Abstract
The subunit composition of the NAD(P)H dehydrogenase complex of Thermosynechococcus elongatus was analyzed by different types of mass spectrometry. All 15 known subunits (NdhA-NdhO) were identified in the purified NDH-1L complex. Moreover, two additional intact mass tags of 4902.7 and 4710.5 Da could be assigned after re-annotation of the T. elongatus genome. NdhP and NdhQ are predicted to contain a single transmembrane helix each and homologues are apparent in other cyanobacteria. Additionally, ndhP is present in some cyanophages in a cluster of PSI genes and it exhibits partial similarity to NDF6, a subunit of the plant NDH-1 complex.
The internal membrane system of cyanobacteria and chloroplasts – the thylakoids – contains a type I NAD(P)H dehydrogenase structurally and functionally similar to complex I (NADH:ubiquinone oxidoreductase) found in mitochondria and eubacteria (1, 2). The subunit composition of complex I-type enzymes varies from 14 (E.coli complex, NuoA-NuoN) (3), which is the minimal set for bioenergetic function (4) to 45 of the bovine complex (5). The cyanobacterial NDH-1 complex is presently believed to consist of 15 different subunits (6-9). Out of these, 11 (NdhA-NdhK) are similar to E. coli proteins, whereas for the E. coli subunits NuoE, NuoF and NuoG no homologues could be found in cyanobacteria and plant chloroplasts (http://www.uniprot.org). These subunits contain the NADH-binding site and carry the FMN cofactor as well as several FeS cluster (10, 11), features that are essential for the function of complex I in eubacteria and mitochondria. To date, it is still a puzzle which proteins are responsible for electron import into the NDH-1 complex of cyanobacteria and plant chloroplasts. The subunits NdhL, NdhM, NdhN and NdhO are unique to organisms performing oxygenic photosynthesis (6, 8, 12) NdhL is important for carbon uptake in cyanobacteria (13), but the functional roles of the others remain unclear. Genes encoding NdhD and NdhF are found in most cyanobacterial genomes in several copies. These gene families represent the basis for the functional variety of NDH-1 complexes in cyanobacteria. Four different types could be defined by reverse genetics (14, 15) and functional proteomics (6-9).
NDH-1L (NdhD1/NdhF1) and NDH-1L' (NdhD2/NdhF1) are suggested to be involved in respiration and cyclic electron flow around PSI (16). NDH-1L is the predominating complex in the thylakoid membrane whereas NDH-1L' has not yet been detected on protein level. Single particle analysis revealed the typical “L-shaped” structure for this type of complex (17) and although, NDH-1L has a striking similarity to the mitochondrial counterpart, there are arguments against its respiratory function in cyanobacteria. The impaired rates of plastoquinone reduction in ndh mutants might be related to low levels of succinate in these strains, which limit the activity of the succinate dehydrogenase, rather than the lack of NDH-1 activity (18). The NDH-1MS (NdhD3/NdhF3) and NDH-1MS' (NdhD4/NdhF4) complexes are responsible for high-affinity and low-affinity CO2 uptake, respectively (9, 19), both unique functions of the cyanobacterial NDH-1 complex. NDH-1MS' is expressed constitutively at low level whereas NDH-1MS is induced under low CO2 conditions (14, 15). The NDH-1MS complex has a “U-shaped” structure caused by additional proteins (CupA/CupS) bound to the cytoplasmic surface (20). Here we focus on the analysis of additional small proteins appearing in the cyanobacterial NDH-1L complex. In-depth mass spectrometry analysis of highly purified material revealed the presence of two new subunits that were designated NdhP and NdhQ.
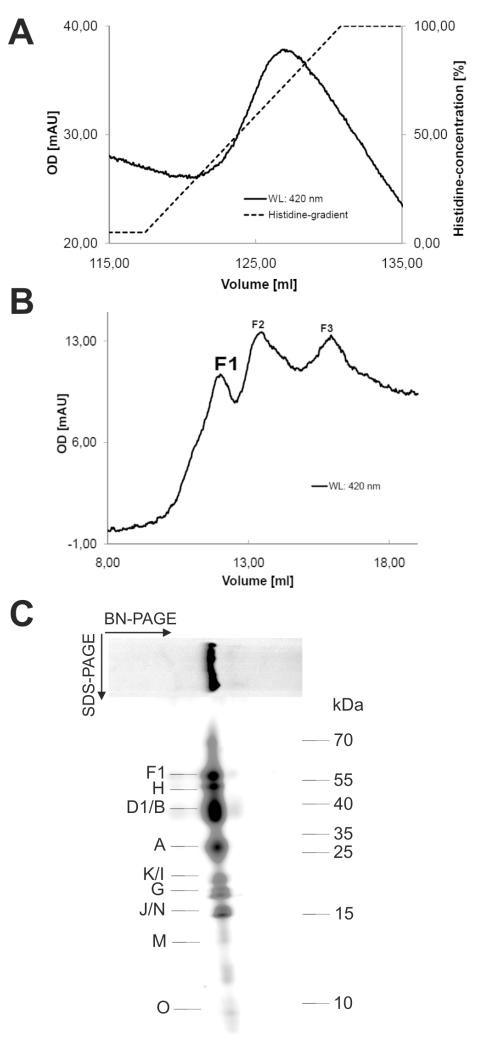
The NDH-1L complex of Thermosynechococcus elongatus was isolated via the “natural his-tag” of the NdhF1 subunit (9) by Ni2+ column chromatography (Figure 1A). In the second step the pre-purified complex was loaded on a size exclusion column (see experimental procedures for details) to separate impurities and breakdown products (Figure 1B). Fractions containing the NDH-1L complex (~500 kDa) corresponding to peak F1 were pooled and concentrated. All further experiments were done with this sample. Figure 1C shows the result of two-dimensional gel electrophoresis with the purified complex. The major band in the BN-page corresponds to the NDH-1L complex and 12 of 15 known subunits could be identified in the spots of the second dimension (Table S1). Additionally, the purified complex was analyzed by tandem mass spectrometry in a shot-gun experiment after digestion with trypsin and/or chymotrypsin and all 15 known subunits of the NDH-1L complex could be identified (Table S2).
Figure 1.
Purification of the NDH-1L complex. NDH-1L complexes were purified by a combination of (A) Ni2+ affinity chromatography and (B) size exclusion chromatography. The purified complexes were analyzed by (C) two-dimensional gel electrophoresis (BN- and SDS-PAGE).
Moreover, the intact complex was analyzed by liquid chromatography with electrospray-ionization mass spectrometry (21) and two masses in the low molecular mass region were identified, which could not be assigned to known NDH-1L subunits. Even after calculation of the theoretical molecular weight of all open reading frames (ORFs) assigned in the genome of Thermosynechococcus elongatus (http://genome.kazusa.or.jp/cyanobase), no concordance was found. Additionally, known post translational modifications of small subunits like N-terminal formylation, acetylation and cleavage of the N-terminal methionine (22, 23) have been considered. However, only after a search for unassigned ORFs in the genome of T. elongatus with GLIMMER (http://bioinformatics.biol.rug.nl/websoftware/orf/orf_start.php) could two new ORFs (Figure S1) be assigned to the unknown masses, named NdhP and NdhQ. The mass of NdhP (4902.7 Da) indicates retention of the formyl group on the initiating N-terminal methionine whereas the measured mass of NdhQ (4710.5 Da) matches the calculated mass based on the amino acid sequence after cleavage of the N-terminal methionine (Table S4), both common post translational modifications in cyanobacteria. MALDI-ToF mass spectrometry shows that both subunits seem to be present in rather equal amounts compared to known NDH-1L subunits (Figure S2), although the exact stoichiometry could not be concluded from the data due to ionization effects. The translated amino acid sequence is predicted to exhibit a single trans-membrane helix (TMH) in both cases (Figure S3). In addition, the identification of the new subunits was verified on the peptide level. For that purpose, the derived amino acid sequences were included in the database for protein identification, which was done by analysis of the spectra generated in the LC-ESI-MS/MS shot-gun experiment. Only one peptide of each protein was identified with high confidence, but this covers the hydrophilic domain almost completely in both cases (Table S3 and Figure 2). Other possible peptides might be not in the right mass range, or they might be lost during sample preparation due to their high level of hydrophobicity.
Figure 2.
Identification of NdhP and NdhQ. Purified NDH-1L complexes were digested with trypsin or chymotrypsin and the peptides were analyzed in a shot-gun experiment by LC-ESI tandem mass spectrometry. The sequence, the predicted topology and the location of the identified peptide (box) of NdhP (A) and NdhQ (B), as well as the corresponding spectra are shown. The topology of NdhP and NdhQ was predicted by SOSUI (http://www.bp.nuap.nagoya-u.ac.jp/sosui/) as indicated in the left figure. TMH: white background color.
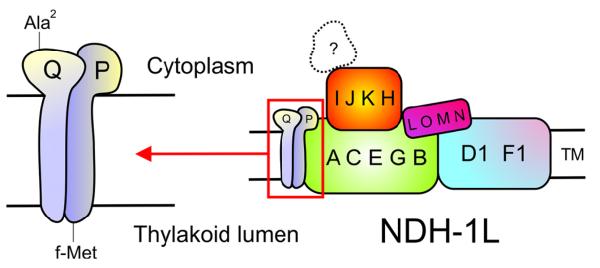
Moreover, the identified post-translational modifications of NdhP and NdhQ might be used to predict their orientation in the membrane. N-terminal processing requires the accessibility of the N-terminus, which is located towards the cytoplasm in this case, whereas a remaining formyl group at the N-terminus indicates an orientation towards the lumen (24). These predictions were included in the model of the NDH-1L complex with NdhP and NdhQ shown in Figure 3. Sequence analysis of ndhP and ndhQ by tblastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that both genes are widespread within the phylum of cyanobacteria (Figure S4). NdhP is present in all 37 cyanobacterial genomes, whereas ndhQ is absent in Gloeobacter violaceus and in some Prochlorococcus strains (http://genome.kazusa.or.jp/cyanobase). Moreover, a domain similar to NdhP could be revealed in the amino acid sequence of NDF6 (Figure S5), a subunit of the NDH-1 complex present in the chloroplasts of Arabidopsis thaliana that was discovered recently (25). However, the relation to NdhP is unclear because NDF6 is much larger and only a small part of it shows some sequence similarity. Interestingly, among the similar sequences present in other organisms, a gene coding for an NdhP homologue could be found in the genome of a cyanophage (PSSM2_253, Figure S4). It is part of an operon with PS1 genes and thought to be a phage-specific ORF with unknown function (26). Here we could show, that this protein is part of the cyanobacterial NDH-1 complex and, in conclusion, its expression by the phage might control the amount of NDH-1 in the host cell or it might support cyclic electron flow around PS1 in order to force the production of ATP. Although they are rather small and sometimes overlooked, single transmembrane domain (STMD) proteins seem to play important roles during biogenesis of membrane protein complexes in general (27) but this has to be proven for NdhP and NdhQ in further experiments.
Figure 3.
Model of the NDH-1L complex. The two new NDH-1 subunits NdhP and NdhQ were included in a model of the NDH-1L complex. Both proteins are predicted to span the membrane by a single TMH and the differences in N-terminal processing suggest an unequal location of the N-termini. The unknown subunits responsible for NAD(P)H oxidation are indicated by the question mark. TM: thylakoid membrane
Supplementary Material
ACKNOWLEDGMENT
We thank G. M. Soriano, A. Pötsch, M. Rögner and A. Trebst for stimulating discussion, M. Völkel for excellent technical assistance and E.-M. Aro and N. Battchikova for the T. elongatus strain used for purification of NDH-1L. This work was supported by the Protein Research Department of the Ruhr-University Bochum (RUB) and funded by a grant from the RUB (Rektoratsprogramm) to M.M.N. The N.I.H. is gratefully acknowledged for support: R01-GM38323, (WAC) and P50-GM-088499 (WAC and JPW).
Footnotes
SUPPORTING INFORMATION PARAGRAPH
Details for experimental procedures, additional tables and figures. This information is available free of charge via the Internet at http://pub.asc.org.
REFERENCES
- 1.Friedrich T, Bottcher B. Bba-Bioenergetics. 2004;1608:1–9. doi: 10.1016/j.bbabio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Friedrich T, Steinmuller K, Weiss H. Febs Lett. 1995;367:107–111. doi: 10.1016/0014-5793(95)00548-n. [DOI] [PubMed] [Google Scholar]
- 3.Friedrich T, Weiss H. J Theor Biol. 1997;187:529–540. doi: 10.1006/jtbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 4.Brandt U. Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 5.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. J Biol Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 6.Battchikova N, Zhang PP, Rudd S, Ogawa T, Aro EM. J Biol Chem. 2005;280:2587–2595. doi: 10.1074/jbc.M410914200. [DOI] [PubMed] [Google Scholar]
- 7.Herranen M, Battchikova N, Zhang PP, Graf A, Sirpio S, Paakkarinen V, Aro EM. Plant Physiol. 2004;134:470–481. doi: 10.1104/pp.103.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prommeenate P, Lennon AM, Markert C, Hippler M, Nixon PJ. J Biol Chem. 2004;279:28165–28173. doi: 10.1074/jbc.M401107200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang PP, Battchikova N, Paakkarinen V, Katoh H, Iwai M, Ikeuchi M, Pakrasi HB, Ogawa T, Aro EM. Biochem J. 2005;390:513–520. doi: 10.1042/BJ20050390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sazanov LA, Hinchliffe P. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 11.Efremov RG, Baradaran R, Sazanov LA. Nature. 2010;465:441–U461. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 12.Rumeau D, Becuwe-Linka N, Beyly A, Louwagie M, Garin J, Peltier G. Plant Cell. 2005;17:219–232. doi: 10.1105/tpc.104.028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa T. Plant Physiol. 1992;99:1604–1608. doi: 10.1104/pp.99.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klughammer B, Sultemeyer D, Badger MR, Price GD. Mol Microbiol. 1999;32:1305–1315. doi: 10.1046/j.1365-2958.1999.01457.x. [DOI] [PubMed] [Google Scholar]
- 15.Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T. P Natl Acad Sci USA. 2001;98:11789–11794. doi: 10.1073/pnas.191258298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battchikova N, Eisenhut M, Aro EM. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Arteni AA, Zhang PP, Battchikova N, Ogawa T, Aro EM, Boekema EJ. Bba-Bioenergetics. 2006;1757:1469–1475. doi: 10.1016/j.bbabio.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 18.Cooley JW, Vermaas WFJ. J Bacteriol. 2001;183:4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, Ogawa T, Pakrasi HB, Mi HL. Plant Cell Physiol. 2008;49:994–997. doi: 10.1093/pcp/pcn074. [DOI] [PubMed] [Google Scholar]
- 20.Folea IM, Zhang P, Nowaczyk MM, Ogawa T, Aro EM, Boekema EJ. Febs Lett. 2008;582:249–254. doi: 10.1016/j.febslet.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Whitelegge JP. P Natl Acad Sci USA. 2002;99:11564–11566. doi: 10.1073/pnas.192449199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Mohsnawy E, Kopczak MJ, Schlodder E, Nowaczyk M, Meyer HE, Warscheid B, Karapetyan NV, Rogner M. Biochemistry-Us. 2010;49:4740–4751. doi: 10.1021/bi901807p. [DOI] [PubMed] [Google Scholar]
- 23.Thangaraj B, Ryan CM, Souda P, Krause K, Faull KF, Weber APM, Fromme P, Whitelegge JP. Proteomics. 2010;10:3644–3656. doi: 10.1002/pmic.201000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa N, Takabayashi A, Ishida S, Hano Y, Endo T, Sato F. Plant Cell Physiol. 2008;49:1066–1073. doi: 10.1093/pcp/pcn083. [DOI] [PubMed] [Google Scholar]
- 26.Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N, Pinter RY, Partensky F, Koonin EV, Wolf YI, Nelson N, Beja O. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zickermann V, Angerer H, Ding MG, Nubel E, Brandt U. Febs Lett. 2010;584:2516–2525. doi: 10.1016/j.febslet.2010.04.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.