Abstract
Integrins are large, membrane-spanning, heterodimeric proteins that are essential for a metazoan existence. All members of the integrin family adopt a shape that resembles a large “head” on two “legs,” with the head containing the sites for ligand binding and subunit association. Most of the receptor dimer is extracellular, but both subunits traverse the plasma membrane and terminate in short cytoplasmic domains. These domains initiate the assembly of large signaling complexes and thereby bridge the extracellular matrix to the intracellular cytoskeleton. To allow cells to sample and respond to a dynamic pericellular environment, integrins have evolved a highly responsive receptor activation mechanism that is regulated primarily by changes in tertiary and quaternary structure. This review summarizes recent progress in the structural and molecular functional studies of this important class of adhesion receptor.
Integrin receptors possess two large extracellular “heads” that mediate ligand binding and subunit association. Two short intracellular “legs” assemble cytoplasmic signaling complexes and transmit structural changes to the heads during activation that allow them to bind ligand.
The name “integrin” was suggested for an integral membrane protein complex first characterized in 1986 (Tamkun et al. 1986). The name was devised because the protein identified linked the extracellular matrix to the cytoskeleton (early developments in this field have been well described [Hynes 2004]). In the 25 years since that first characterization, a vast amount of work has been performed, with consequent increased understanding. The essential role of integrins in tissue organization and cell development, their signal transduction mechanisms (from outside to in and inside to out!), and their potential as therapeutic targets is now established. In this article, we provide an overview of the structure of integrins, the conformational changes that determine activation state, and the mechanisms of ligand binding.
INTEGRIN STRUCTURE
Overall Structure
Integrins are heterodimers of non-covalently associated α and β subunits. In vertebrates, there are 18 α and 8 β subunits that can assemble into 24 different receptors with different binding properties and different tissue distribution (Hynes 2002; Barczyk et al. 2010). The α and β subunits are constructed from several domains with flexible linkers between them. Each subunit has a single membrane-spanning helix and, usually, a short unstructured cytoplasmic tail. The size varies but typically the α- and β-subunits contain around 1000 and 750 amino acids, respectively. Numerous reviews on integrin structure and function have been published (Arnaout et al. 2007; Luo et al. 2007; Askari et al. 2009; Bennett et al. 2009) so here we concentrate mainly on the implications of recent structural work that includes studies of intact ectodomains, membrane spanning regions, cytoplasmic tails and their ligands.
The Ectodomains
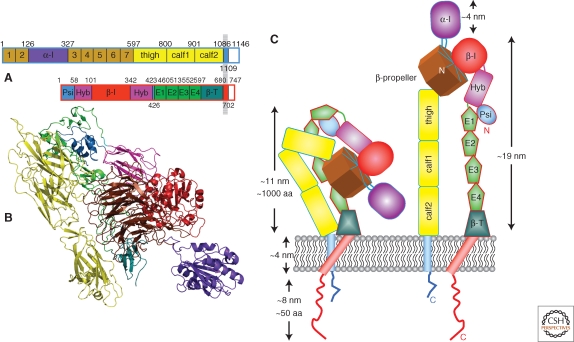
The breakthrough crystal structure of αVβ3 (Xiong et al. 2001) started a deluge of structural information about integrin ectodomains. Structures of αVβ3, with and without ligand (Xiong et al. 2001, 2002), αIIbβ3 (Zhu et al. 2008), and αxβ2 (Xie et al. 2009) are all now available. These crystal structures are all in a similar overall “bent” conformation that would place the ligand binding site near the membrane surface. The overall topology and structure of integrin ectodomains is illustrated in Figure 1 for the case of αxβ2 (Xie et al. 2009), which has an inserted α-I domain.
Figure 1.
Integrin structure. (A) Domain structure of αxβ2 (Xie et al. 2009); (B) structure of αxβ2 using same color code as A (drawn with PyMOL [DeLano Scientific] using PDB coordinates 3K6S); (C) cartoon representation of bent and upright conformations showing approximate dimensions.
Current knowledge of ectodomains has also been enhanced by structures of various integrin fragments including isolated α-I domains (Lee et al. 1995a) and β2-leg fragments (Beglova et al. 2002; Xiao et al. 2004; Shi et al. 2005).
Because of observed flexibility in studies by electron microscopy (EM) and the existence of conformationally sensitive antibody recognition sites (Humphries 2004), there is a general acceptance that conformations other than the bent one are possible and are functionally relevant (see below). The structural studies of intact ectodomains (Xiong et al. 2001; Xie et al. 2009; Zhu et al. 2009) all postulated that an upright structure of the sort illustrated in Figure 1C could exist as well as the bent structure. There has, however, been controversy about whether this large change between bent and upright structures—the “switchblade” model (Luo et al. 2007)—has to take place or if more conservative changes around the bent structure can explain the data—the “deadbolt” model (Arnaout et al. 2005).
The Structure of α Subunit Ectodomains
The α-chain consists of four or five extracellular domains: a seven-bladed β-propeller, a thigh, and two calf domains. Nine of the 18 integrin α chains have an α-I domain of around 200 amino-acids, inserted between blades 2 and 3 of the β-propeller (Larson et al. 1989). The I domain, a copy of which also appears in the β-chain, has five β-sheets surrounded by seven α helices; it is similar to von Willebrand A domains. The last three or four blades of the β-propeller contain domains that bind Ca2+ on the lower side of the blades facing away from the ligand-binding surface. Ca2+ binding to these sites has been shown to influence ligand binding (Oxvig and Springer 1998; Humphries et al. 2003).
The thigh and calf domains have similar, immunoglobulin-like, β-sandwich folds (Xiong et al. 2001). They have 140–170 residues with more β-strands than typical Ig-like domains (∼100 residues). There are two main regions of interdomain flexibility. One is the linker between the β-propeller and the thigh; the other is the “genu” or knee at the bend between the thigh and the calf-1 domain. The α-subunit genu is located close to the similar bend in the β subunit, thereby allowing extension by a hinging at the knees. The α-I domain in αxβ2 is inserted in the β-propeller domain with flexible linkers (Fig. 1C). Unlike the other four α-leg domains, which have relatively rigid structures, I domains show conformational changes within the domain that are important for regulating binding affinity (see below and Fig. 2).
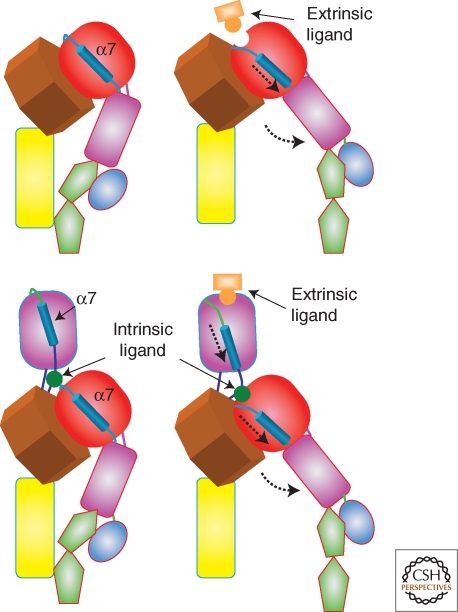
Figure 2.
An illustration of the movement of α7 helix in the I domains and the swing-out of the hybrid domain (the domains are defined in Fig. 1). The top pair corresponds to the closed and open conformations of an integrin without an inserted α-I domain whereas the lower pair represents the situation when there is an α-I domain present. The intrinsic ligand is a glutamate (E310 in αL).
The Structure of β Subunit Ectodomains
The β-leg has seven domains with flexible and complex interconnections. A β-I domain is inserted in a hybrid domain, which is, in turn, inserted in a plexin-semaphorin-integrin (PSI) domain. These domains are followed by four cysteine-rich epidermal growth factor (EGF) modules and a β-tail domain. The hybrid domain in the upper β-leg has a β-sandwich construction. The β-I domain, which is homologous to the α-I domain, is inserted into the hybrid domain. The small PSI domain, with an α/β fold, is also split into two portions (Xiao et al. 2004; Xiong et al. 2004) connected, in β3, by a long-range Cys-13 to Cys-435 disulfide bond.
Unusual EGF module boundaries were first proposed in the αVβ3 structure (Xiong et al. 2004), but recent crystal structures suggest that each EGF module has an even number of eight cysteines, bonded in a C1-C5, C2-C4, C3-C6, and C7-C8 pattern except for EGF1, which lacks the C2-C4 disulfide. The αIIbβ3 structure shows that all 56 cysteines in the integrin β3 subunit are disulfide bonded (Zhu et al. 2008).
The β-tail domain has an α + β fold (Xiong et al. 2001). The weak electron density of this domain observed in the αIIbβ3 crystal structure was taken to suggest a flexible connection to other regions of the β-leg by a mobile “ankle” (Zhu et al. 2008). A contact between the CD loop of the β-tail domain and the α7 helix of the β-I domain has been proposed to inhibit integrin activation—the “deadbolt” model (Arnaout et al. 2005). This contact is, however, small and no such contact is observed in the αIIbβ3 or αxβ2 structures.
In general, the β-leg seems to be more flexible than the α-leg. Evidence for this comes from the ten different structures of αXβ2 observed in three different crystal lattices (Xie et al. 2009). The EGF domain region is relatively plastic, especially between EGF1 and EGF2, the β knee, and at the PSI/hybrid and hybrid/I-EGF1 junctions. There is evidence for important conformational changes occurring in the β-I/hybrid region. A transition from a “closed” to an “open” conformation of the β-I domain has been observed when the β-I α7-helix moves toward the hybrid domain (Xiao et al. 2004). The connecting rodlike motion of the α7-helix causes the hybrid domain to swing-out by ∼60o (see Fig. 2).
Cation Binding Sites
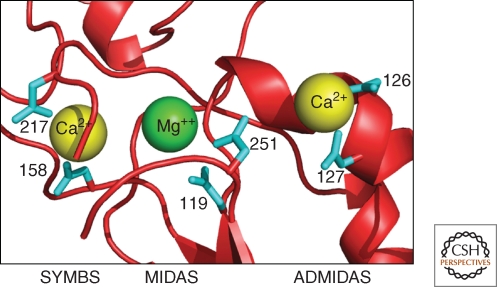
As described below, ligand binding in α-I less integrins takes place at the largest interface between the two subunits (the β-propeller/β-I domain interface); binding is dependent on the cations Mg++, Ca2+, and Mn++. From a structure of an α-I domain it was suggested that integrin ligand binding involves a Mg++ ion, a “metal-ion-dependent adhesion site” (MIDAS) (Lee et al. 1995b); a crystal form with bound Mn++ also showed considerable movement of the α7-helix on activation (Lee et al. 1995a). In the recent αIIbβ3 structure (Zhu et al. 2008) strong electron densities were ascribed to cations at three sites formed by loops in the β-I domain (Fig. 3); Mg++ was assigned to the central MIDAS site with Ca2+ at the two flanking sites. One of these adjacent sites (ADMIDAS) binds an inhibitory Ca2+ ion; binding of Mn++ here results in a structural change that gives an active integrin (Humphries et al. 2003). The second Ca2+-binding site has been called the synergistic metal ion binding site (SyMBS) (Zhu et al. 2008). Mutational studies show that the SyMBS site is responsible for Ca2+ synergy (Chen et al. 2003; Mould et al. 2003a).
Figure 3.
Three β-I domain metal-binding sites in αXβ2 (Zhu et al. 2008); aspartate ligands to the metal ions are shown in cyan (from PDB:3FCS); figure drawn using PyMOL (DeLano Scientific).
As mentioned above, the β-I domain has distinct closed and open conformational states, involving movement of the α7 helix, in α-I-less integrins (Xiao et al. 2004; Luo et al. 2007). Similar conformational changes are seen in α-I-domains. When integrin ligands, such as Arg-Gly-Asp, bind the open state, the MIDAS Mg++ ion coordinates the Asp side chain of the ligand. In α-I integrins it has been suggested that the β-I MIDAS may bind an intrinsic ligand, an invariant Glu, Glu-318 in αX (Xie et al. 2009). Support for this model comes from Glu mutations that abolish integrin activation (Huth et al. 2000; Alonso et al. 2002); the observed flexibility of the αx α-I domain would also facilitate such interdomain interactions (Xie et al. 2009).
The Membrane Spanning Helices
The current view is that association of integrin α and β transmembrane (TM) segments, results in an inactive resting receptor (Wegener and Campbell 2008). Evidence for this includes experiments using EM (Adair and Yeager 2002), disulfide cross-linking (Luo et al. 2004), activating mutations (Partridge et al. 2005), and FRET of labeled cytoplasmic tails (Kim et al. 2003). Recent studies have given new insight into the structure of the resting state. The structures of β3 and αIIb TM segments in phospholipid bicelle model membranes have been solved by NMR separately (Lau et al. 2008a,b) and in complex (Lau et al. 2009). A similar structure was obtained for the TM region in intact αIIbβ3 using disulfide-based distance restraints combined with protein modeling (Zhu et al. 2009). A recent structure of a complex in organic solvents has been described (Yang et al. 2009) but the relevance of a system without a phospholipid/water interface is questionable. A bacterial reporter system has been used to define the sequence motif required for TM helix-helix interactions in β1 and β3 integrin subfamilies (Berger et al. 2010). Several modeling studies of the TM regions have also been published (Gottschalk 2005; Metcalf et al. 2009; Wang and Luo 2010). [Note that whereas most evidence supports the heterodimeric form, there is also evidence that homomeric TM oligomers can form in vitro (Li et al. 2003; Parthasarathy et al. 2008); nevertheless, the evidence for homodimeric forms mainly comes from experiments or simulations performed without the ectodomains, whose presence would be expected to favor the heterodimeric form.]
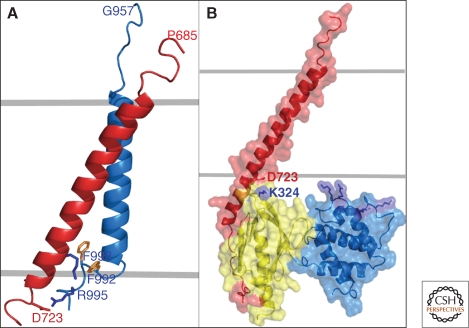
The NMR structure of the αIIbβ3 TM complex is shown in Figure 4. The αIIb helix is perpendicular to the membrane whereas the β3 helix is tilted. There are glycines at the helix-helix interface in the membrane and an unusual αIIb backbone reversal that packs a consecutive pair of Phe residues against the β3 TM helix, promoting electrostatic interactions between αIIb(D723) and β3(R995). The two TM segments have essentially the same structure when studied separately suggesting that the topological features of the TM segments will remain unchanged in the separated, active state.
Figure 4.
(A) NMR structure of the complex between the αIIb (blue) and β3 (red) TM domains (PDB: 2K9J). The approximate position of the membrane glycerol backbone is shown by gray lines. (B) The talin F2 (blue)/F3 (yellow) domain pair in complex with a β integrin tail (red). The salt bridge that forms between K324 on F3 and D723 in the tail is shown; some key Lys and Arg residues are indicated in blue near the putative membrane interface with the F2 domain. B was constructed from a composite of coordinates of the talin/β complex (PDB: 3G9W; [Anthis et al. 2009]) and the membrane complex (PDB:2K9J [Lau et al. 2009]). Images made using PyMOL (DeLano Scientific).
The ectodomain-TM linkers seem to be flexible (Lau et al. 2009; Zhu et al. 2009) and are thus unlikely to constrain the orientation between the ectodomain and the membrane. Although αXβ2 is bent in a similar way to integrins without an α-I domain, the terminal domains of the α-legs and β-legs, calf-2, and β-tail domains, are oriented differently (Xie et al. 2009). These observations are all consistent with the flexible transmembrane domain separation model of activation rather than stiff pistons or levers. The TM complex is also likely to be stabilized by the resting ectodomain.
The Cytoplasmic Tails
Several NMR studies of cytoplasmic tails have been published, although there is little agreement among them. Some studies could not detect an interaction between α and β tails, whether they were connected by a coiled coil construct (Ulmer et al. 2001) or inserted in a membrane with TM segments (Li et al. 2001). A study of isolated mixed peptides found two distinct structures of the αβ complex (Weljie et al. 2002), both significantly different from the most detailed published structure of the αβ tail complex (Vinogradova et al. 2002). The latter structure does not seem to be consistent with the recently solved structures of the TM regions (Lau et al. 2009; Zhu et al. 2009). The most obvious explanation for these discrepancies is that the tails are rather flexible, only forming transient structures in solution in the absence of a protein interaction partner and that complexes between the tails and their binding proteins are likely to provide the most significant insights into transduction events.
Cytoplasmic Tail Ligands
Because they are extended and flexible, the cytoplasmic tails, especially β, can “flycast” (Shoemaker et al. 2000) and reach out to form “hub” interactions with a number of proteins (Wegener and Campbell 2008; Legate and Fassler 2009). In particular, PTB domains bind to one of the two conserved NPXY motifs in β-tails. Especially important for activation of integrins from inside the cell are the proteins talin and kindlin. The talin PTB domain (the head F3 subdomain) binds the first β-NPXY and the membrane proximal helix (Wegener and Campbell 2008) whereas kindlin binds the second NPXY (Moser et al. 2008). It has been proposed that the F2.F3 subdomains of talin make a defined contact with the membrane surface via numerous lysine and arginine residues especially in the F2 domain (Fig. 4B; Anthis et al. 2009). Binding of F3 to the β-tail promotes tail dissociation by breaking the salt bridge between the α and β tails (αIIb[D723]-β3[R995]); a new salt bridge is formed with K324 on the F3 domain. Binding to β plus the F2.F3 contact with the membrane can also influence the orientation of the β-helix, again helping promote separation of the TM and cytoplasmic domains.
Structural Studies of Intact Integrins
Although crystallography of ectodomains and NMR of TM domains have provided detailed information about integrin structure, other techniques have been applied to intact integrins in attempts to distinguish among current models, such as “switchblade” and “deadbolt.” In principle this should be relatively easy because the switchblade model predicts a near doubling in molecular height on activation (Fig. 1C), whereas the deadbolt model predicts a modest change. In general, however, the various studies have not been in good agreement, possibly because of multiple integrin conformations in solution.
Cryoelectron tomography of ice-embedded specimens was used to obtain three-dimensional images of full-length αIIbβ3 incorporated into small liposomes (Ye et al. 2008). No significant height change was observed between the inactive state, and the active state induced by Mn++. In a FRET study, Mn++ activation caused an increase of 5 nm in the separation between membrane and ligand binding site, consistent with a conformational change to the upright configuration (Chigaev et al. 2003). However, FRET between fluorescently labeled Fab fragments and the β3 β-I domain indicated only small changes on platelet activation (Gupta et al. 2007). A small angle neutron scattering (SANS) study of intact αIIbβ3 in Ca2+/detergent solutions found “arched” and “handgun” forms (Nogales et al. 2010) although a Mn++ activated form was not studied. Analytical ultracentrifugation and EM were used to investigate αIIbβ3; measurement of frictional changes under different conditions suggested considerable plasticity in the structure (Rocco et al. 2008).
EM studies of ectodomains are also not entirely consistent although, on balance, they favor the switchblade model. Single-particle reconstructions of the negatively stained αvβ3 ectodomain bound to a fibronectin (FN) fragment suggested that the bent conformation can bind its physiological ligand (Adair et al. 2005). EM of negatively stained αvβ3 ectodomain with a cleavable clasp engineered into the carboxyl terminus showed a majority of molecules in the bent conformation when inactive, and a majority in the upright conformation when active (Takagi et al. 2002). Negative stain EM of a shorter ectodomain construct of α5β1 bound to FN showed similar results (Takagi et al. 2003). The recent x-ray structure papers of intact ectodomains contain EM results consistent with the switchblade model (Zhu et al. 2008; Xie et al. 2009).
A recent study of integrin activation using EM and other studies gives useful insight (Ye et al. 2010). Membrane nanodiscs were synthesized with a single lipid-embedded integrin. The majority (∼90%) of the class-averaged integrin nanodisc EM images in the absence of ligand and the talin head domain had a compact structure with a height of 11±1 nm; i.e., corresponding to the bent conformation in Figure 1C. In the presence of talin, ∼25% of unliganded integrins had an extended structure with a height of 19±1 nm. In contrast, at least 40% of the fibrin-bound integrins were extended. This study provides evidence that talin binding is sufficient to activate and extend membrane-embedded integrin αIIbβ3 without applied force or clustering.
There is evidence that mechanical force is important for regulating integrin adhesiveness (Alon and Dustin 2007; Evans and Calderwood 2007). Talin contains both integrin and actin binding sites (Critchley 2009), and therefore the cytoskeleton could exert a lateral force on the β subunit. Steered molecular dynamics was applied to a complete ectodomain to mimic effects of cell generated tension. Evidence was found that lateral force could be transmitted through the β leg to the hybrid domain and promote the active form (Zhu et al. 2008).
LIGAND BINDING
Historically, the pairing of integrins and their ligands has been uncovered either by ligand affinity chromatography or through the use of subunit-specific monoclonal antibodies (mAbs) to block ligand-mediated cell adhesion. In most cases, protein-protein binding assays have confirmed the associations established by these biochemical or cell biological approaches. A characteristic feature of most integrin receptors is their ability to bind a wide variety of ligands. Conversely, many extracellular matrix (ECM) and cell surface adhesion proteins bind to multiple integrin receptors (Humphries 2000; Plow et al. 2000; van der Flier and Sonnenberg 2001). One molecular explanation for this complexity is the evolutionary selection of common acidic peptide motifs in ECM proteins that mediate integrin binding via coordination to a divalent cation-containing binding pocket.
Integrin-ligand combinations can be clustered into four main classes, based on the nature of the molecular interaction. All five αV integrins, two β1 integrins (α5, α8), and αIIbβ3 recognize ligands containing an RGD tripeptide active site. Crystal structures of αVβ3 and αIIbβ3 complexed with RGD ligands have been reported and they reveal an identical atomic basis for this interaction (Xiong et al. 2002; Xiao et al. 2004). RGD binds at an interface between the α and β subunits, with the basic residue fitting into a cleft in a β-propeller module in the α subunit, and the acidic residue coordinating a cation bound in the β-I-domain. RGD-binding integrins bind to a large number of ECM and soluble vascular ligands, but with different affinities that presumably reflect the preciseness of the fit of the ligand RGD conformation with the specific α,β active site pockets. Although RGD is an essential element of the ligand binding process, macromolecular ligands can contain other binding sites, the best characterized of which is a synergy sequence in fibronectin that also binds the α5 β-propeller (Mould et al. 1997, 2003c).
α4β1, α4β7, α9β1, the four members of the β2 subfamily, and αEβ7 recognize related sequences in their ligands. α4β1, α4β7, and α9β1 bind to an acidic motif, termed “LDV,” that is functionally related to RGD. Fibronectin contains the prototype LDV ligand in its type III connecting segment region, but other ligands (such as VCAM-1 and MAdCAM-1) employ related sequences. Structures of this integrin subfamily are lacking, but it is highly likely that LDV peptides bind similarly to RGD at the junction between the α and β subunits. The β2 family employs a similar mode of ligand binding, but the major interaction takes place via an inserted I-domain in the α subunit (Shimaoka et al. 2003). Despite this fundamental mechanistic difference, the characterized sites within ligands that bind β2 integrins are structurally homologous to the LDV motif (Shimaoka et al. 2003). The major difference is that β1/β7 ligands employ an aspartate residue for cation coordination, whereas β2 integrins use glutamate.
Four α subunits containing an α-I-domain (α1, α2, α10, and α11) combine with β1, and form a distinct laminin/collagen-binding subfamily. A crystal structure of a complex between the α2 I-domain and a triple-helical collagenous peptide has revealed the structural basis of the interaction, with a critical glutamate within a collagenous GFOGER motif providing the key cation-coordinating residue (Emsley et al. 2000). The mechanism of laminin binding is less well understood, although a recent study has suggested that the extreme carboxyl terminus of the γ chain and an undefined site within α subunit laminin G domains together constitute an integrin-binding site (Ido et al. 2007). Three β1 integrins (α3, α6, and α7), plus α6β4, are highly selective laminin receptors. Analysis of laminin fragments indicates that these receptors and the α-I-domain-containing β1 integrins bind to different regions of the ligands. In neither case has the active site been narrowed down to a particular sequence or residue.
ACTIVATION
For the interaction of integrins with their ligands to be meaningful for cellular function, the binding event must be able to regulate signal transduction. However, adhesion is highly dynamic, with cells continuously sampling their pericellular environment, and responding by rapidly changing their position and state of differentiation, and therefore a highly responsive receptor activation mechanism is required. As integrins lack enzymatic activity, signaling is instead induced by the assembly of signaling complexes on the cytoplasmic face of the plasma membrane. Formation of these complexes is achieved in two ways; first, by receptor clustering, which increases the avidity of molecular interactions thereby increasing the on-rate of binding of effector molecules, and second, by induction of conformational changes in receptors that creates or exposes effector binding sites. Current evidence suggests that conformational regulation is the primary mode of affinity regulation of integrins. In turn, this demands a mechanism for conveying conformational changes between the cytoplasmic tails and the ligand-binding head domain over a relatively large distance (∼20 nm).
Evidence for Conformational Regulation
Gross conformational changes in integrins have been monitored by a variety of techniques, and for almost all of these studies, αIIbβ3 has served as a prototype. Treatment with RGD peptides elicited alterations in sedimentation coefficient and Stokes radius (Parise et al. 1987), and receptor activation on platelets triggered changes in intramolecular FRET and cross-linking (Sims et al. 1991). mAbs have proven particularly useful probes of integrin function. Early studies reported activation-dependent changes in mAb binding to αIIbβ3 that were attributed to conformational changes (McEver and Martin 1984; Coller 1985), and these were followed by the identification of a subset of anti-αIIbβ3 mAbs, the epitopes for which were induced in response to ligand binding (Frelinger et al. 1990, 1991). The acronym LIBS was coined to describe these epitopes as ligand-induced binding sites. In most cases that have been examined, activating mAbs appear to function by increasing the affinity of ligand binding. Most LIBS mAbs have epitopes that are regulated by divalent cations, and because cations also regulate ligand binding, it appears that many cation-responsive, activating mAbs recognize naturally occurring conformers of integrins. These mAbs may therefore displace a conformational equilibrium in favor of these forms that leads to an increase in the proportion of high affinity integrin. Some other activating mAb epitopes are unaffected by either ligand or cation binding and here the most likely mechanism of action is through inducing an activated conformation in the integrin (Chen et al. 1999).
The location of LIBS epitopes has contributed significantly to our understanding of the process of receptor activation. The overwhelming majority of activating mAbs recognize the β subunit, and their epitopes are distributed throughout the polypeptide (Humphries 2000; Byron et al. 2009). This is suggestive of a large-scale alteration in the conformation of the whole integrin during activation. The regions recognized include the β-I-domain, the extreme amino terminus of the β subunit in the PSI domain, the hybrid domain, the β-subunit knee region, and distal EGF-like repeats. A few activating anti-α subunit mAbs have been reported, the epitopes for which are found in the β-propeller, the heavy-light chain border and close to the transmembrane domain, suggestive of conformational changes in these regions of the molecule (Loftus et al. 1987; Keizer et al. 1988).
How Are Conformational Changes Coupled?
As discussed above, the various structural studies in the last 10 years have greatly stimulated functional analyses. The relevance of the observed bend in the legs of the integrin dimer has been a highly contentious issue. It has been proposed that integrins are always bent, but several lines of evidence indicate that bent integrins are inactive, and extended integrins are primed. In the original crystal structure of αVβ3, the integrin was bent at an angle of 135°. Locking integrins in this state through disulphide bond engineering abolishes ligand binding by cell surface-expressed receptors (Takagi et al. 2002). Furthermore, when the gross structure of integrins was examined by electron microscopy under conditions in which ligand binding was low, e.g., in Ca2+-containing buffers or following the introduction of intersubunit covalent bond constraints, predominantly bent structures were observed (Takagi et al. 2002; Nishida et al. 2006). In bent integrins, the ligand-binding pocket may be oriented toward the plasma membrane, thereby impeding ligand engagement, but flexibility at the juxtamembrane domain could enable a “breathing” movement for the conversion of bent to extended integrin (Beglova et al. 2002; Takagi et al. 2002). In this context, a cryo-EM reconstruction of unstimulated αIIbβ3 indicated a partially extended conformation (Adair and Yeager 2002). The binding of stimulatory mAbs might then displace a conformational equilibrium, leading to activation. Similarly, breaking the interactions between the α and β cytoplasmic tails appears to lead to a loss of the interactions among the leg regions, disruption of an interface between the head and legs, and a repositioning of the head to point away from the cell surface. Major support for this model comes from studies of soluble recombinant integrins by electron microscopy (Takagi et al. 2002) and from the large number of epitopes of stimulatory mAbs that have now been shown to lie in the knee or leg regions (Humphries et al. 2003). Exposure of these epitopes is low in the bent state of the integrin (where they are masked) but high in the extended state (Beglova et al. 2002).
The pathway of conformational change from the interior of the cell to the ligand-binding site of the integrin is incompletely understood, but movement of the hybrid domain appears to be a central feature of the conformational changes accompanying unbending (Fig. 2). By EM, an acute angle between the hybrid domain and β-I was observed in the bent state, and an obtuse angle in the extended, ligand-occupied state (Takagi et al. 2002). In the bent conformation, any movement of the hybrid domain relative to the β-I domain is prevented, and therefore unbending is probably an essential prerequisite to hybrid domain motion. A central role for hybrid domain movement in affinity regulation has been established by a number of approaches. Activating mAb epitopes in the hybrid domain map close to an interface between the hybrid domain and the α subunit β-propeller (Mould et al. 2003b). These epitopes would be masked in bent integrins, but would become exposed when the hybrid domain swings away from the propeller. Furthermore, engineering of glycosylation sites between the hybrid domain and the β-I-domain produces a putative wedge that leads to integrin activation (Luo et al. 2003).
Conformational changes in the head are the key determinant of ligand-binding activity, specifically, the conformation of the β-I domain, which, in turn, is determined by the position of the hybrid domain. Thus, a swing-out of the hybrid domain away from the α-subunit pulls downward on the α7 helix of the β-I domain and favors the upward movement of the α1 helix (Xiao et al. 2004). The motion of these two helices shifts the β-I domain from a low-affinity into a high-affinity conformation by backbone movements of loops that contain cation-coordinating residues. Mutations that favor a downward shift of the α7 helix (Mould et al. 2003b; Hato et al. 2006; Cheng et al. 2007) also result in a high-affinity state.
Although current models of integrin function strongly suggest that conversion to a high-affinity receptor requires extension, there is some evidence to suggest that ligand-bound integrin can adopt a bent conformation. Crystallized αVβ3 can bind a cyclic RGD peptide in the bent conformation (Xiong et al. 2002) and electron microscopy images also show bent αVβ3 in complex with a fragment of fibronectin (Adair et al. 2005). In addition, studies that have either used FRET or competition ELISA to measure the distance between a fluorescently tagged ligand peptide and labeled cell membrane or between mAbs directed against the head piece and leg regions of αIIbβ3 on platelets have revealed partial unbending (Calzada et al. 2002; Chigaev et al. 2003, 2007). Nevertheless, when FRET-FLIM is employed to analyze the conformation of α5β1 in adherent cells, by measuring FRET between a fluorescently labeled Fab bound to α5β1 and fluorescent dye intercalated into the cell membrane, it has been shown that integrins are extended in focal adhesions and bent elsewhere (Askari et al. 2010).
Integrin Antagonists as Therapeutic Agents
The short acidic peptides that serve as ligand active sites are essentially pro-drugs, and both RGD and LDV peptides have been converted into small molecules therapeutics. RGD-based, peptidomimetic antagonists of αIIbβ3, such as eptifibatide (from 1998) and tirofiban (from 1998), are now used widely as antithrombotic agents (Hanson et al. 2004), and LDV-based compounds are in development for treatment of asthma and multiple sclerosis. In addition, mAbs that block integrin function and cell adhesion have been developed as therapeutic agents. These agents were originally assumed to compete with ligands for receptor binding, but this now appears not to be the case, with many anti-integrin mAbs having been shown to function via allosteric mechanisms. Current evidence suggests that mAbs inhibit ligand binding either by stabilizing the unoccupied state of the receptor or by preventing a conformational change necessary for ligand occupancy. In turn, the allosteric inhibition of ligand binding by anti-functional anti-integrin mAbs implies that it may be feasible to synthesize small molecule inhibitors that function in the same way. Such inhibitors could have advantages over competitive inhibitors in that a partial inhibition of function may be obtained and therefore adhesion may be more easily controlled, and they may not possess the agonistic properties of ligand mimetics, and may therefore not suffer from mechanism-related side-effects. Small molecule allosteric inhibitors that bind to α-I-domains have now been reported. These molecules appear to stabilize the low affinity conformation of the α-I-domain by blocking downward movement of the terminal α7-helix and thereby preventing rearrangements at the ligand-binding pocket necessary for high affinity ligand binding (Kallen et al. 1999).
CONCLUSION
There has been remarkable progress in our understanding of integrin structure and function in the last 10 years. The basis of much previous work on conformation, which was performed with conformationally sensitive antibodies, and ligand binding, which was largely based on mutational analyses, can now be modeled at atomic resolution. A unifying biophysical model of integrin function, which incorporates features such as catch bonds, extreme flexibility at the knees and the on- and off-rates of ligand and effector binding is therefore within reach. The process of integrin activation from inside the cell is also now quite well understood at a structural level. However, a number of major questions remain unresolved. These include outside-in signaling, which is much less well understood compared with inside-out signaling and it is unclear how similar or different the two processes are. We still do not know how an integrin allows a cell to interpret the binding of different ligands, and therefore how microenvironmental sensing is achieved at a molecular level. Looking further ahead, the process of “inactivation,” where integrins return to their resting state, is not understood, and we are just starting to develop approaches to measure force transduction at adhesion sites.
Footnotes
Editors: Richard Hynes and Kenneth Yamada
Additional Perspectives on Extracellular Matrix Biology available at www.cshperspectives.org
REFERENCES
- Adair BD, Yeager M 2002. Three-dimensional model of the human platelet integrin αIIbβ3 based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci 99: 14059–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair BD, Xiong JP, Maddock C, Goodman SL, Arnaout MA, Yeager M 2005. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J Cell Biol 168: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R, Dustin ML 2007. Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity 26: 17–27 [DOI] [PubMed] [Google Scholar]
- Alonso JL, Essafi M, Xiong JP, Stehle T, Arnaout MA 2002. Does the integrin αA domain act as a ligand for its βA domain? Curr Biol 12: R340–R342 [DOI] [PubMed] [Google Scholar]
- Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, et al. 2009. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J 28: 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout MA, Goodman SL, Xiong JP 2007. Structure and mechanics of integrin-based cell adhesion. Curr Opin Cell Biol 19: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP 2005. Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol 21: 381–410 [DOI] [PubMed] [Google Scholar]
- Askari JA, Buckley PA, Mould AP, Humphries MJ 2009. Linking integrin conformation to function. J Cell Sci 122: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari JA, Tynan CJ, Webb SE, Martin-Fernandez ML, Ballestrem C, Humphries MJ 2010. Focal adhesions are sites of integrin extension. J Cell Biol 188: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczyk M, Carracedo S, Gullberg D 2010. Integrins. Cell Tissue Res 339: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglova N, Blacklow SC, Takagi J, Springer TA 2002. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat Struct Biol 9: 282–287 [DOI] [PubMed] [Google Scholar]
- Bennett JS, Berger BW, Billings PC 2009. The structure and function of platelet integrins. J Thromb Haemost 7: 200–205 [DOI] [PubMed] [Google Scholar]
- Berger BW, Kulp DW, Span LM, DeGrado JL, Billings PC, Senes A, Bennett JS, DeGrado WF 2010. Consensus motif for integrin transmembrane helix association. Proc Natl Acad Sci 107: 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron A, Humphries JD, Askari JA, Craig SE, Mould AP, Humphries MJ 2009. Anti-integrin monoclonal antibodies. J Cell Sci 122 (Pt 22): 4009–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada MJ, Alvarez MV, Gonzalez-Rodriguez J 2002. Agonist-specific structural rearrangements of integrin αIIbβ3. Confirmation of the bent conformation in platelets at rest and after activation. J Biol Chem 277: 39899–39908 [DOI] [PubMed] [Google Scholar]
- Chen J, Salas A, Springer TA 2003. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat Struct Biol 10: 995–1001 [DOI] [PubMed] [Google Scholar]
- Chen LL, Whitty A, Lobb RR, Adams SP, Pepinsky RB 1999. Multiple activation states of integrin α4β1 detected through their different affinities for a small molecule ligand. J Biol Chem 274: 13167–13175 [DOI] [PubMed] [Google Scholar]
- Cheng M, Foo SY, Shi ML, Tang RH, Kong LS, Law SK, Tan SM 2007. Mutation of a conserved asparagine in the I-like domain promotes constitutively active integrins αLβ2 and αIIbβ3. J Biol Chem 282: 18225–18232 [DOI] [PubMed] [Google Scholar]
- Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA 2003. FRET detection of cellular α4-integrin conformational activation. Biophys J 85: 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A, Waller A, Zwartz GJ, Buranda T, Sklar LA 2007. Regulation of cell adhesion by affinity and conformational unbending of α4β1 integrin. J Immunol 178: 6828–6839 [DOI] [PubMed] [Google Scholar]
- Coller BS 1985. A new murine monoclonal antibody reports an activation-dependent change in the conformation and/or microenvironment of the platelet glycoprotein IIb/IIIa complex. J Clin Invest 76: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley DR 2009. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys 38: 235–254 [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC 2000. Structural basis of collagen recognition by integrin α2β1. Cell 101: 47–56 [DOI] [PubMed] [Google Scholar]
- Evans EA, Calderwood DA 2007. Forces and bond dynamics in cell adhesion. Science 316: 1148–1153 [DOI] [PubMed] [Google Scholar]
- Frelinger AL III, Cohen I, Plow EF, Smith MA, Roberts J, Lam SC, Ginsberg MH 1990. Selective inhibition of integrin function by antibodies specific for ligand-occupied receptor conformers. J Biol Chem 265: 6346–6352 [PubMed] [Google Scholar]
- Frelinger AL III, Du XP, Plow EF, Ginsberg MH 1991. Monoclonal antibodies to ligand-occupied conformers of integrin αIIbβ3 (glycoprotein IIb-IIIa) alter receptor affinity, specificity, and function. J Biol Chem 266: 17106–17111 [PubMed] [Google Scholar]
- Gottschalk KE 2005. A coiled-coil structure of the αIIbβ3 integrin transmembrane and cytoplasmic domains in its resting state. Structure 13: 703–712 [DOI] [PubMed] [Google Scholar]
- Gupta V, Gylling A, Alonso JL, Sugimori T, Ianakiev P, Xiong JP, Arnaout MA 2007. The β-tail domain (βTD) regulates physiologic ligand binding to integrin CD11b/CD18. Blood 109: 3513–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, de Leval X, David JL, Supuran C, Pirotte B, Dogne JM 2004. Progress in the field of GPIIb/IIIa antagonists. Curr Med Chem Cardiovasc Hematol Agents 2: 157–167 [DOI] [PubMed] [Google Scholar]
- Hato T, Yamanouchi J, Yakushijin Y, Sakai I, Yasukawa M 2006. Identification of critical residues for regulation of integrin activation in the β6-α7 loop of the integrin β3 I-like domain. J Thromb Haemost 4: 2278–2280 [DOI] [PubMed] [Google Scholar]
- Humphries MJ 2000. Integrin structure. Biochem Soc Trans 28: 311–339 [PubMed] [Google Scholar]
- Humphries MJ 2004. Monoclonal antibodies as probes of integrin priming and activation. Biochem Soc Trans 32: 407–411 [DOI] [PubMed] [Google Scholar]
- Humphries MJ, Symonds EJ, Mould AP 2003. Mapping functional residues onto integrin crystal structures. Curr Opin Struct Biol 13: 236–243 [DOI] [PubMed] [Google Scholar]
- Huth JR, Olejniczak ET, Mendoza R, Liang H, Harris EA, Lupher ML Jr, Wilson AE, Fesik SW, Staunton DE 2000. NMR and mutagenesis evidence for an I domain allosteric site that regulates lymphocyte function-associated antigen 1 ligand binding. Proc Natl Acad Sci 97: 5231–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO 2002. Integrins: Bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Hynes RO 2004. The emergence of integrins: A personal and historical perspective. Matrix Biol 23: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido H, Nakamura A, Kobayashi R, Ito S, Li S, Futaki S, Sekiguchi K 2007. The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin γ chains in integrin binding by laminins. J Biol Chem 282: 11144–11154 [DOI] [PubMed] [Google Scholar]
- Kallen J, Welzenbach K, Ramage P, Geyl D, Kriwacki R, Legge G, Cottens S, Weitz-Schmidt G, Hommel U 1999. Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain. J Mol Biol 292: 1–9 [DOI] [PubMed] [Google Scholar]
- Keizer GD, Visser W, Vliem M, Figdor CG 1988. A monoclonal antibody (NKI-L16) directed against a unique epitope on the α-chain of human leukocyte function-associated antigen 1 induces homotypic cell-cell interactions. J Immunol 140: 1393–1400 [PubMed] [Google Scholar]
- Kim M, Carman CV, Springer TA 2003. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301: 1720–1725 [DOI] [PubMed] [Google Scholar]
- Larson RS, Corbi AL, Berman L, Springer T 1989. Primary structure of the leukocyte function-associated molecule-1 α subunit: An integrin with an embedded domain defining a protein superfamily. J Cell Biol 108: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Dua V, Ulmer TS 2008a. Structure of the integrin αIIb transmembrane segment. J Biol Chem 283: 16162–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Kim C, Ginsberg MH, Ulmer TS 2009. The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J 28: 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Partridge AW, Ginsberg MH, Ulmer TS 2008b. Structure of the integrin β3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry 47: 4008–4016 [DOI] [PubMed] [Google Scholar]
- Lee JO, Bankston LA, Arnaout MA, Liddington RC 1995a. Two conformations of the integrin A-domain (I-domain): A pathway for activation? Structure 3: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Lee JO, Rieu P, Arnaout MA, Liddington R 1995b. Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18). Cell 80: 631–638 [DOI] [PubMed] [Google Scholar]
- Legate KR, Fassler R 2009. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J Cell Sci 122: 187–198 [DOI] [PubMed] [Google Scholar]
- Li R, Babu CR, Lear JD, Wand AJ, Bennett JS, DeGrado WF 2001. Oligomerization of the integrin αIIbβ3: Roles of the transmembrane and cytoplasmic domains. Proc Natl Acad Sci 98: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Mitra N, Gratkowski H, Vilaire G, Litvinov R, Nagasami C, Weisel JW, Lear JD, DeGrado WF, Bennett JS 2003. Activation of integrin αIIbβ3 by modulation of transmembrane helix associations. Science 300: 795–798 [DOI] [PubMed] [Google Scholar]
- Loftus JC, Plow EF, Frelinger AL III, D'Souza SE, Dixon D, Lacy J, Sorge J, Ginsberg MH 1987. Molecular cloning and chemical synthesis of a region of platelet glycoprotein IIb involved in adhesive function. Proc Natl Acad Sci 84: 7114–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA 2007. Structural basis of integrin regulation and signaling. Annu Rev Immunol 25: 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Springer TA, Takagi J 2003. Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. Proc Natl Acad Sci 100: 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Takagi J, Springer TA 2004. Locking the β3 integrin I-like domain into high and low affinity conformations with disulfides. J Biol Chem 279: 10215–10221 [DOI] [PubMed] [Google Scholar]
- McEver RP, Martin MN 1984. A monoclonal antibody to a membrane glycoprotein binds only to activated platelets. J Biol Chem 259: 9799–9804 [PubMed] [Google Scholar]
- Metcalf DG, Kulp DW, Bennett JS, DeGrado WF 2009. Multiple approaches converge on the structure of the integrin αIIb/β3 transmembrane heterodimer. J Mol Biol 392: 1087–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R 2008. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med 14: 325–330 [DOI] [PubMed] [Google Scholar]
- Mould AP, Askari JA, Aota S, Yamada KM, Irie A, Takada Y, Mardon HJ, Humphries MJ 1997. Defining the topology of integrin α5β1-fibronectin interactions using inhibitory anti-α5 and anti-β1 monoclonal antibodies. Evidence that the synergy sequence of fibronectin is recognized by the amino-terminal repeats of the α5 subunit. J Biol Chem 272: 17283–17292 [DOI] [PubMed] [Google Scholar]
- Mould AP, Barton SJ, Askari JA, Craig SE, Humphries MJ 2003a. Role of ADMIDAS cation-binding site in ligand recognition by integrin α5β1. J Biol Chem 278: 51622–51629 [DOI] [PubMed] [Google Scholar]
- Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ 2003b. Conformational changes in the integrin βA domain provide a mechanism for signal transduction via hybrid domain movement. J Biol Chem 278: 17028–17035 [DOI] [PubMed] [Google Scholar]
- Mould AP, Symond EJ, Buckley PA, Grossmann JG, McEwan PA, Barton SJ, Askari JA, Craig SE, Bella J, Humphries MJ 2003c. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J Biol Chem 278: 39993–39999 [DOI] [PubMed] [Google Scholar]
- Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA 2006. Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity 25: 583–594 [DOI] [PubMed] [Google Scholar]
- Nogales A, Garcia C, Perez J, Callow P, Ezquerra TA, Gonzalez-Rodriguez J 2010. Three-dimensional model of human platelet integrin αIIbβ3 in solution obtained by small angle neutron scattering. J Biol Chem 285: 1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxvig C, Springer TA 1998. Experimental support for a β-propeller domain in integrin α-subunits and a calcium binding site on its lower surface. Proc Natl Acad Sci 95: 4870–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise LV, Helgerson SL, Steiner B, Nannizzi L, Phillips DR 1987. Synthetic peptides derived from fibrinogen and fibronectin change the conformation of purified platelet glycoprotein IIb-IIIa. J Biol Chem 262: 12597–12602 [PubMed] [Google Scholar]
- Parthasarathy K, Lin X, Tan SM, Law SK, Torres J 2008. Transmembrane helices that form two opposite homodimeric interactions: An asparagine scan study of αM and β2 integrins. Protein Sci 17: 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH 2005. Transmembrane domain helix packing stabilizes integrin αIIbβ3 in the low affinity state. J Biol Chem 280: 7294–7300 [DOI] [PubMed] [Google Scholar]
- Plow EF, Haas TA, Zhang L, Loftus J, Smith JW 2000. Ligand binding to integrins. J Biol Chem 275: 21785–21788 [DOI] [PubMed] [Google Scholar]
- Rocco M, Rosano C, Weisel JW, Horita DA, Hantgan RR 2008. Integrin conformational regulation: Uncoupling extension/tail separation from changes in the head region by a multiresolution approach. Structure 16: 954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Sundramurthy K, Liu B, Tan SM, Law SK, Lescar J 2005. The crystal structure of the plexin-semaphorin-integrin domain/hybrid domain/I-EGF1 segment from the human integrin β2 subunit at 1.8 {angstrom} resolution. J Biol Chem 280: 30586–30593 [DOI] [PubMed] [Google Scholar]
- Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, McCormack A, Zhang R, Joachimiak A, Takagi J, et al. 2003. Structures of the αL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell 112: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG 2000. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci 97: 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims PJ, Ginsberg MH, Plow EF, Shattil SJ 1991. Effect of platelet activation on the conformation of the plasma membrane glycoprotein IIb-IIIa complex. J Biol Chem 266: 7345–7352 [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110: 599–511 [DOI] [PubMed] [Google Scholar]
- Takagi J, Strokovich K, Springer TA, Walz T 2003. Structure of integrin α5β1 in complex with fibronectin. EMBO J 22: 4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, DeSimone DW, Fonda D, Patel RS, Buck C, Horwitz AF, Hynes RO 1986. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 46: 271–282 [DOI] [PubMed] [Google Scholar]
- Ulmer TS, Yaspan B, Ginsberg MH, Campbell ID 2001. NMR analysis of structure and dynamics of the cytosolic tails of integrin αIIbβ3 in aqueous solution. Biochemistry 40: 7498–7508 [DOI] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A 2001. Function and interactions of integrins. Cell Tissue Res 305: 285–298 [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas T, Plow E, Qin J 2002. A structural mechanism of integrin αIIbβ3 ‘inside-out’ activation as regulated by its cytoplasmic face. Cell 110: 587–597 [DOI] [PubMed] [Google Scholar]
- Wang W, Luo BH 2010. Structural basis of integrin transmembrane activation. J Cell Biochem 109: 447–452 [DOI] [PubMed] [Google Scholar]
- Wegener KL, Campbell ID 2008. Transmembrane and cytoplasmic domains in integrin activation and protein–protein interactions. Mol Membr Biol 25: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weljie AM, Hwang PM, Vogel HJ 2002. Solution structures of the cytoplasmic tail complex from platelet integrin αIIb- and β3-subunits. Proc Natl Acad Sci 99: 5878–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Coller BS, Wang JH, Springer TA 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhu J, Chen X, Mi L, Nishida N, Springer TA 2009. Structure of an integrin with an αI domain, complement receptor type 4. EMBO J 29: 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA 2001. Crystal structure of the extracellular segment of integrin αVβ3. Science 294: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Goodman SL, Arnaout MA 2004. A novel adaptation of the integrin PSI domain revealed from its crystal structure. J Biol Chem 279: 40252–40254 [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA 2002. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science 296: 151–155 [DOI] [PubMed] [Google Scholar]
- Yang J, Ma YQ, Page RC, Misra S, Plow EF, Qin J 2009. Structure of an integrin αIIbβ3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc Natl Acad Sci 106: 17729–17734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH 2010. Recreation of the terminal events in physiological integrin activation. J Cell Biol 188: 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Liu J, Winkler H, Taylor KA 2008. Integrin αIIbβ3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J Mol Biol 378: 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA 2009. The structure of a receptor with two associating transmembrane domains on the cell surface: Integrin αIIbβ3. Mol Cell 34: 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell 32: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]