Abstract
Immunogenicity arises via many synergistic mechanisms, yet the overall dissimilarity of pathogenic proteins versus the host proteome has been proposed as a key arbiter. We have previously explored this concept in relation to Bacterial antigens; here we extend our analysis to antigens of viral and fungal origin. Sets of known viral and fungal antigenic and non-antigenic protein sequences were compared to human and mouse proteomes. Both antigenic and non-antigenic sequences lacked human or mouse homologues. Observed distributions were compared using the non-parametric Mann-Whitney test. The statistical null hypothesis was accepted, indicating that antigen and non-antigens did not differ significantly. Likewise, we could not determine a threshold able meaningfully to separate non-antigen from antigen. We conclude that viral and fungal antigens cannot be predicted from pathogen genomes based solely on their dissimilarity to mammalian genomes.
Background
Immunogenicity manifests itself through both humoral (mediated through the binding of whole protein antigens by antibodies) and cellular immunology (mediated by the recognition of proteolytically cleaved peptides by T-cells). Mechanistically, protein immunogenicity arises from many synergistic factors. These include host-side properties and pathogenside properties. Properties mediated by the host include the density of Tcell or B-cell epitopes and how easily it is degraded; while pathogen mediated properties include protein expression levels and sub-cellular location. Thus a viable candidate vaccine should be highly expressed, available for immune surveillance, and possess epitopes that the host recognises. Other properties, mostly intrinsic to the protein, include molecular size, aggregation status, and possession of so-called dangersignals, typically chemical modifications, post-translational in origin, which are recognised by pattern recognition receptors of the innate immune system. An alternative hypothesis replaces such complexity with the simple, straightforward idea of phylogenetic distance as the chief determinant of immunogenicity. This notion finds its most eloquent advocate in the work of Kundac et al. [1]. In previous work [2], we have examined a restated variant of this concept: that non-redundancy at the sequence level is crucial to the accurate prediction of antigen-based candidate vaccine antigens. When examining bacterial antigens, both antigens and non-antigens lacked equally similarity to host proteomes. Moreover, both were as dissimilar as the background distribution offered by a complete bacterial genome. We were unable to find an appropriate and useable threshold capable of differentiate antigens from non-antigens on the basis of similarity to the host's proteome. In the present work, we extend our analysis of “immunogenicity as phylogenetic distance” to embrace the full scope of life, pushing to the limits of Woese's three domains of life and beyond. As there is little or no evidence for archaea acting as pathogens, though some are implicated as commensals with pathogen-mediated disease, we look at two other kinds of widely phyletically distributed pathogenic organism, namely viruses and fungi.Viruses are structurally-simple, quasi-organisms that straddle the boundary between complex chemical entities and true living organisms. Most would agree that bacteria are alive yet a piece of DNA is not. Viruses give rise to diverse infections: for example, over 300 distinct viruses cause upper respiratory infections bracketed as the common cold, while others are responsible for diseases as different as chickenpox, influenza, HCV, and AIDS. Fungi are complex, eukaryotic organisms distinct from plants and animals. Some fungal species are very pathogenic, and can infect all exposed individuals, i.e. Coccidioides immitis and Histoplasma capsulatum. Others are opportunities pathogens, such as Candida Albicans, which give rise to disease only in immunocompromised hosts. Importantly both have very different mechanisms of pathogenesis when compared to each other and also to bacteria. Thus both are distinct, and distinct also from prokaryotic bacteria. Compared to viral infections, fungal diseases are less harmful but more tenacious. Fungi such as Candida typically colonise external and mucosal surfaces, such as the intestines. Overtly harmless, such fungi can induce local infections of the skin, nails, mouth, vagina, or sinuses: they are seldom serious, except in the immune deficient. Occasionally, infections occur when habitual suppression of fungi is disturbed. For example, bacteria found in the vagina or alimentary canal usually limits fungal growth. Antibiotics can kill such helpful bacteria, allowing fungi to grow unchecked, and thus cause symptoms, which are typically mild. Certain fungal infections, such as blastomycosis, may be more serious.By using the sequence similarity search tool BLAST [3] we expand our sequence similarity discrimination of non-antigen from antigen, again also seeking a value that can separate these classes. More specifically, we analysed the similarity between sets of known fungal and viral antigens and the Human Genome and Mouse Genome, compared with a control group of non-antigens, and likewise made an assessment versus the similarity evinced by a background distributions, namely fungal genomes and large sets of viral sequences. Our primary outcome establishes a systematic benchmark for the success of “ immunogenicity as phylogenetic distance” as a concept useful in antigen hunting.
Methodology
Datasets of known antigens obtained previously from the literature were analysed [2, 4, 5, 6]. Non-antigens were selected from SWISSPROT so that they mirrored the antigen sets, being selected at random from the same set of species[5, 6]. Fungal and Viral antigens used here are listed below in Figure 1. Additionally, genomes corresponding to Human, Mouse, and Fungus were downloaded from FTP sites at National Center for Biotechnology Information (NCBI) [ http://www.ncbi.nlm.nih.gov/] European Bioinformatics Institute (EBI) [ http://www.ebi.ac.uk/] and Ensembl [ http://www.ensembl.org/ ] Since viral genomes rarely exceed a few hundred sequences, and are thus of a comparable size to our datasets, we obtained a larger compilation of viral sequences from the Viral Bioinformatics Resource Center (http://www.biovirus.org/). All the peptide sequences obtained were in FASTA format.
Figure 1.

Protein sequences compiled and annotated in-house as fungal and viral antigens
As before [2], antigen datasets, non-antigen datasets and various pathogenic genome sequences to the Human and Mouse Genome were analysed using a local, standalone version of BLAST [3] which afforded full management of E-value cut-offs. E-value thresholds were raised from 10 to 6000 to identify best matches even when these lacked statistical significance. We also analysed (log10E-value)+1 values obtained from BLAST. By using the statistical package Minitab, Release 14.1, we compared antigen and non-antigen sets, as random samples of two larger, independent populations, utilising a Mann‐Whitney test.
Discussion
Following a protocol established earlier for analysing the similarity of bacterial and eukaryotic proteomes [2] sets of antigens and non-antigens from both fungi and viruses were compared to the Human and Mouse proteomes, producing lists of highest scoring similarities. Proteomes and pseudo-proteomes of fungal and viral origin were also compared to the human and mouse proteomes. Antigen sets derived from the literature contained 100 viral antigens and 33 fungal antigens respectively; similarly sized sets of non-antigens for the two types of pathogen were also obtained, selected at random from the same sets of species.
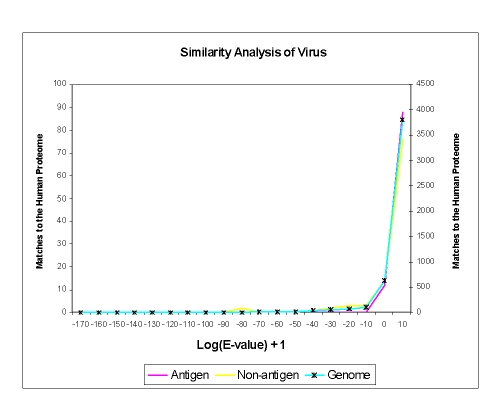
Results obtained for viruses were even more extreme than those observed previously for bacteria [2]. For the case of viruses, apart from rare anomalies such as mimivirus, few available genomes contain more than a few hundred sequences, which is not sufficient to give a reference background to the analysis. Instead, we used a composite viral pseudogenome comprising sequences from many viral species. Score histograms for antigen and non-antigen sets, and the viral pseudo-proteome, versus the human proteome are shown in Figure 2. Overall, viral antigens show no overt similarity to the human or mouse genomes. All the protein sequences were found to be in the range of 0 to 10 when compared the human proteome, while comparison to the mouse proteome yielded sequences with values between -10 to 10, indicating high dissimilarity of viral antigens compared to host proteomes. Values for non-antigenic protein sequences were found to lie between -40 to 10 when compared to human and mouse proteomes. Only 2 non-antigenic protein sequences were less than -40. Again, any signal here is too weak to stand as a useful threshold.
Figure 2.
Viral Sequence Comparison. A sequence similarity comparison with the E-value as 6000 and BLOSUM 62 matrix, between the Antigen, Non-antigen and viral genome sequences. Two separate scales were used as the number of matches to the Human Genome varied from the antigen and non-antigen datasets to the genome. The blue line with the star marker corresponding to the genome is plotted on the scale to the right hand axis (Y axis).
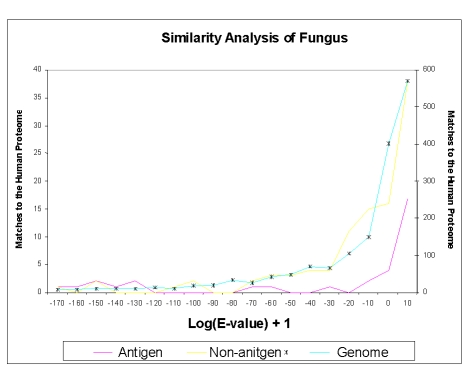
Results obtained for fungal antigens and non-antigens were very similar to those obtained for Bacteria and viruses, despite the smaller sample size. This reduced prevalence reflects the small number of known fungal antigens available at the time of the study. A similar analysis for four fungal proteomes (Aspergillus fumigatus, Candida albicans, Encephalitozoon cuniculi and Saccharomyces cerevisiae) versus Human and Mouse proteomes was also undertaken. All sets analysed were similarly distant to both mouse and human genomes. Score histograms for antigen and non-antigen sets, and the Aspergillus fumigatus proteome versus the human proteome are shown in Figure 3. It is clear that the three distributions strongly overlap, and that no clear cut-off point presents itself. However, we did observe five fungal antigens with high similarity to human and mouse proteomes. These proteins were characteristically highly-conserved over evolution, such as enzymes or heat shock proteins. Looked at overall, the observed ranges of score (–70 and 10 for antigens versus -80 to 10 for non-antigens) could not be easily differentiated thus stymieing attempts to use this as a tool for predicting whole-protein antigens.
Figure 3.
Fungal Sequence Comparison. A sequence similarity comparison with the E-value as 6000 and BLOSUM 62 matrix, between the Antigen, Non-antigen and Aspergillus fumigatus genome sequences. Two separate scales were used as the number of matches to the Human Genome varied from the antigen and non-antigen datasets to the genome. The blue line with the star marker represents the genome is plotted on the scale to the right hand axis (Y axis).
We also used statistical significance testing to quantify the commonality between observed score distributions. Since our distributions were not normally distributed, we used the straightforward, non-parametric Mann‐ Whitney Test for this assessment, since it is more robust for skewed distributions such as these, rather than using, say, the 2 sample Kolmogorov-Smirnov test to compare distributions. The Mann-Whitney test was undertaken at the 95% confidence interval. The p-value for comparison of the viral distributions was 0.322. The corresponding p-value for fungal sequences was 0.381. As the p-value was greater than 0.05 in both cases, the null hypothesis was accepted for both viruses and fungi. Note that the p-value for the bacterial comparison [2]was 0.239. Thus there was no statistically significant difference in any of the three cases.
We cannot use these values to determine a reliable threshold able to distinguish antigen from non-antigen when compared to against the background distribution for whole genomes: what difference there is statistically insignificant. BLAST is seen by many as very much a blunt instrument, particularly for the fine-detail analysis of large and/or complex protein families. Even if a wholly correct and validated match can be discovered, BLAST can struggle to distinguish orthologues from paralogues. Future work could thus include more sophisticated approaches to quantifying sequence similarity beyond that offered by BLAST.
Conclusion
Despite the negative outcomes observed in this study, which though disappointing are not altogether unexpected, given prior results [2] there is still an urgent and pressing need for the development of robust techniques for the identification of whole protein antigens. The problem is not that bacterial or viral or fungal proteins are not clearly different from the human genome: indeed they are. No, for both fungi and viruses, the problem is that antigens and non-antigens are different to the same degree; as with bacteria [2] we cannot calibrate a useful threshold able to separate them.
Lack of transparent variation in measured similarity and the inability to locate thresholds that split positives from negatives is by no means unique to the present problem. An illuminating example is chemical similarity searching [7]. Bit-string representations of molecular structure have long been applied to small molecule similarity searching. Despite the simplicity of the approach, such bit-string patterns have been unexpectedly successful. Yet, we currently have no generally applicable similaritymetrics and structural similarity thresholds able to indicate reliably equivalent biological activity.
It is an old maxim in management consultancy that 20% of the total effort generates 80% of the total result; how true this assertion is, in quantitative terms, we leave to your discretion, yet the sentiment it embodies carries with it much sagacity. Our current results show the inadequacy of this approach, despite the gallimaufry of countervailing caveats of which one could conceive to counter this conjecture. The clear lack of a signal apparent here is not likely to be modulated much by fiddling round with parameters or even using different but similar approaches.
Footnotes
Citation:Ramakrishnan et al Bioinformation 5(1):35-38 (2010)
References
- 01.Kanduc D, et al. Autoimmun Rev. 2007;6:290. doi: 10.1016/j.autrev.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 02.Ramachrisnan K, Flower D. Bioinformation. 2010;4 [Google Scholar]
- 03.Altschul S. Nucleic Acids Research. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 04.Doytchinova I, Flower D. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 05.Doytchinova I, Flower D. Vaccine. 2007;25:856. doi: 10.1016/j.vaccine.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 06.Doytchinova I, Flower D. The Open Vaccine journal. 2008;1:22. [Google Scholar]
- 07.Flower D. Journal or Chemical Information and Computer Science. 1998;38:379. [Google Scholar]