this would suggest that SNP j follows a model in which A is dominant and Ā is recessive. Further clustering of the A A and A Ā multi-locus genotypes with their corresponding Ā Ā multi-locus genotype implies that SNP j is not associated with the disease, and therefore we should remove it from consideration. In a similar manner, by continuously clustering multi-locus genotypes of more than one locus, the method is able to select an interaction model. The algorithm is also flexible enough to incorporate biological information. For example, if at SNP j clustering two multi-locus genotypes with A A and Ā Ā, but not the corresponding one with A Ā, is not considered biologically plausible, we might exclude this possible clustering from the algorithm, and hence help improve the algorithm’s performance. A simple example of the clustering algorithm is given in Web Appendix B.
4. Simulations
We conducted two sets of simulations to investigate the performance of the proposed optimal robust ROC curve method. The first simulation is conducted under a well studied disease scenario for which we know the causal loci and their mode of inheritance, the second one is simulated under a scenario in which we have no such prior knowledge.
4.1. Simulation I
We assume we are interested in building a predictive genetic test from three independent diallelic SNP loci (i.e., three diallelic SNPs in linkage equilibrium) with the disease susceptibility allele frequencies 0.15, 0.1 and 0.2, respectively. In a real situation, we would have other factors – e.g., age, gender and environmental factors – but here, for simplicity, we concentrate on genetic factors only. Let rj1 (rj2) denotes the risk associated with genotype Aj Aj (Aj Āj) relative to Āj Āj, where Aj and Āj respectively denote the risk and non-risk allele. We assumed that at the first locus the rarer allele is recessive(r11=2.5; r12=1), at the second locus multiplicative (r21 =2; r22=1.4), and at the third dominant (r31=r32=2).
We sampled equal numbers n of cases and controls (n = 250,500,1000) and investigated the logistic regression model, the classification tree and the optimal robust ROC curve method described in section 3.1. Because the mode of inheritance of each of the three loci is known, we incorporated that information into the analysis by coding the genetic variables as follows:
| (6) |
where c is an arbitrary positive number greater than 1. Since there is no gene-gene interaction, we only considered the main effects in the logistic regression analysis and fitted the model logit(μ) = α+ Zβ̰, where β̰ = (β1, β2, β3) are the coefficients for the three genetic variables. To implement the classification tree, we built a tree that fits the data perfectly (i.e., the largest possible tree). The AUC estimates from the three methods were then compared with the true AUC value, which was calculated by using the method in Web Appendix A. The average biases and standard deviations of the AUC estimates are summarized in Table 1, based on 1000 replicate samples.
Table 1.
Comparison of the bias (BIAS), standard deviation (SD) and mean squared error (MSE) of the AUC among the optimal robust ROC curve method (OPT-ROC), logistic regression (LOG REG) and classification tree (CLA-TREE) in the ideal scenario where we know the causal loci and the modes of inheritance.
| Cases:Controls | 250:250 | 500:500 | 1000:1000 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BIAS | SD | MSE | BIAS | SD | MSE | BIAS | SD | MSE | |
| OPT-ROC | 0.0001 | 0.0219 | 0.00048 | 0.0009 | 0.0165 | 0.00027 | −0.0001 | 0.0114 | 0.00013 |
| LOG-REG | −0.0001 | 0.0225 | 0.00050 | 0.0000 | 0.0169 | 0.00029 | −0.0003 | 0.0116 | 0.00013 |
| CLA-TREE | 0.0065 | 0.0216 | 0.00051 | 0.0030 | 0.0165 | 0.00028 | 0.0009 | 0.0115 | 0.00013 |
As expected, the methods perform better when the sample size is larger. For all three simulated settings, the performance of the optimal robust ROC curve method is comparable to that of logistic regression and classification tree, and the AUC estimates from all three methods closely approximate the true AUC value. Therefore, for well studied disease scenarios, it seems that logistic regression, classification tree and the optimal robust ROC curve methods are equally appropriate tools for constructing predictive genetic tests.
4.1. Simulation II
To mimic a more complex disease scenario, we simulated three disease loci and five “noise” loci. The simulation for the three disease loci was similar to that in the previous simulation, but with an additional interaction effect between locus two and locus three. If we denote by H the high risk group with all possible genotype combinations having at least one of the disease-susceptibility alleles at each of the two loci (i.e., A2, A3) and denote by C the low risk group with all the other genotype combinations, a simple interaction effect can be introduced by doubling the risk for all genotypes in the high risk group,
where pGl is the probability of disease for the low risk group. The allele frequency for each noise locus was independently sampled from a uniform distribution. pj Uniform(0.1,0.9), j = 4, …,8. Following the same procedure, we created the simulated data (Ysamp, Xsamp) and first fitted the data with logistic regression, classification tree and the optimal ROC curve method described in section 3.2.
For the logistic regression, we considered all possible single locus and two-way interaction effects, and implemented backward selection to choose the most parsimonious model with the smallest value of Akaike’s A Information Criterion (AIC) (Akaike, 2001). The logistic regression model and backward selection were performed using the glm and step functions in R. Based on the selected model, we formed the ROC curve and estimated the AUC. Since the underlying mode of inheritance of the disease markers was unknown, we assumed all loci followed either multiplicative, dominant, or recessive models, and coded the predictors using the corresponding formulas in equation (6). To perform the classification tree analysis, we used the tree package in R and chose the deviance as the criterion to guide cost-complexity pruning. For the optimal robust ROC curve analysis, we implemented the robust approach described in section 3.2, and only considered as possible clusters those consistent with a plausible biological model (i.e., we did not allow clustering together any multi-locus genotypes containing Aj Aj and Āj Āj unless the corresponding genotypes containing Aj Āj was also included in the cluster). Using 10-fold cross-validation, we chose the best model and estimated the AUC. We compared the AUC estimates from our proposed method, logistic regression and the classification tree, and summarize the results in Table 2.
Table 2.
Comparison among the optimal robust ROC curve method (OPT-ROC), logistic regression with backward selection (Mul LOG-REG, Dom LOG-REG, and Rec LOG-REG), and classification tree (CLA-TREE)
| Cases:Controls | 250:250 | 500:500 | 1000:1000 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BIAS | SD | MSE | BIAS | SD | MSE | BIAS | SD | MSE | |
| OPT-ROC | 0.0158 | 0.0338 | 0.00139 | 0.0010 | 0.0205 | 0.00042 | 0.0007 | 0.0154 | 0.00024 |
| Mul LOG-REG | 0.0418 | 0.0250 | 0.00237 | 0.0210 | 0.0170 | 0.00073 | 0.0119 | 0.0122 | 0.00029 |
| Dom LOG-REG | 0.0433 | 0.0247 | 0.00249 | 0.0190 | 0.0174 | 0.00067 | 0.0123 | 0.0121 | 0.00030 |
| Rec LOG-REG | −0.0546 | 0.0242 | 0.00357 | −0.0717 | 0.0176 | 0.00546 | −0.1046 | 0.0108 | 0.01106 |
| CLA-TREE | −0.0195 | 0.0362 | 0.00169 | −0.0229 | 0.0220 | 0.00101 | −0.0225 | 0.0153 | 0.00074 |
For the sample sizes 500 and 1000, in this limited simulation, the performance of the optimal robust ROC curve is slightly better than the other two approaches in terms of mean squared error. The estimates from the optimal robust ROC curve also approximate the underlying AUC values better than those from the logistic regression and classification tree approaches, while the estimates from logistic regression usually have smaller variances than those of the other two methods. For a sample size 2000, which is now commonly used for GWA studies, the performance of the logistic regression that assumes a multiplicative or dominant model could be comparable to the performance of the optimal robust ROC curve in terms of mean squared error. However the estimates from the logistic regression that assumed a recessive model seriously underestimated the underlying AUC value. Thus the performance of logistic regression depends on how well the mode of inheritance is specified. By assuming the “right” mode of inheritance, logistic regression can outperform the classification tree, given a large enough sample. These results are consistent with the findings of Austin (2007).
Using the same simulated data, we then investigated two original nonparametric approaches, the unordered optimal method and the jagged ordered method, introduced by Baker (2000). Simply applying these methods to the genetic marker data without considering any disease model and marker selection can cause over-fitting. With the simulated data, the average biases of the AUC estimates from the unordered optimal method based on 1000 replicates were 0.2922, 0.2339, and 0.2011 for the sample sizes 250, 500, and 1000, respectively, while the average biases of the AUC estimates from the jagged ordered method were 0.2155, 0.2015, and 0.1778.
5. Application to Type 1 Diabetes Data from the Wellcome Trust Consortium
The Wellcome Trust Consortium genome-wide association (GWA) study is one of the largest and most comprehensive studies aimed at discovering the genetic contributions to seven common complex diseases (Wellcome Trust Case Control Consortium, 2007). The study was undertaken in the British population and consists of approximate 2,000 cases of each of seven diseases and 3,000 shared controls. We used the Type 1 diabetes data in the Wellcome Trust GWA dataset that comprises 1963 Type 1 diabetes patients and 2938 controls from both the 1958 British Birth Cohort and the UK Blood Services. Five diallelic SNPs were detected in that study as showing strong association with Type 1 diabetes at a significance level of 10−7, and later replicated in an independent study (Todd et al., 2007). To evaluate the impact of these novel findings for clinical use, we constructed predictive genetic tests using the three different methods: the optimal robust ROC curve approach, logistic regression and classification tree. To perform the logistic regression analysis, we coded genetic variants with multiplicative, dominant, and recessive models, and implemented backward selection as indicated above to choose the most parsimonious model from all five possible single locus main effects and all their possible higher order (i.e. 26) interaction effects.
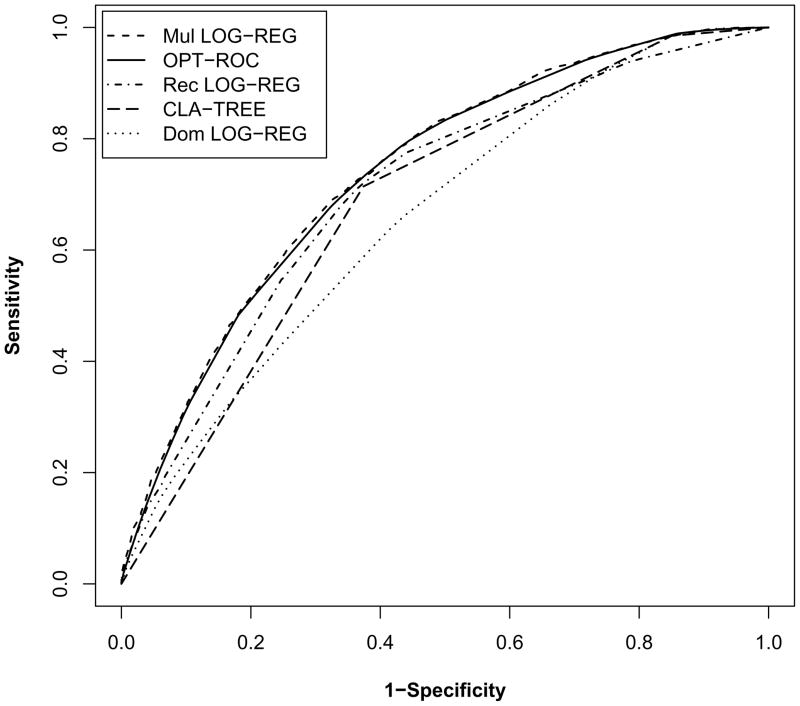
The classification accuracy of the predictive genetic test from the optimal robust ROC curve method (AUC=0.7373) is comparable to that from the logistic regression that assumes a multiplicative model (AUC=0.7420), but is superior to those from the other approaches - in particular, the classification tree (AUC=0.6868) and logistic regression that assumes a dominant model (AUC=0.6596) (Figure 1).
Figure 1.
The five lines in the plot, from top to bottom, correspond to the ROC curves derived from: logistic regression assuming a multiplicative effect, the optimal robust ROC curve, logistic regression assuming a recessive effect, classification tree, and logistic regression assuming a dominant effect. The estimated AUC values of these five approaches are 0.7420, 0.7373, 0.7079, 0.6868 and 0.6596, respectively.
We applied replicated split-sample validation (Austin, 2007; Baker and Kramer, 2006; Michiels, Koscielny, and Hill, 2005) to assess the tests’ performance in an independent sample. To perform the replicated split-sample validation, we randomly chose 2/3 of the data as the training dataset and used the remaining data as the validation dataset. The models were first built on the training dataset using the various methods, and then applied to the validation dataset to calculate the AUC. For instance, when applying the optimal robust ROC curve method, we implemented ten-fold cross validation to select the most parsimonious model in the training dataset, and used that model to estimate the prediction AUC in the validation dataset. We repeated this process 1000 times and the AUCs calculated from the validation datasets were then averaged. The prediction AUC values of these five tests obtained this way were respectively 0.7349, 0.7213, 0.7038, 0.6888, and 0.6530 (Table 3), results consistent with our previous finding. The standard deviations of the AUC estimates are also reported in table 3, and the ROC curves based on 100 repeated split samples are plotted in Web Figure 1 to give a graphical view of the variability of the ROC curves. It should be noted that in general the choice of SNPs to include in the test should be part of the cross validation procedure. However, we did not do this in view of the fact that an independent study (Todd et al., 2007) replicated the association of Type 1 diabetes with the five diallelic SNPs, and this would explain our finding that the AUC for the validation dataset is only slightly lower than the AUC for the training data set.
Table 3.
AUC estimates and standard deviations (SD) for the five methods based on 1000 repeated split samples from the Wellcome Trust dataset.
| Training Dataset | Validation Dataset | |||
|---|---|---|---|---|
| AUC | SD | AUC | SD | |
| Mul LOG-REG | 0.7427 | 0.0051 | 0.7349 | 0.0100 |
| OPT-ROC | 0.7388 | 0.0100 | 0.7213 | 0.0103 |
| Rec LOG-REG | 0.7092 | 0.0052 | 0.7038 | 0.0104 |
| CLA-TREE | 0.6908 | 0.0083 | 0.6888 | 0.0105 |
| Dom LOG-REG | 0.6607 | 0.0052 | 0.6530 | 0.0106 |
Using the Type 1 diabetes data, we also investigated the unordered optimal method and the jagged ordered method proposed by Baker (2000) (Web Table 1). Although both methods gave slightly over-fitted AUC estimates in the training dataset, the predictive genetic tests built by these two methods performed well in the validation dataset. Compared to the unordered optimal method, the jagged ordered method appeared to result in less over-fitting and hence a more robust performance, results consistent with the findings by Baker (2000).
Note that we evaluated the performance of the methods in terms of the AUC. Because the AUC is a global measure of a test’s discriminative ability, it might not be a good measure from a decision-theoretic viewpoint. For the purpose of decision making, we are only interested in the clinically important part of the ROC curve, and should therefore use a measure for the relevant part of the ROC curve (e.g., a partial AUC). The clinically important region of the ROC curve can be determined based on the disease prevalence and the ratio of profit to loss incurred on using the test (Baker and Kramer, 2007; Baker, 2000). In our case, Type 1 diabetes is a rare disease with an estimated prevalence of 0.0017, and we expect a large ratio of loss to profit on using the test owing to the lack of a successful disease prevention approach. Therefore, as discussed by Baker and Kramer (2007), the part of the ROC curve we are interested in would be the left portion of the ROC curve where the FPR is low. If we compare the methods based on the region where the FPR is low (e.g., less than 0.1), then the above conclusion for the comparison of the three methods will still hold - except that now dominant logistic regression performs better than the classification tree.
6. Discussion
In this article we proposed two robust forms of the optimal ROC curve-based method for building predictive genetic tests on the basis of genetic markers, one for the situation when there is prior knowledge of the disease model and one for the situation when there is no such prior knowledge. They can be looked upon as illustrations of the original optimality theory based on the likelihood ratio (Egan, 1975). Such theory simply indicates that a decision rule based on the likelihood ratio is best (Egan, 1975; McIntosh and Pepe, 2002). We have shown elsewhere that a test built using the LR can attain the highest classification accuracy in terms of the AUC (Lu and Elston, 2008).
Through simulations, we evaluated the new method and compared it with commonly used approaches, such as logistic regression and classification tree. If we know the underlying model (i.e., the causal loci, their modes of inheritance and interactions), the ROC curves built with all three methods approach the underlying ROC curve very well and there are no significant differences among them. However, their performances can be quite different if we have no prior knowledge of the disease model, which is usually the case for complex diseases. If we assume the “right” disease model, logistic regression could have a comparable or even slightly better performance than the optimal robust ROC curve method. However, when we have no prior knowledge of the disease model, we might assume the “wrong” disease model and this could seriously underestimate the predictive genetic test’s classification accuracy. As illustrated in simulation II, logistic regression assuming a recessive model had poor performance when the disease loci followed a dominant or multiplicative model. In another scenario, we have shown (Web Appendix C) that when all three disease loci followed a recessive model the test build by logistic regression assuming a dominant model had poor classification accuracy. In contrast to logistic regression, the optimal robust ROC curve method does not require the assumption of a particular disease model; i.e., it is genetic model free and is robust to a variety of underlying disease models. The performance of the optimal robust ROC curve method is always comparable to that of the best logistic regression model (i.e., the one that assumes the “right” disease model). Note that in the two simulations, although logistic regression assuming a multiplicative model had a slightly worse performance than the optimal robust ROC curve method, overall it gave quite robust AUC estimates. Thus, similarly to the optimal ROC curve method, logistic regression assuming a multiplicative model could potentially be used for scenarios where limited knowledge of the underlying disease model is known.
Another advantage of the optimal robust ROC curve method is that it is nonparametric. For traditional parametric methods, such as logistic regression, the number of parameters grows exponentially as we try to model higher order interactions among multiple genetic variables. For instance, 31 (25 − 1) and 255 (28 − 1) parameters are required in order to model all possible gene-gene interactions among the 5 loci of the real data example and the 8 loci used for simulation II, respectively. Fitting a logistic regression model with 255 parameters and then using backward selection to choose the most parsimonious model is computationally difficult. Moreover, having too many parameters in the model can lead to biased estimates and increased type I error (Peduzzi et al., 1996). To avoid these issues, Hosmer and Lemeshow (2000) suggested that the number of parameters in the model should be less than or equal to min (n1, n0)/(10 − 1), where n1 and n0 are the numbers of cases and controls, respectively. If we apply their formula to the dataset with 250 cases and 250 controls, the number of parameters should be less than 28, so that fitting a logistic regression model with all possible gene-gene interactions (i.e., with 255 parameters) is inappropriate. Therefore, compared with logistic regression, nonparametric methods such as the optimal robust ROC curve and classification tree methods avoid the issue of an increasing number of parameters, but nevertheless have the advantage of identifying the high order interactions.
Similar to a classification tree, the optimal robust ROC curve method gives an easily interpretable result because the high risk group individuals will always appear on the left part of the ROC curve and the low risk group will appear on the right part of the curve. However, as illustrated by the simulation and real data examples, the tests formed by the optimal robust ROC curve have better classification accuracy than that of the classification tree in terms of the AUC.
The approach proposed in section 3.2 can also be looked upon as an extension of Baker’s unordered optimal method that includes model selection, and we have illustrated the importance of model selection when building predictive genetic tests. As we have shown in simulation II, simple implementation of Baker’s unordered optimal method for genetic data without considering model selection can bias the AUC estimate. Baker’s approach was originally proposed in order to combine multiple tumor biomarkers for prediction proposes. Although our method is derived for genetic marker data, it could also be used for tumor biomarkers and clinical predictors, especially in those situations where we need to select the predictors and are interested in investigating interactions among them.
The important role of predictive genetic tests on health care has been recognized by both researchers and the public (Epstein, 2006; Evans, Skrzynia, and Burke, 2001; Jones M, 2000), and searching for successful predictive genetic tests has already been initiated for several diseases (Weedon et al., 2006). With the recent completion of genome-wide association studies, many novel associated genetic variants have been discovered. By using the proposed method, we are able to form a powerful predictive genetic test with these new discoveries, and therefore translate these findings into potential clinical use. We formed a predictive genetic test for Type 1 diabetes based on 5 novel loci discovered from a GWA study. The test reached a mid level of classification accuracy that is much higher than those of the tests for other common diseases (e.g., Type 2 diabetes). With the discovery of more disease risk variants, and eventually their interactions, we might be able to form a predictive genetic test for Type 1 diabetes that could be implemented in clinical use.
Supplementary Material
Acknowledgments
This work was supported by U.S. Public Health Service Resource grant (RR03655) from the National Center for Research Resources, Research grant (GM28356) from the National Institute of General Medical Sciences, and Cancer Center Support Grant P30CAD43703 from the National Cancer Institute. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for that project was provided by the Wellcome Trust under award 076113. We would like to thank the anonymous Associated Editor and Reviewer for their suggestions that greatly improved our presentation.
Footnotes
Web Appendices A, B, and C, referenced in Sections 3.1, 3.2 and 6, are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
References
- 1.Akaike H. A new look at the statistical model identification. IEEE Transactions Automatic Control. 2001;AC-19:716–723. [Google Scholar]
- 2.Austin PC. A comparison of regression trees, logistic regression, generalized additive models, and multivariate adaptive regression splines for predicting AMI mortality. Statistics in Medicine. 2007;26 (15):2937–2957. doi: 10.1002/sim.2770. [DOI] [PubMed] [Google Scholar]
- 3.Baker SG. Identifying combinations of cancer markers for further study as triggers of early intervention. Biometrics. 2000;56 (4):1082–1087. doi: 10.1111/j.0006-341x.2000.01082.x. [DOI] [PubMed] [Google Scholar]
- 4.Baker SG, Kramer BS. Identifying genes that contribute most to good classification in microarrays. BMC Bioinformatics. 2006;7:407. doi: 10.1186/1471-2105-7-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker SG, Kramer BS. Peirce, Youden, and receiver operating characteristic curves. American Statistician. 2007;61 (4):343–346. [Google Scholar]
- 6.Egan JP. Signal Detection Theory and ROC Analysis. New York: Academic Press; 1975. [Google Scholar]
- 7.Epstein CJ. Medical genetics in the genomic medicine of the 21st century. American Journal of Human Genetics. 2006;79 (3):434–438. doi: 10.1086/507610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nature Reviews Cancer. 2003;3 (4):243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 9.Evans JP, Skrzynia C, Burke W. The complexities of predictive genetic testing. British Medical Journal. 2001;322 (7293):1052–1056. doi: 10.1136/bmj.322.7293.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosmer DW, Lemeshow S. Applied logistic regression. John Wiley & Sons; New York: 2000. [Google Scholar]
- 11.Jones M. The genetic report card. New York Times Magazine. 2000;11:80. [Google Scholar]
- 12.Lu Q, Elston RC. Using the optimal receiver operating characteristic curve to design a predictive genetic test, exemplified with type 2 diabetes. American Journal of Human Genetics. 2008;82 (3):641–651. doi: 10.1016/j.ajhg.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClish DK. Analyzing a portion of the ROC curve. Medical Decision Making. 1989;9 (3):190–195. doi: 10.1177/0272989X8900900307. [DOI] [PubMed] [Google Scholar]
- 14.McIntosh MW, Pepe MS. Combining several screening tests: optimality of the risk score. Biometrics. 2002;58 (3):657–664. doi: 10.1111/j.0006-341x.2002.00657.x. [DOI] [PubMed] [Google Scholar]
- 15.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365 (9458):488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 16.Peduzzi P, Concato J, Kemper E, Holford TR, Feinstein AR. A simulation study of the number of events per variable in logistic regression analysis. Journal of Clinical Epidemiology. 1996;49 (12):1373–1379. doi: 10.1016/s0895-4356(96)00236-3. [DOI] [PubMed] [Google Scholar]
- 17.Pepe MS, Cai T, Longton G. Combining predictors for classification using the area under the receiver operating characteristic curve. Biometrics. 2006;62 (1):221–229. doi: 10.1111/j.1541-0420.2005.00420.x. [DOI] [PubMed] [Google Scholar]
- 18.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, Zeggini E, Rayner NW, Shields B, Owen KR, Hattersley AT, Frayling TM. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. Plos Medicine. 2006;3 (10):e374. doi: 10.1371/journal.pmed.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447 (7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clinical Chemistry. 1993;39 (4):561–577. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.