Abstract
The neuronal Na+-dependent glutamate transporter, excitatory amino acid carrier 1 (EAAC1, also called EAAT3), has been implicated in the control of synaptic spillover of glutamate, synaptic plasticity, and the import of cysteine for neuronal synthesis of glutathione. EAAC1 protein is observed in both perisynaptic regions of the synapse and in neuronal cell bodies. Although amino acid residues in the carboxyl terminal tail have been implicated in the dendritic targeting of EAAC1 protein, it is not known if mRNA for EAAC1 may also be targeted to dendrites. Sorting of mRNA to specific cellular domains provides a mechanism by which signals can rapidly increase translation in a local environment; this form of regulated translation has been linked to diverse biological phenomena ranging from establishment of polarity during embryogenesis to synapse development and synaptic plasticity. In the present study, EAAC1 mRNA sequences were amplified from dendritic samples that were mechanically harvested from low-density hippocampal neuronal cultures. In parallel analyses, mRNA for histone deacetylase 2 (HDAC-2) and glial fibrillary acidic protein (GFAP) was not detected, suggesting that these samples are not contaminated with cell body or glial mRNAs. EAAC1 mRNA also co-localized with Map2a (a marker of dendrites) but not Tau1 (a marker of axons) in hippocampal neuronal cultures by in situ hybridization. In control rats, EAAC1 mRNA was observed in soma and proximal dendrites of hippocampal pyramidal neurons. Following pilocarpine- or kainate-induced seizures, EAAC1 mRNA was present in CA1 pyramidal cell dendrites up to 200 μm from the soma. These studies provide the first evidence that EAAC1 mRNA localizes to dendrites and suggest that dendritic targeting of EAAC1 mRNA is increased by seizure activity and may be regulated by neuronal activity/depolarization.
Keywords: glutamate transport, EAAC1, EAAT3, epilepsy, pilocarpine, seizure, mRNA targeting
1. Introduction
Extracellular concentrations of the excitatory amino acids (EAAs) are controlled by a family of Na+-dependent glutamate transporters that include GLAST, GLT-1, EAAC1 (also called EAAT1, EAAT2, and EAAT3 respectively), EAAT4, and EAAT5 (for review, see Danbolt, 2001; Sheldon and Robinson, 2007). EAAC1 protein is found in inhibitory inter-neurons, excitatory neurons, and oligodendroglia (Conti et al., 1998; Rothstein et al., 1994). EAAC1 is enriched in pyramidal neurons in the forebrain where it is localized to both cell bodies and peri-synaptic regions of post-synaptic elements (He et al., 2000; He et al., 2001). In inhibitory cells, EAAC1 is thought to provide glutamate as a precursor for the synthesis of GABA (Liang et al., 2006; Mathews and Diamond, 2003; Sepkuty et al., 2002). There is also evidence that EAAC1 limits the spillover of excitatory neurotransmitter between synapses (Diamond, 2001; Scimemi et al., 2009) and contributes to neuronal uptake of cysteine, a critical precursor for glutathione synthesis (see Aoyama et al., 2006 for discussion). Together these studies suggest that EAAC1 has a plurality of functions in the CNS (for review, see Nieoullon et al., 2006).
Excitatory synaptic transmission is extremely plastic with increases or decreases that can be either short- or long-lasting. In many cases, this synaptic plasticity is thought to be synapse-specific (i.e. not all inputs to a given cell are modified). There is strong evidence that receptor redistribution on or off the plasma membrane contributes to this plasticity (for review, see Newpher and Ehlers, 2008). As observed for several ligand-gated glutamate receptors (for reviews, see Elias and Nicoll, 2007; Greger and Esteban, 2007), the levels of EAAC1 protein at the cell surface can be rapidly (within min) regulated by diverse stimuli, including protein kinase C, growth factor receptor activation, and NMDA receptor activation (for review, see Robinson, 2006). In fact, the plasma membrane expression of EAAC1 changes after induction of long term potentiation (LTP) or after contextual fear conditioning (Levenson et al., 2001; Pita-Almenar et al., 2006). In addition to receptor redistribution, there is evidence that regulation of translation of mRNAs in dendrites also contributes to synaptic plasticity (for reviews, see Kelleher et al., 2004; Wang et al., 2010).
Although many groups have examined the distribution of EAAC1 mRNA in tissue sections by in situ hybridization (Berger and Hediger, 1998; Kugler and Schmitt, 1999; Shibata et al., 1996; Simantov et al., 1999; Velaz-Faircloth et al., 1996), none have reported localization of EAAC1 mRNA in dendrites. In the present study, we provide evidence that EAAC1 mRNA is selectively targeted to dendrites in hippocampal neurons in vitro. EAAC1 mRNA is restricted to the cell body and proximal dendrites of hippocampal pyramidal neurons in vivo under control conditions. Finally, the amount of EAAC1 mRNA detected in dendrites increases dramatically in CA1 pyramidal neurons after a seizure induced by either pilocarpine or kainate.
2. Materials and Methods
2.1. Materials
Dulbecco's modified Eagle Medium (DMEM), trypsin-EDTA, Neurobasal A medium, B27 supplement, L-glutamine, normal goat serum, and penicillin-streptomycin were purchased from Life Technologies (Grand Island, NY). Culture plates were purchased from Corning (Cambridge, MA). Pilocarpine hydrochloride, kainic acid monohydrate, and scopolamine methyl nitrate were bought from Sigma-Aldrich (St. Louis, MO). Monoclonal anti-microtubule associated protein-2a and b (Map2a), and monoclonal anti-tau-1 protein (Tau-1) antibodies were purchased from Millipore (Billerica, MA).
2.2. Primary neuronal cultures
Primary neuron-enriched cultures were prepared from embryonic Sprague-Dawley rat hippocampi at 18 or 19 days of gestation (see Miyashiro et al., 1994 for original references). Briefly, pregnant rats were sacrificed by CO2 overdose and embryos were removed by C-section. After removal of the meninges, hippocampi were isolated and incubated in trypsin for 20 min at 37°C. Dissociated cells were obtained by gentle mechanical trituration using a flame polished Pasteur pipette. Cells (100,000 cells/ml) were plated on poly-D-lysine coated coverslips (0.5 mg/ml). Cells were maintained in defined Neurobasal A Medium (Invitrogen, Eugene, OR) with Glutamax (Invitrogen) and 2% B-27 supplement (Invitrogen) in 5% CO2 at 37°C. After 14-16 days, these cultures are very low density, providing easy access to isolate dendrites with a micro-pipette. Greater than 90% of cells are neurons, based on the use of cell specific markers, and less than 5% are glial fibrillary acidic protein (GFAP)-positive.
2.3. Isolation and amplification of dendritic RNA
RNA was harvested from transected dendrites of cultured hippocampal neurons. These samples were pooled and linearly amplified as previously described (Eberwine, 1996). Briefly, anti-RNA was amplified using oligo-dT-primers designed with an extended T7 RNA promoter at the 5′ end, followed by incubation with T7 RNA polymerase. RNA products underwent reverse transcription using oligo-dT primers to derive the first strand cDNA products, and then tested for the presence of specific gene products by PCR using coding primers for each gene. The primer specificity was then verified with MacVector software (Cary, NC).
2.4. Fluorescence in situ hybridization
Primary hippocampal neurons were washed twice in phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde (PFA) for 10 min. Cells were rinsed three times in Tris-buffered saline and blocked in 5% normal goat serum and 0.1% Triton X-100 for 30 min. To generate digoxigenin-labeled antisense and sense riboprobes, cDNA fragments of EAAC1 (GenBank accession number NM_013032, coding nucleotides 1092-1569), were synthesized by PCR and cloned into the pBluescript SK vector (Clontech, Mountain View, CA). Linearized plasmids served as templates for in vitro transcription using the digoxigenin (DIG) RNA labeling Kit (Sp6/T7/T3) from Roche (Welwyn Garden City, UK) and hybridized to fixed cultured neurons as described previously (Bassell et al., 1998). Sheep anti-digoxigenin antibody (Invitrogen, 1:100) in blocking solution was added to the cells and incubated overnight at 4°C. The cells were rinsed with blocking solution three times. Fluorescein isothiocyanate (FITC)-labeled donkey anti-sheep IgG (1:1500; Jackson ImmunoResearch Labs, West Grove, PA) was warmed to 37°C for 30 min and centrifuged at 100 × g for 5 min to remove any precipitate; then the cells were incubated with secondary antibody for 2 hr. A FITC labeled mouse anti-FITC antibody (1:2000; Jackson ImmunoResearch Labs) was included to amplify the signal. For the immunolabeling of Map2a or Tau-1, the incubation with primary monoclonal anti-Map2a (1:100) or Tau-1 (1:100) was followed by rhodamine-X labeled rabbit anti-mouse antibody (1:2000; Jackson ImmunoResearch Labs). After three rinses with PBS, cells were post-fixed with cold methanol for 8 min, and dried. Coverslips were mounted on glass slides with Vectashield containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) and sealed with clear nail polish.
2.5. Chemoconvulsant-Induced Seizure models
Adult male Sprague-Dawley rats (8 to 10 weeks) from Charles River (Malvern, PA) were used for these studies. Animals were maintained for a minimum of two days for acclimatization in a temperature- and light-controlled environment in accordance with the principles and procedures of the National Institutes of Health Guidelines for Care and Use of Laboratory Animals. The experimental design was approved by the Institutional Animal Care and Use Committee of the Children's Hospital of Philadelphia. Rats in the pilocarpine model, (sham or seizure) were pretreated with an intraperitoneal (i.p.) injection of scopolamine methyl nitrate (1 mg/Kg in sterile saline) to suppress peripheral cholinergic effects. After 30 mins, rats were injected with pilocarpine hydrochloride to induce seizures (385 mg/Kg, i.p.) or a 1/10 dose of pilocarpine (sham). In the kainate model, rats were either injected with kainic acid (12 mg/Kg, i.p. in PBS) or vehicle (control). Animals were continuously monitored after injection and seizures were classified using a previously published behavioral scale (Racine, 1972). Within the first hour after injection, approximately 80% of animals developed seizures evolving into recurrent generalized convulsive seizures stage III-IV, defined as status epilepticus (SE). Approximately 20% of the treated animals either did not seize or died within the first 3 hr and were not included in the study. Animals were euthanized at different times after initiation of SE (3 or 6 hrs). Rats were perfused transcardially with 0.1M PBS (pH 7.4) followed by 4% PFA in PBS under deep anesthesia with ketamine/xylazine. Brains were extracted from the skull, post-fixed with 4% PFA overnight, and then transferred to 30% sucrose/0.1M PBS at 4° C for dehydration. Tissue was frozen and sectioned (10 ! m) using a Leica cryostat.
2.6. In situ hybridization (ISH)
Tissue sections were fixed for 30 min with 4% PFA in PBS and washed extensively with PBS. Postfixed brain sections were washed in 1× PBS, and rehydrated sequentially in 100, 95, 70, 50% ethanol and 2× SSC (20× SSC containing 3.0M sodium chloride, 0.3M sodium citrate buffer at pH 7.0). They were treated for 10 min at room temperature with 50! g/ml proteinase K (Roche) in proteinase K buffer containing 5mM ethylenediaminetetraacetic acid (EDTA), 50mM Tris-HCl, (pH 7.2) and reacted with acetic anhydride in 0.1M RNase-free triethanolamine, pH 8.0. Then the brain sections were dehydrated using sequential washes of 2× SSC containing 0, 50, 70, and 95% ethanol followed by 100% ethanol. Section were first incubated for 30 min at 60° C in hybridization buffer composed of 50% formamide, 5× SSC, 0.3mg/ml yeast tRNA, 100! g/ml heparin, 2× Denhardt's solution (0.02% bovine serum albumin, 0.02% polyvinylpyrrolidone, and 0.02% Ficoll 400), 0.1% Triton X-100, and 5mM EDTA. After size-reduction of the riboprobes to ∼0.1 kb with alkaline hydrolysis, sections were incubated with 0.5-1 ! g/ml probes in hybridization solution for 12-16 h at 60°C. After hybridization, the sections were sequentially washed in 2× SSC, 1× SSC, and 0.2× SSC for 10, 10, and 60 min at 37, 60, and 60°C. To remove unmatched cRNA hybrids, the sections were incubated with RNase A (0.1! g/ml) in 1× SSC 30 min at 37°C followed by a wash with 0.2× SSC. After blocking with 20% heat-inactivated sheep serum in PBS with Triton X-100 (PBS, 2mg/ml BSA, and 0.1% Triton X-100) (PBT) for 3 h at room temperature, sections were incubated overnight at 4° C with preabsorbed anti-digoxigenin antibody coupled to alkaline phosphatase (AP) (Roche, Indianapolis, IN, USA) diluted 1:3000 in blocking solution. Sections were washed in PBT and AP buffer (100mM Tris, pH 9.5, 50mM MgCl2, 100mM NaCl, and 0.1% Triton X-100) and then visualized after reacting with nitroblue-tetrazolium-chloride (1μg/ml) and 5-bromo-4-chlor-indolyl-phosphate (3.5! l/1ml) in the dark for 8-10 hr.
2.7. Immunofluorescent and brightfield microscopy
Slides were examined by confocal microscopy with a Leica Inverted DM IRE2 HC fluo TCS 1-B-UV microscope coupled to a Leica TCS SP2 spectral confocal system/UV (Exton, PA). Immunostained cultures were sectioned optically at 0.5 ! m intervals with a 64× oil objective; these sections were assembled as Z-stacks. Sections through the center of the cell were identified by examining the optical cross-section of the DAPI nuclear staining. Some slides were examined with a Leica DMR epifluorescent microscope at 40× and 64× magnifications, using an Orca digital camera (Hamamatsu Inc, Japan), and Openlab software for analysis. In addition to using excess unlabelled probe or a sense probe, appropriate omissions of primary and/or secondary antibody were used in every experiment to confirm that the signals were dependent upon primary antibody and that the secondary antibody was specific for the appropriate primary antibody.
2.8 Preparation of Synaptoneurosomes
Synaptoneurosomes were prepared as previously described (Hollingsworth et al., 1985). All steps were conducted on ice or 4°C and all solutions were made under RNAase free conditions using diethylpyrocarbonate-treated or nuclease-free water. Briefly, the dissected hippocampus was gently homogenized at 4°C in 10 volumes of isolation media (0.32M sucrose, 10 mM Tris-HCl, 1 mM EDTA). After a low speed centrifugation step to remove cell bodies (P1), the resulting supernatant was centrifuged at 12,500 RPM for 20 min using a Beckman JA-17 rotor. The resulting pellet (P2) was gently suspended in a small volume of isolation media and then brought to 12% Ficoll. After layering 7% Ficoll over this solution, followed by a layer of isolation media, the samples underwent ultracentrifugation at 27,000 RPM for 30 min using a Beckman SW-41ti. Synaptoneurosomes were isolated at the 7%/12% interface and washed four times in isolation media. The final pellets containing synaptoneurosome preparations were suspended (1 mL) in ice-cold aerated buffer (containing (mM) 118 NaCl, 4.7 KCl, 1.2 MgSO4, 2.5 CaCl2, 1.53 KH2PO4, 212.7 glucose, and 33 Tris-HCl, pH 7.4), supplemented with 30 U/ml human placental RNase inhibitor (Amersham Biosciences, Uppsala, Sweden).
2.9 Western Blot Analysis
Equal amounts of protein from each sample were resolved on 8% SDS-polyacrylamide gels. Proteins were transferred to polyvinylidene diflouride (PVDF) membranes and blocked with Tris buffered saline containing (0.1%) Tween 20, and 1% nonfat milk. The blots were probed with specific antibodies against histone H-3 antibody (Cell Signaling Technology) followed by horseradish peroxidase-conjugated secondary antibodies (donkey anti-rabbit IgG, Amersham Bioscience). Immunoreactive bands were visualized using enhanced chemiluminescence according to the manufacturer's instructions (GE Healthcare UK limited, Little Chalfont, UK) and captured on autoradiography Amersham Bioscience films. Immunoreactivity was quantified using NIH Image J (National Institutes of Health, Bethesda, MD) software (http://rsb.info.nih.gov/ij/).
3.0 Quantitative measurement of mRNAs by real-time PCR
The absolute levels of selected mRNAs were determined in subcellular fractions according to manufacturer's instructions. RNA was isolated from the subcellular fractions using a MicroRNA isolation kit (Ambion Austin, TX), and cDNA was produced from equal amounts of RNA using random hexamers. Equivalent amounts of RNA and/or cDNA were quantified using a NanoDrop ND-1000 spectrophotmeter (Wilmington, DE). Real-time PCR was performed with primers designed by Applied Biosystems Taqman gene expression assays. The crossing threshold (Ct) values for each dilution were plotted against the assigned copy numbers. For the experimental sample, the relative copy number of the mRNA was calculated using its Ct value and standard curve obtained for that mRNA using the same set of primers. The list of Applied Biosystems Taqman gene expression assays tested are GluR2 (Gria2, Rn00568514_m1), HDAC3 (Rn00584926_m1), EAAC1 (SLC1A1, Rn00564704_m1), CamKII! (Rn00563883_m1), and Cyclophilin A (PPIA, Rn00690933_m1). Data are presented as the mean ± standard error of the mean and were compared by ANOVA using a Bonferroni post hoc correction for multiple comparisons.
3. Results
3.1. Amplification of EAAC1 mRNA from hippocampal dendritic samples by PCR
Two approaches were used to determine if EAAC1 mRNA might be localized to neuronal dendrites in low-density hippocampal neuron-enriched cultures maintained in vitro. First, dendrites were mechanically harvested and RNA was linearly amplified (see methods). Products for EAAC1 were amplified from these specimens using two different pairs of oligonucleotide primers (Fig. 1A & B). These products migrated to the same size as that observed in a control specimen of RNA obtained from C6 glioma, a cell line that has been previously shown to endogenously express EAAC1 (Dowd et al., 1996; Palos et al., 1996). To provide confirmation of PCR products identity, all bands were sequenced and the resulting sequences corresponded to those published. As predicted (Kacharmina et al., 2000; Miyashiro et al., 1994), GluR2 mRNA was present in these samples (Fig. 1A). To reduce the likelihood that the presence of EAAC1 mRNA was due to contamination with cell body mRNA, we tested for the presence of the cell body specific mRNA, histone deacetylase-2 (HDAC2) (Zeng et al., 1998). Although the primers employed amplified a product of the predicted size from a control mRNA specimen (from SHSY5Y cells), no product was observed in the RNA samples generated from hippocampal dendritic aRNA samples (Fig. 1A). These cultures contain very few (<5%) glial fibrillary acidic protein (GFAP)-positive cells, but it is still possible that astrocytic RNAs could contaminate these aRNA samples. To rule out this possibility, GFAP-specific primers were used to test for the presence of GFAP in these samples. Although a product for GFAP was amplified from mRNA harvested cultures of astrocytes, no signal was detected in dendritic aRNA samples.
Figure 1. PCR Amplification of mRNAs in isolated dendrites from primary hippocampal neuronal cultures.
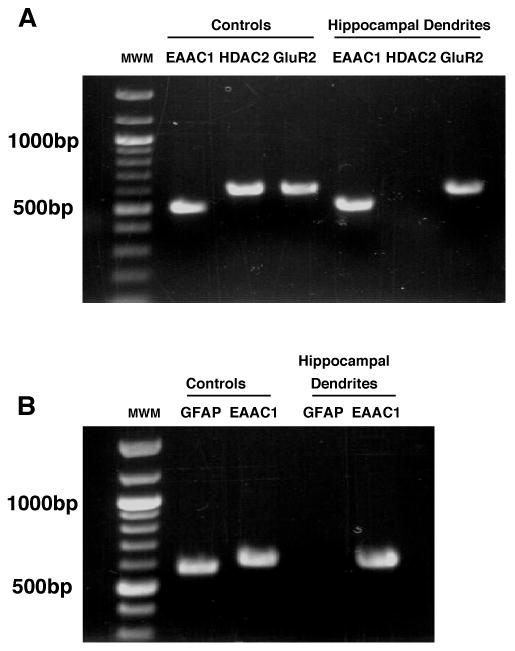
Dendritic samples were isolated from cultured hippocampal neurons and linearly amplified. A. RT-PCR (reverse transcriptase-PCR) was used to amplify EAAC1, HDAC2, and GluR2. Histone deacetylase 2 (HDAC2) mRNA (a soma-localized mRNA) was used as negative control and was not found in these samples. Using the same protocols, mRNA prepared from the C6 glioma and SH-SY5Y cell lines were used as controls to verify the EAAC1, GluR2, and HDAC2 primers. Note: the primers used to amplify HDAC2 were designed to work with either rodent or human sequences. B. Using a different set of primer oligos, EAAC1 was detected in the hippocampal dendrites and glial fibrillary acidic protein (GFAP) was not found. GFAP (a glial mRNA) was obtained from astrocyte cultures and used as negative control to determine if the samples were contaminated with glial elements. All bands were extracted and sequenced to confirm identity. This experiment was repeated in an independent sample.
3.2. Analysis of EAAC1 mRNA distribution in low-density hippocampal neuronal cultures using in situ hybridization
To further test for the presence of EAAC1 mRNA in dendrites, a digoxigenin labeled anti-sense oligonucleotides complementary to the coding region of EAAC1 combined with anti-digoxigenin antibodies were used to test for the presence of EAAC1 mRNA in low-density neuron-enriched cultures. In these cultures, signal was observed in cell bodies and dendrites; this was not observed in studies using a control probe (sense sequence) or in studies using 100-fold excess unlabeled probe (Fig. 2, top row). Based on bright field illumination, it appeared that the signal was in both cell bodies and dendrites (data not shown). To confirm this localization, we tested for co-localization of EAAC1 mRNA signal with the somato-dendritic neuronal marker, microtubule associated protein-2 a (Map2a). In these studies, clear staining for EAAC1 mRNA (green) was observed in processes that also stain for Map2a protein (red) (Fig. 2, middle row). Furthermore, to confirm that the EAAC1 mRNA signal was specific for only cell bodies and dendrites, and not axons, we tested for co-localization of EAAC1 mRNA with the axonal marker, Tau-1. In these studies, EAAC1 mRNA and Tau-1 protein never co-localized (Fig. 2, bottom row). Together, these data provide strong evidence that EAAC1 mRNA is localized in dendrites in vitro.
Figure 2. In Situ analyses of EAAC1 mRNA in primary hippocampal neuronal cultures and co-localization with neuronal markers.
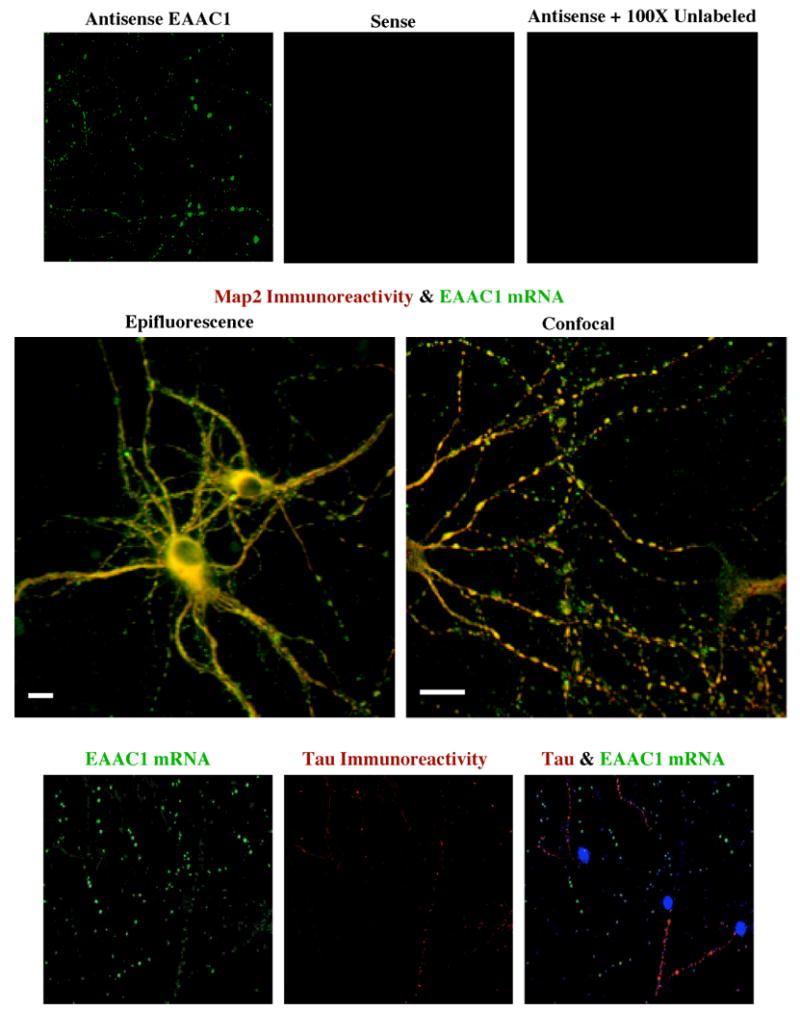
Top row: RNA-RNA in situ hybridization was used to probe for EAAC1 mRNA in dendrites of hippocampal neurons maintained in culture. Sheep anti-digoxigenin antibody and donkey anti-sheep-FITC were used to detect digoxigenin-labeled EAAC1 riboprobes. No signal was observed in sense controls or in experiments with anti-sense and 100× unlabeled riboprobe. Middle row: Analysis of co-localization of EAAC1 mRNA (antisense EAAC1) and Map2a. Mouse anti-Map2ab antibody and goat anti-mouse-rhodamine-X were used to determine if the dendritic marker, Map2a, co-localized with EAAC1 mRNA. The signals for both EAAC1 and Map2a overlap in some of the same processes (yellow-orange). The scale bars represent 50 μm. Bottom row: Distribution of EAAC1 mRNA (antisense EAAC1) and Tau-1 protein. Mouse anti-Tau antibody and goat anti-mouse rhodamine-X were used to determine if EAAC1 mRNA co-localizes with Tau. The signals for Tau protein and EAAC1 mRNA never overlapped in the same processes. All of the data presented in this figure is representative of at least 3 independent experiments.
3.3 Distribution of EAAC1 mRNA in tissue sections from control animals or animals injected with chemoconvulsants
In initial studies, the distribution of EAAC1 mRNA was examined in naïve control animals. For these studies, digoxigenin-labeled anti-sense probes combined with alkaline phosphatase-labeled anti-digoxigenin antibodies were used to test for the presence of EAAC1 mRNA. Although a clear, specific signal for EAAC1 was observed in cell bodies, signal for EAAC1 mRNA did not extend beyond the proximal dendrites.
Some mRNAs display a pronounced signal in dendrites after electrical activation such as that observed with activity regulated cytoskeleton associated protein (Arc/Arc3.1) after induction of long-term potentiation (Steward et al., 2007) or brain-derived neurotrophic factor (BDNF) after seizures (Tongiorgi et al., 2004) or BDNF and TrkB after depolarization with high potassium (Tongiorgi et al., 1997). Therefore, the effects of seizures on the distribution of EAAC1 mRNA were examined using the muscarinic agonist, pilocarpine to induce status epilepticus (Gibbs et al., 1997). For these analyses, only animals that displayed continuous recurrent stage III-IV seizures for 3 (or 6) hrs were tested for mRNA localization. In coronal sections from sham animals, there was a strong signal in CA1 pyramidal cell bodies and a modest signal in proximal dendrites within the stratum radiatum (SR) (Fig. 3); this distribution was indistinguishable from that observed in naïve animals (not shown). Three hrs after induction of SE, there appeared to be an increase in total signal. In these same animals, there was a clear, robust signal in CA1 pyramidal cell dendrites in stratum radiatum, extending up to ∼200 ! m from the cell soma. In area CA3, labeling projected into stratum lucidum (the terminal field of mossy fibers), with low levels of labeling extending into distal dendritic laminar regions (data not shown). In the dentate gyrus, labeling was limited to the granule cell bodies (not shown). Although a more thorough time course for the effect was not examined, the dendritic signal was more modest 6 hr after induction of a seizure (Fig. 3). In these experiments, there was essentially no labeling with the sense riboprobes or when 100× unlabeled antisense riboprobes was combined with the labeled anti-sense probe, providing evidence that this signal is specific. In both CA1 and CA3, seizures did not lead to labeling in the stratum oriens, which contains both the basal dendrites and axons of these pyramidal neurons. Although the processes of pyramidal cells in the cortex are not organized in lamellae like those observed in stratum radiatum of the hippocampus, EAAC1 mRNA was also observed in processes of cells in the cortex of animals that experienced a seizure and not sham animals (data not shown). We also examined the effects of 3h of status on EAAC1 protein in total hippocampus and observed an increase to 122 ± 4% (n= 7 independent experiments, p<0.05) as observed by Western blot. There was no change in actin observed in these same experiments.
Figure 3. Analysis of EAAC1 mRNA distribution in hippocampus of control (sham) and pilocarpine-treated animals.
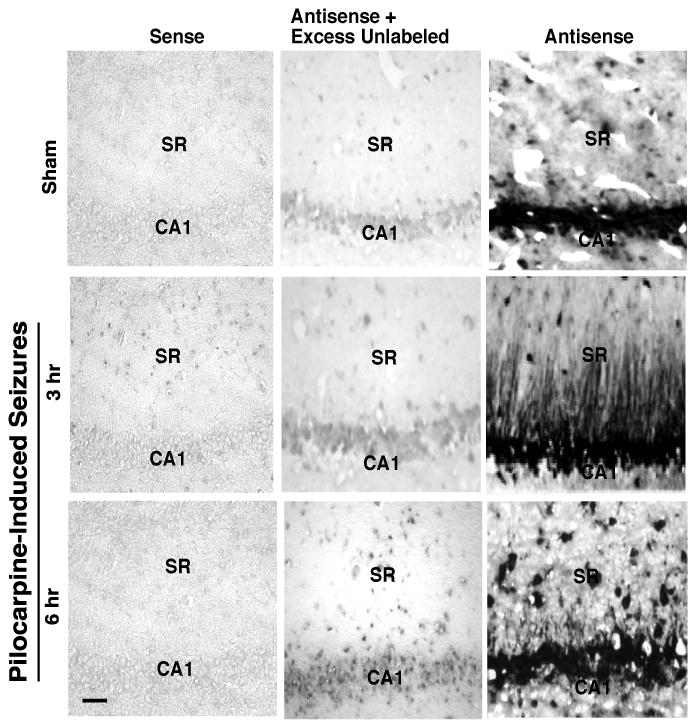
RNA-RNA in situ hybridization with alkaline phosphatase was used to detect EAAC1 mRNA in coronal sections through the hippocampus. This figure shows EAAC1 mRNA signal in the occasional proximal dendrite (10-20 μm) in sham animals (top row). After pilocarpine-induced seizures, EAAC1 mRNA was detected in CA1 pyramidal cell dendrites up to ∼200 μm from the soma. This EAAC1 mRNA signal decreases at 6 h after the onset of SE. These processes are morphologically consistent with dendrites (note there was no evidence of signal projecting into stratum oriens). This result is representative of at least three independent experiments. The scale bar in the lower left panel represents 100 μm.
These studies suggest that seizures increase the transport of EAAC1 mRNA into dendrites (or the unmasking of mRNA signal), but one cannot exclude the possibility that muscarinic receptor activation, independent of seizures, is sufficient to increase the signal for EAAC1 mRNA. Therefore, the distribution of EAAC1 mRNA was also examined in animals treated with a mechanistically distinct chemoconvulsant, kainic acid, which activates a subtype of glutamate receptor (Simantov et al., 1999). Although the signal was not quite as robust as that observed with pilocarpine, a specific signal for EAAC1 mRNA was observed in dendrites after a kainate-induced seizure. This signal extended ∼100 μm from the CA1 pyramidal cells (Fig. 4). Interestingly, labeling was not increased in other hippocampal subfields (data not shown).
Figure 4. Analysis of EAAC1 mRNA distribution in hippocampus of control (sham) and kainate-treated animals.
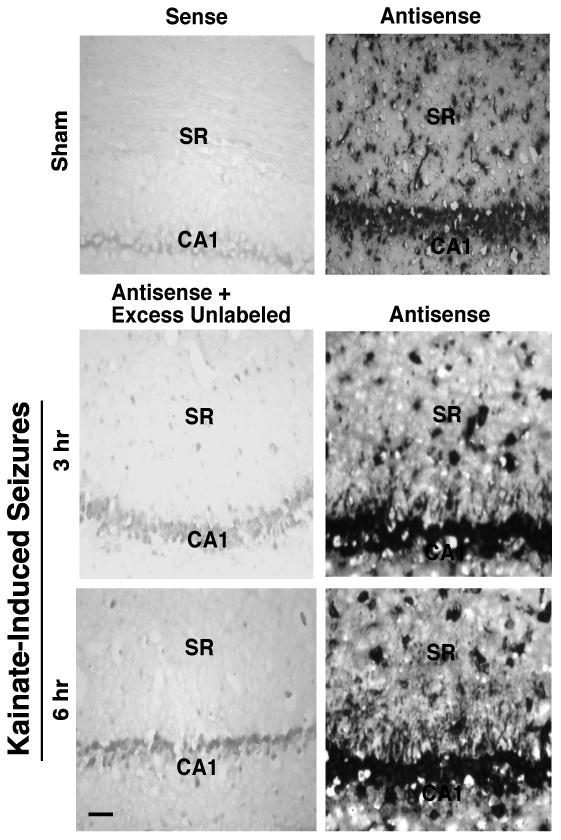
RNA-RNA in situ hybridization with alkaline phosphatase was used to detect EAAC1 mRNA in coronal sections through the hippocampus. This figure shows EAAC1 mRNA signal extending from the cell bodies of CA1 pyramidal cells into the dendritic areas within the stratum radiatum (SR) 3 hr or 6 hr after the induction of kainate-induced seizures. This result is representative of three independent experiments. The scale bar in the lower left panel represents 100 μm.
Animals subjected to an acute chemoconvulsant dose of pilocarpine experience a latent period of no seizures lasting several days and then start to experience spontaneous seizures. This has been used as a model of temporal lobe epilepsy (Gibbs et al., 1997). To determine if the increase in EAAC1 mRNA signal is only present after an acute chemoconvulsant-induced seizure, a subset of animals were allowed to develop spontaneous seizures and tested for the presence of dendritic EAAC1 mRNA. Three hrs after a spontaneous seizure stage III or greater, as identified by behavioral criteria using video recording, a specific signal was observed in dendrites (Fig. 5). We did not attempt to determine if dendritic EAAC1 mRNA was observed in animals that were free of behavioral seizures for 24 hrs because video monitoring is not sufficiently sensitive to rule out sub-convulsive electrographic seizures. These studies show spontaneously epileptic animals have higher levels of dendritic EAAC1 mRNA.
Figure 5. Analysis of EAAC1 mRNA distribution in hippocampus of control (sham) and spontaneous seizure animals.
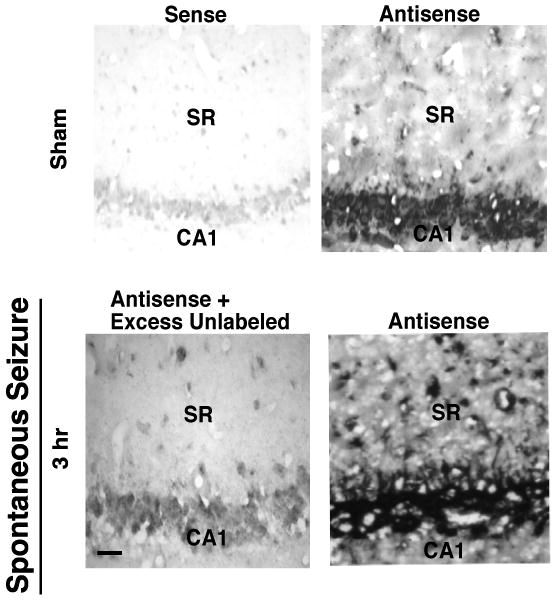
RNA-RNA in situ hybridization with alkaline phosphatase was used to detect EAAC1 mRNA in coronal sections through the hippocampus. This figure shows EAAC1 mRNA signal extending from the cell bodies of CA1 pyramidal cells into the dendritic areas 3 hr after a spontaneous seizure class IV-V. This result is representative of three independent experiments. The scale bar in the lower left panel represents 100 μm.
3.4. Pilocarpine-induced seizure increase EAAC1 mRNA levels in synaptoneurosomes and soma
To determine if this seizure-induced increase in dendritic targeting of EAAC1 mRNA might be a more generalized phenomenon and to reduce the likelihood that the increase in signal might simply be related to increased detection by in situ, rather than an increase in mRNA per se, quantitative real-time PCR was used to analyze the relative mRNA levels in soma and synaptoneurosomes obtained from hippocampi of sham and pilocarpine-treated animals (3 hr of recurrent SE). The levels of GluR2 and calmodulin kinase 2 (CamK2α) were examined in parallel, as it is generally accepted that both of these mRNAs are found in dendritic compartments (for reviews, see Smith, 2004; Steward and Schuman, 2003). Histone deacetylase-3 (HDAC3) and cyclophilin A (PPIA) were also examined. As might be predicted, the levels of all of these mRNAs were higher in the subcellular fraction that is enriched in cell soma than in the synaptoneurosome fraction; however the relative enrichment of HDAC3 mRNA was roughly 3-fold higher than that observed for the other mRNAs (Table 1). The relative amounts of GluR2 and CamK2! mRNA are consistent with the literature (Poon et al., 2006; Zhong et al., 2006). The levels of HDAC3 and PPIA mRNAs were unchanged following acute seizures, the levels of GluR2 mRNA were significantly elevated in the synaptoneurosomes, and the levels of EAAC1 and CamK2α mRNA were significantly increased in both fractions after a seizure (Fig. 6). The data were normalized to the average values obtained in sham animals and changes for each mRNA were compared in the fractions enriched in soma and synaptoneurosomes. EAAC1 mRNA was the only mRNA to increase significantly more in the synaptoneurosome fraction than in the soma/cell body fraction following SE.
Table 1. Levels of mRNAs found in soma and synaptoneurosomes of hippocampi from sham animals.
| mRNA | Soma | Synaptoneurosome | Ratio Soma/Synapto. |
|---|---|---|---|
| HDAC3 | 0.45 ± 0.08 | 0.030 ± 0.007 | 15 |
| PPIA | 0.71 ± 0.12 | 0.140 ± 0.016 | 5.1 |
| GluR2 | 1.93 ± 0.06 | 0.317 ± 0.027 | 6.1 |
| EAAC1 | 3.07 ± 0.47 | 0.587 ± 0.040 | 5.2 |
| CamK2α | 2.87 ± 0.32 | 0.761 ± 0.098 | 3.8 |
The levels of various mRNAs were analyzed by quantitative PCR from fractions obtained after subcellular fractionation as described in the methods. The samples from each animal was analyzed twice; these values were averaged. Data presented are the mean ± SEM from six different animals.
Figure 6.
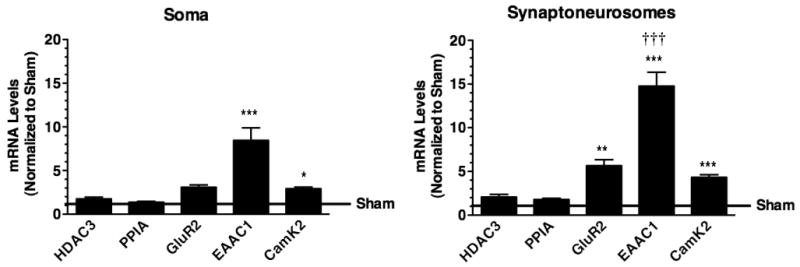
Quantitative analysis of EAAC1 and other mRNAs in cell body and synaptoneurosome subcellular fractions obtained from control (sham) or pilocarpine-treated animals. Three hrs after induction of seizures with pilocarpine, hippocampi were harvested from sham or pilocarpine-treated animals. After homogenization and subcellular fractionation, quantitative PCR was used to measure the levels of HDAC3, cyclophilin A (PPIA), GluR2, EAAC1, and CamK2 mRNAs. The levels of mRNA in each specimen were analyzed twice and averaged. Data represent the mean ± SE of six animals. The absolute levels of each mRNA measured in the pilocarpine-treated animals was compared to those obtained in sham animals by ANOVA with Bonferroni correction. * indicates a P < 0.05, ** indicates a P < 0.01, and *** indicates a P < 0.001. After the data were normalized to the mean sham values, the changes observed in synaptoneurosomes were compared to the changes in the soma fraction by ANOVA with Bonferroni correction. ††† indicates a P < 0.001 compared to the change observed in soma. No other changes were significantly different in the two fractions.
To test the relative contamination of the synaptoneurosome fractions with cell nuclei, we examined the distribution of the nuclear protein, histone H3. The following fractions were analyzed and probed on western blots: total hippocampal lysate, P1 soma fraction, P2 crude synaptoneurosome fraction, and P3 purified synaptoneurosome fraction (Fig. 7). There was a robust immunoreactive band for this histone H3 protein in hippocampal lysates and for the P1 fractions obtained during the subcellular fractionation procedure when less than 0.1% of the total preparation was loaded on a western blot. Even when 20% of the total synaptoneurosome P3 fraction was loaded onto a western blot, histone H3 protein was not detected. These studies provide strong evidence that these synaptoneurosomes are at most, minimally contaminated with cell bodies/nuclei.
Figure 7. Analysis of Histone H3 levels in subcellular fractions obtained from control (sham) and pilocarpine-treated animals.
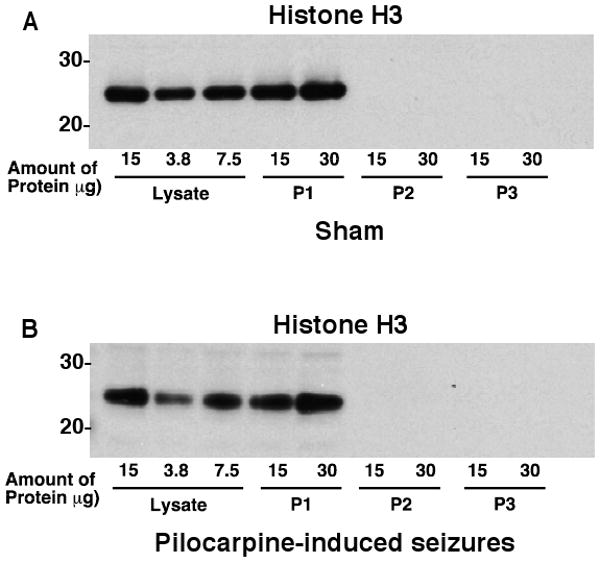
During the subcellular fractionation the different fractions were collected, analyzed for protein, and resolved by SDS-PAGE electrophoresis. The amounts of histone H3 protein were examined in lysates, P1 (should contain intact cell bodies/nuclei), P2 (myelin, mitochondria, and synaptoneurosomes), and P3 (synaptoneurosomes used in these analyses). These studies show that the synaptoneurosomes are relatively free of the nuclear protein, histone H3. This analysis is representative of three independent experiments.
4. Discussion
EAAC1 protein has been found in peri-synaptic regions by immunogold labeling combined with electron microscopy (He et al., 2000; He et al., 2001). Although Cheng and colleagues demonstrated that carboxyl terminal amino acid motif of EAAC1 is necessary for dendritic sorting and conferred partial dendritic sorting to a glutamate transporter that was not normally preferentially sorted to dendrites (Cheng et al., 2002), it seemed reasonable that EAAC1 mRNA might also be targeted to dendrites and contribute to polarized sorting. mRNAs for other gene products linked to synaptic plasticity have been found in dendrites (see introduction). Therefore in the present study, we tested for the presence of EAAC1 mRNA in dendrites to better understand the mechanisms that might control EAAC1 expression and localization. We found that EAAC1 mRNA can be amplified from dendritic samples harvested from primary hippocampal cultures. Furthermore, a specific EAAC1 mRNA signal was identified in Map2a-positive processes, but not in processes that are Tau-1-positive by in situ hybridization. Together, these data suggest that mRNA for EAAC1 is actively targeted to dendrites close to synapses and a potential source for EAAC1 protein production.
Although EAAC1 mRNA was observed in proximal dendrites of some CA1 pyramidal cell dendrite in naïve (see Fig. 5 for an example) or sham control animals, a robust signal was observed in dendrites of animals injected with either pilocarpine or kainate to induce SE. Similar seizure-induced increases are also observed for brain-derived neurotrophic factor (BDNF) (Tongiorgi et al., 2004), Arc/Arg 3.1 (Akiyama et al., 2008), and matrix metalloproteinase-9 (Konopacki et al., 2007). This suggests that the transport of a subclass of dendritically targeted mRNA might be regulated by activity/depolarization. Since we found no evidence for an increase in CAMKIIα and others have found no increase in TrkC (Tongiorgi et al., 2004), this implies that depolarization selectively increases dendritic localization of subsets of mRNAs.
This localization of EAAC1 mRNA to dendrites implies that proteins bind to EAAC1 mRNA to both facilitate trafficking and to control translation (for discussions, see Bear et al., 2004; Job and Eberwine, 2001; Smith, 2004; Wang and Tiedge, 2004). A wide range of proteins bind to dendritically targeted mRNAs, such as heterogenous nuclear ribonucleoproteins (hnRNPs), cytoplasmic polyadenylation element binding protein, and fragile X mental retardation protein (for discussions, see Darnell et al., 2001; Huang et al., 2003; Kelleher and Bear, 2008; Steward and Schuman, 2003). At present, it is not clear which if any of these proteins bind to EAAC1 mRNA. In many cases, the 3′ non-coding region contributes to dendritic sorting of mRNAs (for recent discussions, see Andreassi and Riccio, 2009; Smith, 2004). Again, this has not been examined for EAAC1, however, it is interesting to note that two independent studies have associated single nucleotide polymorphisms (SNPs) in the 3′ non-coding region of EAAC1 with obsessive compulsive disorder (OCD) in males (Arnold et al., 2006; Dickel et al., 2006). It will be interesting to determine if these SNPs regulate either targeting or regulated translation of EAAC1. In unpublished studies, we have found that incubation of synaptoneurosomes prepared from sham or pilocarpine-injected animals with the Group 1 mGluR agonist, dihydroxyphenylglycine, increases EAAC1 protein levels (Ross et al., unpublished). The increase in EAAC1 protein is much greater in synaptoneurosomes prepared from pilocarpine treated animals and is blocked by two different inhibitors of translation, but not by inhibitors of transcription. Together, these studies suggest that translation of EAAC1 may also be regulated in dendrites, especially following a seizure.
Several other studies have examined the relationship of glutamate transporters to seizure disorders. In fact, genetic deletion of GLT-1 (EAAT2) causes seizures (Tanaka et al., 1997). Although mice genetically deleted of EAAC1 do not appear to express any overt behavioral manifestations of seizures (Peghini et al., 1997), chronic intraventricular administration of oligonucleotides to knock down expression of EAAC1 cause a seizure phenotype that initially involves facial twitches and freezing but then progresses to tonic forepaw extension and clonic seizures (Rothstein et al., 1996). The levels of EAAC1 mRNA and protein have been examined in several studies using animal models or tissue (post-mortem or surgical resections) from humans with various epilepsies (for review, see Sheldon and Robinson, 2007). While many of these studies have reported increases in EAAC1 protein and/or mRNA (Crino et al., 2002; Miller et al., 1997; Proper et al., 2002; Ueda et al., 2001; Voutsinos-Porche et al., 2006), others have documented either no change or decreases (Ghijsen et al., 1999; Simantov et al., 1999). Some of these differences are invariably related to the fact that some of these agents cause cell death (e.g. kainate) that ultimately results in a decrease in EAAC1 protein (Simantov et al., 1999). Overall, these studies are consistent with our observed increases in EAAC1 mRNA. Interestingly kainate stimulates endocytosis of EAAC1 and subsequent lysosomal degradation (Yu et al., 2006). It is not known if pilocarpine has a similar effect, but it would interesting the study the dynamics of changes in mRNA relative to this internalization. If degradation precedes the increases in mRNA, it is possible that EAAC1 becomes limiting under these conditions. Based on functions associated with EAAC1 (see introduction), it is possible that the seizure-induced increases in targeting and regulated translation of EAAC1 may provide a mechanism to dampen excitability by clearing excitatory neurotransmitter or to limit oxidant damage by increasing cysteine uptake for glutathione synthesis.
In summary, these studies provide the first evidence that EAAC1 mRNA can be found in dendrites in vitro and in vivo and that EAAC1 mRNA increases in dendrites of hippocampal pyramidal cells following an acute prolonged seizure and in chronically epileptic animals. Neuronal activity appears to increase EAAC1 mRNA production and targeting to dendrites. Furthermore, these studies suggest that both protein targeting (Cheng et al., 2002) and mRNA targeting may provide complementary mechanisms for sorting of EAAC1 to the dendritic compartment of neurons.
Research Highlights.
EAAC1 mRNA is observed in hippocampal neuronal dendrites in vitro
EAAC1 mRNA is observed in dendrites in vivo
Pilocarpine-induced seizures increase dendritic localization of EAAC1 mRNA
Kainate-induced seizures increase in dendritic localization of EAAC1 mRNA
Acknowledgments
This work was supported by NIH grant RO1 HD060132 to MBR, NIH Grant RO1 NS056222 to BEP, NIH grant RO1 AG9900 to JHE, JR was partially supported by NRSA F31 NS049690, and support for cores is partially derived from IDDRC P30 HD26979. The authors would also like to thank Margie Maronski from Dr. Marc Dichter's laboratory for her provision of the neuronal cultures. They would like to thank the members of the Robinson laboratory for their helpful discussions and suggestions for editing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama K, Ishikawa M, Saito A. mRNA expression of activity-regulated cytoskeleton-associated protein (arc) in the amygdala-kindled rats. Brain Res. 2008;1189:236–46. doi: 10.1016/j.brainres.2007.10.102. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–74. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Aoyama K, Suh SW, Hamby AM, Liu J, Chan WY, Chen Y, Swanson RA. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat Neurosci. 2006;9:119–26. doi: 10.1038/nn1609. [DOI] [PubMed] [Google Scholar]
- Arnold PD, Sicard T, Burroughs E, Richter MA, Kennedy JL. Glutamate transporter gene SLC1A1 associated with obsessive-compulsive disorder. Arch Gen Psychiatry. 2006;63:769–76. doi: 10.1001/archpsyc.63.7.769. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–65. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends in Neurosciences. 2004;27:370–7. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Berger UV, Hediger MA. Comparative analysis of glutamate transporter expression in rat brain using differential double in situ hybridization. Anat Embryol (Berl) 1998;198:13–30. doi: 10.1007/s004290050161. [DOI] [PubMed] [Google Scholar]
- Cheng C, Glover G, Banker G, Amara SG. A novel sorting motif in the glutamate transporter excitatory amino acid transporter 3 directs its targeting in Madin-Darby canine kidney cells and hippocampal neurons. Journal of Neuroscience. 2002;22:10643–10652. doi: 10.1523/JNEUROSCI.22-24-10643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti F, DiBiasi S, Minelli A, Rothstein JD, Melone M. EAAC1, a high-affinity glutamate transporter, is localized to astrocytes and gabaergic neurons besides pyramidal cells in the rat cerebral cortex. Cerebral Cortex. 1998;8:108–116. doi: 10.1093/cercor/8.2.108. [DOI] [PubMed] [Google Scholar]
- Crino PB, Jin H, Shumate MD, Robinson MB, Coulter DA, Brooks-Kayal AR. Increased expression of the neuronal glutamate transporter (EAAT3/EAAC1) in hippocampal and neocortical epilepsy. Epilepsia. 2002;43:211–8. doi: 10.1046/j.1528-1157.2002.35001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Progress in Neurobiology. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–99. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Diamond JS. Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. Journal of Neuroscience. 2001;21:8328–8338. doi: 10.1523/JNEUROSCI.21-21-08328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Veenstra-VanderWeele J, Cox NJ, Wu X, Fischer DJ, Van Etten-Lee M, Himle JA, Leventhal BL, Cook EH, Jr, Hanna GL. Association testing of the positional and functional candidate gene SLC1A1/EAAC1 in early-onset obsessive-compulsive disorder. Arch Gen Psychiatry. 2006;63:778–85. doi: 10.1001/archpsyc.63.7.778. [DOI] [PubMed] [Google Scholar]
- Dowd LA, Coyle AJ, Rothstein JD, Pritchett DB, Robinson MB. Comparison of Na+-dependent glutamate transport activity in synaptosomes, C6 glioma, and Xenopus Oocytes expressing excitatory amino acid carrier 1 (EAAC1) Molecular Pharmacology. 1996;49:465–473. [PubMed] [Google Scholar]
- Eberwine J. Amplification of mRNA populations using aRNA generated from immobilized oligo(dT)-T7 primed cDNA. BioTechniques. 1996;20:584–91. doi: 10.2144/19962004584. [DOI] [PubMed] [Google Scholar]
- Elias GM, Nicoll RA. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell Biol. 2007;17:343–52. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Ghijsen WE, da Silva Aresta, Belo AI, Zuiderwijk M, Lopez da Silva FH. Compensatory change in EAAC1 glutamate transporter in rat hippocampus CA1 region during kindling epileptogenesis. Neuroscience Letters. 1999;276:157–60. doi: 10.1016/s0304-3940(99)00824-1. [DOI] [PubMed] [Google Scholar]
- Gibbs JW, 3rd, Shumate MD, Coulter DA. Differential epilepsy-associated alterations in postsynaptic GABA(A) receptor function in dentate granule and CA1 neurons. J Neurophysiol. 1997;77:1924–38. doi: 10.1152/jn.1997.77.4.1924. [DOI] [PubMed] [Google Scholar]
- Greger IH, Esteban JA. AMPA receptor biogenesis and trafficking. Curr Opin Neurobiol. 2007;17:289–97. doi: 10.1016/j.conb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- He Y, Janssen WGM, Rothstein JD, Morrison JH. Differential synaptic locallization of the glutamate transporter EAAC1 and glutamate receptor subunit GluR2 in the rat hippocampus. Journal of Comparative Neurology. 2000;418:255–269. [PubMed] [Google Scholar]
- He Y, Hof PH, Janssen WGM, Rothstein JD, Morrison JH. Differential synaptic localization of GluR2 and EAAC1 in the macaque monkey entorhinal cortex: a postembedding immunogold study. Neuroscience Letters. 2001;311:161–164. doi: 10.1016/s0304-3940(01)02180-2. [DOI] [PubMed] [Google Scholar]
- Hollingsworth EB, McNeal ET, Burton JL, Williams RJ, Daly JW, Creveling CR. Biochemical characterization of a filtered synaptoneurosome preparation from guinea pig cerebral cortex: cyclic adenosine 3′:5′-monophosphate-generating systems, receptors, and enzymes. Journal of Neuroscience. 1985;5:2240–53. doi: 10.1523/JNEUROSCI.05-08-02240.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes and Development. 2003;17:638–53. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C, Eberwine J. Identification of sites for exponential translation in living dendrites. Proc Natl Acad Sci U S A. 2001;98:13037–42. doi: 10.1073/pnas.231485698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacharmina JE, Job C, Crino P, Eberwine J. Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc Natl Acad Sci U S A. 2000;97:11545–50. doi: 10.1073/pnas.97.21.11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Konopacki FA, Rylski M, Wilczek E, Amborska R, Detka D, Kaczmarek L, Wilczynski GM. Synaptic localization of seizure-induced matrix metalloproteinase-9 mRNA. Neuroscience. 2007;150:31–9. doi: 10.1016/j.neuroscience.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Kugler P, Schmitt A. Glutamate transporter EAAC! is expressed in neurons and glial cells in the rat nervous system. Glia. 1999;27:129–142. doi: 10.1002/(sici)1098-1136(199908)27:2<129::aid-glia3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Levenson J, Weeber E, Selcher JC, Kategaya LS, Sweatt JD, Eskin A. Long-term potentiation and contextual fear conditioning increase neuronal glutamate uptake. Nature Neuroscience. 2001;5:155–161. doi: 10.1038/nn791. [DOI] [PubMed] [Google Scholar]
- Liang SL, Carlson GC, Coulter DA. Dynamic regulation of synaptic GABA release by the glutamate-glutamine cycle in hippocampal area CA1. Journal of Neuroscience. 2006;26:8537–48. doi: 10.1523/JNEUROSCI.0329-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews GC, Diamond JS. Neuronal glutamate uptake contributes to GABA synthesis and inhibitory synaptic strength. Journal of Neuroscience. 2003;23:2040–2048. doi: 10.1523/JNEUROSCI.23-06-02040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HP, Levey AI, Rothstein JD, Tzingounis AV, Conn PJ. Alterations in glutamate transporter protein levels in kindling-induced epilepsy. Journal of Neurochemistry. 1997;68:1564–1570. doi: 10.1046/j.1471-4159.1997.68041564.x. [DOI] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci U S A. 1994;91:10800–4. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Glutamate receptor dynamics in dendritic microdomains. Neuron. 2008;58:472–97. doi: 10.1016/j.neuron.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieoullon A, Canolle B, Masmejean F, Guillet B, Pisano P, Lortet S. The neuronal excitatory amino acid transporter EAAC1/EAAT3: does it represent a major actor at the brain excitatory synapse? J Neurochem. 2006;98:1007–18. doi: 10.1111/j.1471-4159.2006.03978.x. [DOI] [PubMed] [Google Scholar]
- Palos TP, Ramachandran B, Boado R, Howard BD. Rat C6 and human astrocytic tumor cells express a neuronal type of glutamate transporter. Molecular Brain Research. 1996;37:297–303. doi: 10.1016/0169-328x(95)00331-l. [DOI] [PubMed] [Google Scholar]
- Peghini P, Janzen J, Stoffel W. Glutamate transporter EAAC-1-deficient mice develop dicarboxylic aminoaciduria and behavioral abnormalities but no neurodegeneration. EMBO Journal. 1997;16:3822–3832. doi: 10.1093/emboj/16.13.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita-Almenar JD, Sol Collado M, Colbert CM, Eskin A. Different mechanisms exist for the plasticity of glutamate reuptake during early long-term potentiation (LTP) and late LTP. Journal of Neuroscience. 2006;26:10461–71. doi: 10.1523/JNEUROSCI.2579-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–9. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proper EA, Hoogland G, Kappen SM, Jansen GH, Rensen MG, Schrama LH, van Veelen CW, van Rijen PC, van Nieuwenhuizen O, Gispen WH, de Graan PN. Distribution of glutamate transporters in the hippocampus of patients with pharmaco-resistant temporal lobe epilepsy. Brain. 2002;125:32–43. doi: 10.1093/brain/awf001. [DOI] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation II. Motor seizure. Electroencephalography and Clinical Neurophysiology. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- Robinson MB. Acute regulation of sodium-dependent glutamate transporters: A focus on constitutive and regulated trafficking. Handbook of Experimental Pharmacology. 2006;175:251–275. doi: 10.1007/3-540-29784-7_13. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Martin L, Levey AI, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl RW. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, Kanai Y, Hediger M, Wang Y, Schielke JP, Welty DF. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Scimemi A, Tian H, Diamond JS. Neuronal transporters regulate glutamate clearance, NMDA receptor activation, and synaptic plasticity in the hippocampus. J Neurosci. 2009;29:14581–95. doi: 10.1523/JNEUROSCI.4845-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepkuty JP, Cohen AS, Eccles C, Rafiq A, Behar K, Ganel R, Coulter DA, Rothstein JD. A neuronal glutamate transporter contributes to neurotransmitter GABA synthesis and epilepsy. Journal of Neuroscience. 2002;22:6372–6379. doi: 10.1523/JNEUROSCI.22-15-06372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem Int. 2007 doi: 10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Watanabe M, Tanaka K, Wada K, Inoue Y. Dynamic changes in expression of glutamate transporter mRNAs in developing brain. NeuroReport. 1996;7:705–709. doi: 10.1097/00001756-199602290-00006. [DOI] [PubMed] [Google Scholar]
- Simantov R, Crispino M, Hoe W, Broutman G, Tocco G, Rothstein JD, Baudry M. Changes in expression of neuronal and glial glutamate transporters in rat hippocampus following kainate-induced seizure activity. Brain Res Mol Brain Res. 1999;65:112–23. doi: 10.1016/s0169-328x(98)00349-0. [DOI] [PubMed] [Google Scholar]
- Smith R. Moving molecules: mRNA trafficking in Mammalian oligodendrocytes and neurons. Neuroscientist. 2004;10:495–500. doi: 10.1177/1073858404266759. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- Steward O, Huang F, Guzowski JF. A form of perforant path LTP can occur without ERK1/2 phosphorylation or immediate early gene induction. Learn Mem. 2007;14:433–45. doi: 10.1101/lm.554607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, Okuyama S, Kawashima N, Hori S, Takimoto M, Wada K. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. Journal of Neuroscience. 1997;17:9492–505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Armellin M, Giulianini PG, Bregola G, Zucchini S, Paradiso B, Steward O, Catteneo A, Simonanto M. Brain-derived neurotrophic factor mRNA and protein are targted to discrete dendritic laminas by events that trigger epileptogenesis. Journal of Neuroscience. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Doi T, Tokumaru J, Yokoyama H, Nakajima A, Mitsuyama Y, Ohya-Nishiguchi H, Kamada H, Willmore LJ. Collapse of extracellular glutamate regulation during epileptogenesis: down-regulation and functional failure of glutamate transporter function in rats with chronic seizures induced by kainic acid. J Neurochem. 2001;76:892–900. doi: 10.1046/j.1471-4159.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- Velaz-Faircloth M, McGraw TS, alandro MS, Fremeau RT, Jr, Kilberg MS, Anderson KJ. American Journal of Physiology. Vol. 270. 1996. Characterization and distribution of the neuronal glutamate transporter EAAC1 in rat brain; pp. C67–75. [DOI] [PubMed] [Google Scholar]
- Voutsinos-Porche B, Koning E, Clement Y, Kaplan H, Ferrandon A, Motte J, Nehlig A. EAAC1 glutamate transporter expression in the rat lithium-pilocarpine model of temporal lobe epilepsy. J Cereb Blood Flow Metab. 2006;26:1419–30. doi: 10.1038/sj.jcbfm.9600295. [DOI] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010;33:173–82. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tiedge H. Translational control at the synapse. Neuroscientist. 2004;10:456–66. doi: 10.1177/1073858404265866. [DOI] [PubMed] [Google Scholar]
- Yu YX, Shen L, Xia P, Tang YW, Bao L, Pei G. Syntaxin 1A promotes the endocytic sorting of EAAC1 leading to inhibition of glutamate transport. J Cell Sci. 2006;119:3776–87. doi: 10.1242/jcs.03151. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Tang CM, Yao YL, Yang WM, Seto E. Cloning and characterization of the mouse histone deacetylase-2 gene. J Biol Chem. 1998;273:28921–30. doi: 10.1074/jbc.273.44.28921. [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]


