SUMMARY
Embryonal rhabdomyosarcoma (eRMS) shows the most myodifferentiation amongst sarcomas, yet the precise cell of origin remains undefined. Using Ptch1, p53 and/or Rb1 conditional mouse models and controlling prenatal or postnatal myogenic cell of origin, we demonstrate that eRMS and undifferentiated pleomorphic sarcoma (UPS) lie in a continuum, with satellite cells predisposed to giving rise to UPS. Conversely, p53 loss in maturing myoblasts gives rise to eRMS, which have the highest myodifferentiation potential. Irrespective of origin, Rb1 loss modifies tumor phenotype to mimic UPS. In human sarcomas that lack pathognomic chromosomal translocations, p53 loss of function is prevalent whereas Shh or Rb1 alterations likely act primarily as modifiers. Thus, sarcoma phenotype is strongly influenced by cell of origin and mutational profile.
INTRODUCTION
Rhabdomyosarcoma is a soft tissue sarcoma showing myogenic differentiation. The most common form of rhabdomyosarcoma is the embryonal subtype (eRMS)(Breneman et al., 2003). Metastatic eRMS portends only 40% overall survival(Breneman et al., 2003). Studies of mouse and man implicate a number of causative mutations in eRMS that include p53, Shh/Ptch1/Sufu, Ras and Rb1(Kohashi et al., 2008; Langenau et al., 2007; Li and Fraumeni, 1969). Numerous descriptive and animal model studies have given preliminary insight into cell of origin of eRMS, which is postulated to be the muscle stem cell (satellite cell)(Hettmer and Wagers; Tiffin et al., 2003). However, the cell of origin is debatable because eRMS not only express markers of satellite cells (e.g., Pax7(Tiffin et al., 2003)), but also markers of myoblasts (MyoD, Myogenin) and differentiated muscle (Desmin, muscle specific actin)(Morotti et al., 2006).
Undifferentiated pleomorphic sarcomas (UPS), synonymous with either malignant fibrous histiocytoma (MFH) or undifferentiated spindle cell sarcoma (USCS), is a term that is used for the classification of neoplasms that are presumed to be sarcomas yet show no definable line of differentiation by histological, immunohistochemical, ultrastructural or molecular criteria(Weiss, 2007). The field has been presumed that many UPS/MFH/USCS are poorly differentiated variants of well-known sarcomas, which in their more differentiated forms would show a defined line of differentiation as defined by consistent histological, immunohistochemical, ultrastructural or molecular features(Fletcher et al., 2002). However, this assertion is impossible to prove since classification requires characteristics that are dependent on differentiation. It is not unusual to see undifferentiated sarcomas arising from more differentiated sarcomas that, without the differentiated component being present, would merit the classification of UPS. Furthermore, UPS is not a distinct morphological entity. Extensive heterogeneity exists in the appearance of lesions classified as UPS, ranging from cellular spindle cell neoplasms to very pleomorphic neoplasms with epithelioid cytomorphology, suggesting that UPS may arise from a variety of precursors. Anaplasia, characterized by the presence of tumor cells with large, hyperchromatic nuclei with or without large atypical mitotic figures may be seen in both embryonal and alveolar rhabdomyosarcoma, implying that diffusely pleomorphic variants without obvious rhabdomyoblastic differentiation might exist and that these variants would be virtually impossible to classify correctly(Qualman et al., 2008).
In this study, we present a genetic dissection of cell of origin and mutational profile of a series of eRMS models and human tumors, with specific attention given to the p53, Shh/Ptch and Rb1 pathways.
RESULTS
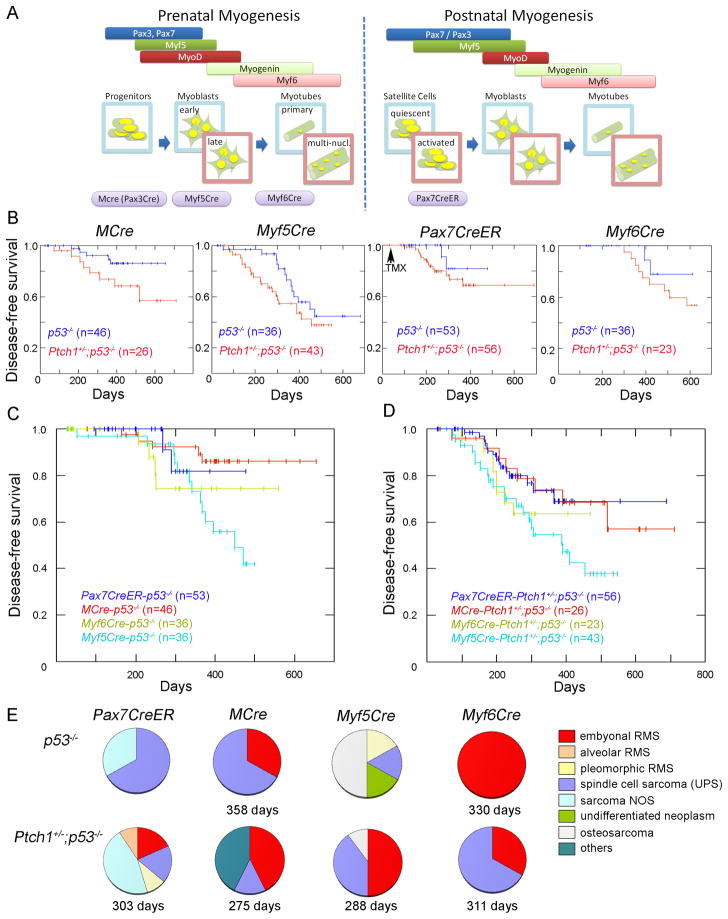
Embryonal rhabdomyosarcoma can arise from muscle stem cells or downstream myogenic precursor lineages, but mutation profile alters proportions of UPS and eRMS
To generate mouse models of embryonal rhabdomyosarcoma, lineage-specific homozygous deletion of p53 with or without concurrent heterozygous Patched1 (Ptch1) deletion was achieved by interbreeding Ptch1 or p53 conditional lines with the myogenic Cre mouse lines MCre(Brown et al., 2005), Myf5Cre(Haldar et al., 2008), Pax7CreER(Nishijo et al., 2009), or Myf6Cre(Keller et al., 2004). A diagram correlating Cre driver expression with stage of prenatal or postnatal myogenesis is given in Figure 1A. MCre is specific for the prenatal and postnatal hypaxial lineage of Pax3 that includes postnatal satellite cells(Brown et al., 2005; Relaix et al., 2006). Myf5Cre is specific for the prenatal and postnatal lineage of Myf5 that includes quiescent and activated satellite cells and early myoblasts(Beauchamp et al., 2000; Cornelison and Wold, 1997; Kuang et al., 2007), as well as mesenchymal precursors of brown fat (Seale et al., 2008). Pax7CreER is specific for the postnatal lineage of Pax7 that includes quiescent and activated satellite cells(Nishijo et al., 2009), and Myf6Cre is specific for the prenatal and postnatal lineage of Myf6 that includes maturing myoblasts(Keller et al., 2004). For activation of Pax7CreER, tamoxifen was administered intraperitoneally 4 weeks after birth for 5 consecutive days as previously described(Nishijo et al., 2009). Results of conditional mouse crosses are shown in Figure 1B. When p53 was homozygously inactivated, tumors arising in the different mouse Cre lines developed at a penetrance rate of 13–56% within a 600 day follow-up period. Myf5Cre-p53−/− or Myf5Cre- Ptch1+/−;p53−/− developed tumors at the highest frequency (60% at 500 days, Figure 1C–D). Tumors amongst most of these mouse lines distributed amongst the extremities, trunk and face, except that tumors did not occur on the face of Myf6Cre mice. A strong propensity for regional lymph node metastasis and distant metastases to the lungs was seen in all of these mouse lines (greater than two-thirds of mice which developed tumors).
Figure 1. Survival analysis and tumor histology of each genotype.
(A) Representation of prenatal and postnatal myogenesis in the context of Cre drivers used to activate tumorigenesis in this study. multi-nucl., multi-nucleated.
(B) Disease-free survival is shown as a Kaplan Meier curve for conditionally homozygous p53 deleted mice with or without conditional Patched1 (Ptch1) haploinsufficiency, using different Cre alleles.
(C–D) Comparison of disease–free survival in different Cre mice when p53 alleles were conditionally, homozygously deleted (B), or when p53 alleles were conditionally, homozygously deleted concurrently with heterozygous conditional Ptch1 deletion (C). The difference between p53 and Ptch1-p53 disease-free survival did not reach statistical significance by the log rank test (p>0.05).
(E) Histological classification. Mean latency of embryonal rhabdomyosarcoma (eRMS; red slices) is shown below each pie chart.
See also Figure S1 and Tables S1, S2 and S3.
Histological assessments were made by a pathologist specialized in pediatric and adult sarcoma pathology (author BPR). The histological appearance of eRMS varied from tumors composed of spindle cells without obvious rhabdomyoblastic differentiation to neoplasms composed of an admixture of primitive appearing to spindle shaped cells with variable numbers of epithelioid or spindle cell shaped rhabdomyoblasts. Occasional cases of eRMS exhibited cross-striations. Invariably, all cases of eRMS expressed Desmin and Myogenin or MyoD. UPS varied in histological appearance but did not contain cells with the histological appearance of rhabdomyoblasts and generally lacked unequivocal strong and or widespread Desmin, Myogenin and MyoD expression by immunohistochemistry. Cases for which the diagnosis was indeterminate between UPS and eRMS with possible focal rhabdomyosarcomatous differentiation were reviewed by two other sarcoma pathologists (authors ZY and DMP). For mice harboring only Cre-driven p53 deletions, tumors were soft tissue sarcomas except that half of Myf5Cre-p53 tumors were osteosarcoma arising from the ribs (Figure 1E). Occurrence of osteosarcoma in Myf5Cre-p53 mice is consistent with a previous report that embryonic Myf5 lineage contributes to rib morphogenesis(Haldar et al., 2008). Ptch1+/−;p53−/− mice of all Cre lineages had a tendency to develop tumors at a higher frequency than did p53−/− mice, although this trend did not reach statistical significance (Figure 1B). All Ptch1+/−;p53−/− tumors were soft tissue tumors except for osteosarcoma. Osteosarcoma rarely arose from Myf5Cre-Ptch1+ −;p53−/− mice (<10%) whereas it occurred in a larger proportion of Myf5Cre-p53−/− mice (Figure 1E). Histological diagnosis of the soft tissue tumors included a spectrum of myogenic malignancies, ranging from alveolar and embryonal RMS to UPS. Embryonal rhabdomyosarcoma (eRMS) arose from all Cre driver mice, suggesting that eRMS could occur in a range of myogenic lineages. Myf6Cre-p53−/− gave rise to the highest percentage of eRMS (100%) in the context of p53 deletion only (Figure 1E), and MCre was the second highest with 31% and 42% of the tumors diagnosed as eRMS in p53−/− and Ptch1+/−;p53−/−, respectively (Figure 1E). eRMS developed in 13% of Pax7CreER-Ptch1+/−;p53−/− mice but in none of Pax7CreER-p53−/− mice, most tumors of which were diagnosed as UPS (Figure 1E). This data suggests that PAX7+ satellite cells can give rise to eRMS, but their degree of myogenic differentiation is lower than tumors that arise from maturing myoblasts (the Myf6 lineage). Representative cases with eRMS histology are shown in Figures 2 and 3. These murine eRMS cases were also found to express markers commonly associated with human eRMS(Davicioni et al., 2006; Grass et al., 2009) that also allowed murine eRMS and murine aRMS to be distinguished by linear discriminate analysis (Figure S1A–C).
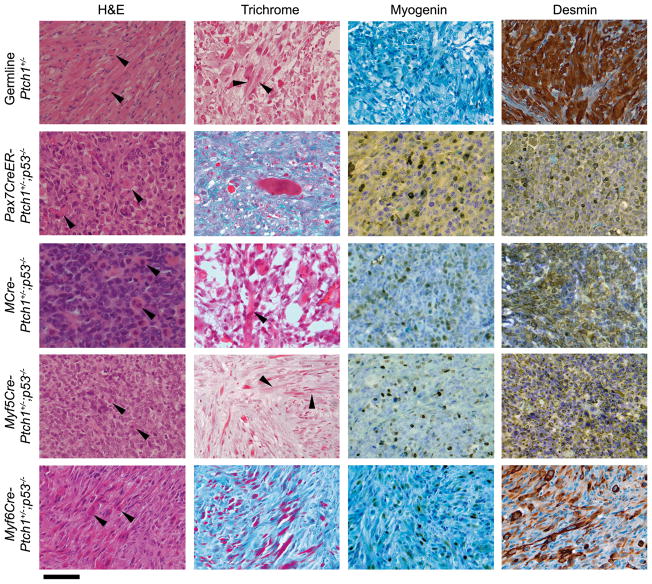
Figure 2. Histological analysis of Ptch1+/−;p53−/− eRMS for each Cre line.
Genotypes are indicated in column headings. A tumor from a mouse with germline deletion of the Ptch1 gene is shown as a typical case of eRMS (left column). H&E staining, Masson s trichrome, and immunohistochemical staining for Myogenin and Desmin were performed for diagnosis of all tumors. Arrowheads point to eosinophilic strap-like epithelioid rhabdomyoblasts in H&E panels. Cross striations are shown on trichrome (some cases; arrowheads). Scale bar, 40 μm.
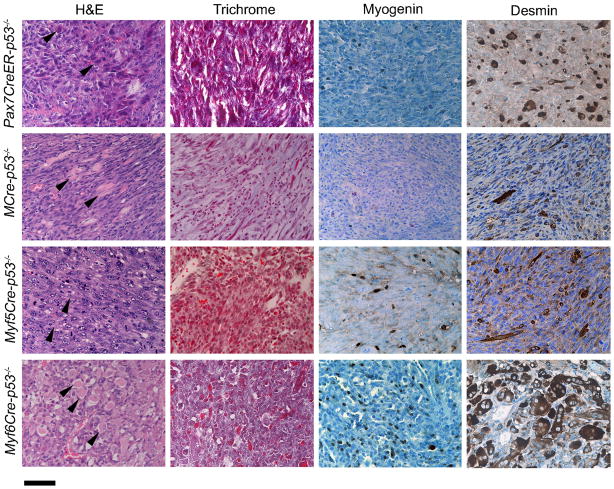
Figure 3. Histological analysis of p53−/− eRMS in each Cre line.
Representative cases with eRMS are presented. H&E staining, Masson s trichrome, and immunohistochemical staining of Myogenin and Desmin were performed for diagnosis of all tumors. Eosinophilic rhabdomyoblasts are highlighted by arrows. Scale bar, 40 μm.
Cell of Origin cannot be inferred from tumor phenotype
To examine whether tumors from a specific lineage continue to express the original myogenic markers when Cre recombinase was triggered, expression of Myf5, Pax7, Myf6, and Pax3 was studied by quantitative RT-PCR (Figure S1D). None of these genes was upregulated in the tumors from the original lineage compared to other groups of tumors (for example, Myf6 is not over-expressed in soft tissue tumors from Myf6Cre-Ptch1+/−;p53−/− or Myf6Cre-p53−/− mice compared to tumors of other mouse lines). Unsupervised global gene expression analysis also failed to demonstrate a relationship between cell of origin and molecular phenotype for fusion-negative (non-aRMS) sarcomas (Figure S1E). Results of pattern recognition analysis were similar (Table S1 showing high classification errors). These findings suggest that expression of myogenic markers inherent to the myogenic lineage that gave rise to tumors may not be retained after tumorigenic transformation. Therefore, expression of these markers does not reflect the cell of origin in rhabdomyosarcoma.
Rb1 absence is a modifier, leading to loss of differentiation of sarcomas
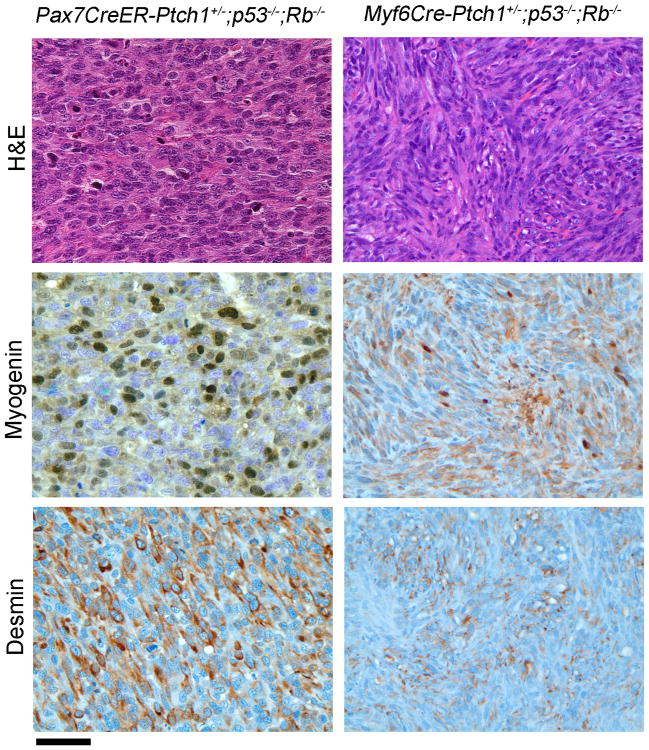
High frequency of retinoblastoma (Rb1) gene mutation has been reported in a subset of human rhabdomyosarcoma (Kohashi et al., 2008). To examine the effect of Rb1 loss on eRMS development, we inactivated both p53 and Rb1, with or without Ptch1 haploinsufficiency. Loss of Rb1 in the Myf5 or hypaxial Pax3 lineages (Myf5Cre and MCre, respectively) led to embryonic lethality, which is consistent with a previous report (Huh et al., 2004). When Rb1 was inactivated in Myf6 or Pax7 lineage using Myf6Cre or Pax7CreER mice, mice were born in normal Mendelian ratios and developed normally throughout adolescence and early adulthood. Unexpectedly, most of these mice developed silent pituitary corticotropinomas instead of rhabdomyosarcoma (Hosoyama et al., 2010), and less than 10% of these mice developed soft tissue tumors (data not shown). On histological analysis, these rare soft tissue tumors were usually composed of undifferentiated spindle cells without rhabdomyoblasts or other myogenic features. Only rare cases fulfilled the diagnostic criteria for eRMS or aRMS with Desmin and Myogenin or MyoD immunoreactivity. Two bona fide cases of eRMS from this cohort are shown in Figure 4. Myogenin and Desmin-positive tumor cells were far less frequent than Ptch1+/−;p53−/ − or p53−/− tumors with intact Rb1 (from any Cre driver). The most strongly staining tumors are shown in Figure 4 to emphasize that, in a subset of these UPS tumors, focal Myogenin and Desmin staining were sometimes present. Overall, these results suggest that Rb1 loss may lead to loss of differentiation in sarcomas. Unsupervised global gene expression analysis demonstrated a strong difference between tumors with intact versus deleted Rb1 (Figure S1F). Pattern recognition analysis similarly showed that the ability to discriminate between fusion-positive aRMS and fusion-negative soft tissue sarcomas was diminished when Rb1 was homozygously deleted (Table S2).
Figure 4. Histology of Ptch1+/−;p53−/−;Rb1−/− eRMS from Pax7CreER and Myf6Cre lines.
Shown are the areas with the most immunoreactivity. The majority of the tumors did not exhibit immunopositivity. Histologically, tumors appeared as either poorly differentiated epithelioid cell (left) or spindle cell (right) neoplasms and only showed rhabdomyoblastic differentiation by immunohistochemistry. Scale bar, 40 μm. See also Figure S2.
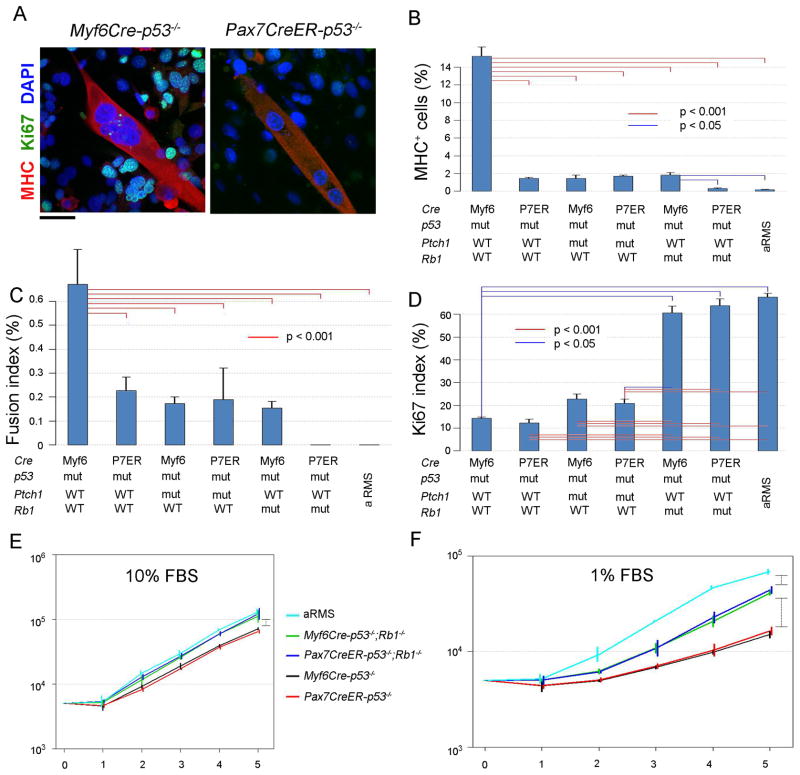
Tumors of the Myf6 lineage show the highest degree of in vitro myodifferentiation, which is negated by Rb1 loss
To examine the myogenic differentiation potential of tumors from different lineages, an in vitro differentiation assay was performed using primary tumor cell cultures (Figure 5). Tumor cells from Myf6Cre-p53−/−, Pax7CreER-p53−/−, Myf6Cre-Ptch1+/−;p53−/−, Pax7CreER-Ptch1+/−;p53−/−, Myf6Cre-p53−/−;Rb1−/−, Pax7CreER-p53−/−;Rb1−/−, and Myf6Cre-Pax3P3F/P3F; p53−/− mice were cultured under myogenic differentiation condition for 5 days. Immunofluorescent staining of Myosin heavy chain (MHC) shows that the number of MHC-positive cells was higher in Myf6Cre-p53−/− cells than in Pax7CreER-p53−/− cells (15 vs. 2%, Figure 5A,B). Fusion index defined as MHC-positive multi-nucleated cells was also higher in Myf6Cre-p53−/− cells than in Pax7CreER-p53−/− cells (Figure 5C). Concurrent Rb1 and p53 inactivation decreased MHC-positive cells and the fusion index in Pax7CreER-p53−/− cells (Figure 5B,C), supporting our previous observation that Rb1 loss may lead to an undifferentiated phenotype (Figure 4). To examine the relationships between growth ability and myodifferentiation, eRMS cells were stained for the S-phase marker Ki67. Myf6Cre-p53−/− and Pax7CreER-p53−/− cells showed similar percentages of Ki67 positivity (Figure 5D). On the other hand, Myf6Cre-p53−/−;Rb1−/− and Pax7CreER-p53−/−;Rb1−/− tumor cells also showed Ki67 positivity similar to each other but significantly higher than tumors with intact Rb1. The difference in cell growth rates between tumors with and without Rb1 deletion was accentuated under low serum conditions (Figure 5E,F). These results suggest that cell of origin does not affect cell proliferation; however, the difference in myogenic differentiation between Myf6Cre and Pax7CreER tumors may be linked to the degree of myogenic differentiation of the cell from which these tumors are derived rather than alterations in growth ability. The results also suggest that loss of Rb1 can increase tumor cell proliferation and reduce myogenic differentiation.
Figure 5. Cellular phenotypes of primary tumor cell cultures vary by cell of origin and mutation profile.
(A) Cell lines from both Myf6Cre-p53 and Pax7CreER-p53 tumors formed multinuclear myotubes, which were positive for myosin heavy chain (MHC). scale bar, 40 μm.
(B) Percentage of MHC-positive cells was calculated in each cell line. Total cells in 5 views of 100x fields were counted in each cell line. WT, wild type. mut, mutant. aRMS, alveolar rhabdomyosarcoma tumors from Myf6Cre-Pax3P3F/P3F;p53−/− mice.
(C) Fusion index (percentage of cells with MHC-positive (MHC+) multi-nuclear myotubes /total cells) was calculated.
(D) Ki67 index (percentage of Ki67-positive cells /total cells) was calculated.
(E–F) In vitro growth properties of each cell line under normal (10% fetal bovine serum (FBS), E) and low (1% FBS, F) serum conditions. Vertically oriented brackets represent comparisons for which p values are < 0.001.
For all panels in this figure, p values represents ANOVA with Tukey's multiple testing correction. For Figure 5B–C, values were log transformed. Rank order was used for Figure 5D. Error bars represent SEM.
An additional interesting result was that all eRMS primary cell cultures showed a greater capacity for terminal myogenic differentiation than Myf6Cre-Pax3P3F/P3F; p53−/− aRMS primary cell cultures. This result is consistent with reports of human RMS patients treated with multimodal therapies (chemotherapy, radiation), whereby eRMS tumors have been demonstrated to show features of myodifferentiation in association with treatment effect, but aRMS fail to undergo cytodifferentiation after therapy(Smith et al., 2002).
eRMS and UPS are part of a tumor continuum
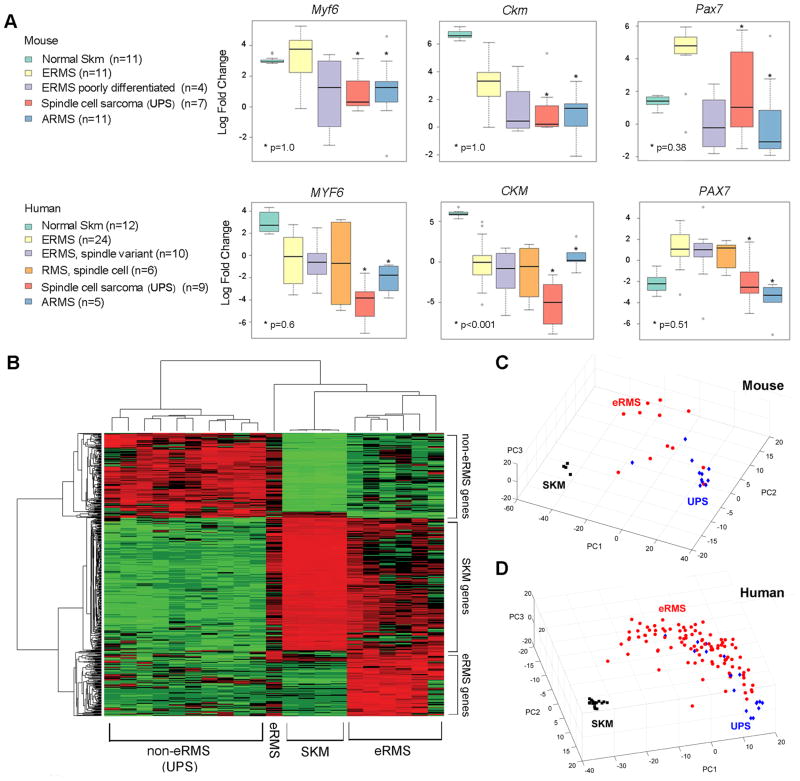
Having compared the myodifferentiation capacity of eRMS by cell of origin relative to aRMS, we next explored in more detail the observation that Pax7CreER-Ptch1+/−;p53−/− tumors were often morphologically similar but showed varying degrees of phenotypic differentiation leading to the pathological diagnosis of either eRMS or UPS. To this end, we examined markers of myodifferentiation among Ptch1+/−;p53−/− or p53−/− mouse tumors from any cell of origin, correlating cohorts of tumors diagnosed by the pathologists as eRMS or undifferentiated spindle cell sarcoma (i.e., UPS), based upon histological features with Myogenin or Desmin staining. UPS tumors expressed significantly lower levels of satellite cell, myoblast, and myofiber marker genes Pax7, Myf6 and Ckm (muscle creatine kinase) than eRMS, although murine aRMS and murine UPS were not statistically different for these genes. Amongst human pediatric fusion-negative soft tissue sarcomas, a decreasing gradient was again seen for these markers in eRMS, eRMS spindle cell variant, non-eRMS spindle cell RMS, and UPS, respectively (Figure 6A). Thus, eRMS and UPS appear to lie in a continuum of myodifferentiation.
Figure 6. Continuum of Gene Expression Features in Mouse and Human Sarcomas.
(A) Expression of myogenic markers in mouse and human SKM, eRMS, UPS and intermediate diagnoses. aRMS is shown in comparison. Mouse genes have the first letter capitalized, whereas human genes have all letters capitalized. Error bars represent SEM.
(B) Supervised clustering identified the genes specifically up-regulated in each group, eRMS, non--eRMS (UPS), and skeletal muscles (SKM) (raw p-value <0.05 & fold change>2). A gene list of eRMS specific genes is shown as Table S2. Tumors analyzed are Ptch1,p53 tumors without Rb1 deletions from Pax7CreER, MCre, Myf5Cre, and Myf6Cre lineages.
(C) Principal component analysis for mouse tumors using the 345 signature genes that differentiated ERMS (red) and UPS (blue) mouse tumors from normal skeletal muscle (black).
(D) Principal component analysis for human RMS, UPS and normal skeletal muscle samples using 283 significant probes that reached significance (p-value < 0.05 & fold change > 1.5).
To further characterize the gene expression profile specific to tumors histologically diagnosed as eRMS, we performed microarray analysis for histologically confirmed murine eRMS, murine UPS (non-eRMS), and skeletal muscle of 4 week old (juvenile) mice. Expression of select genes was validated by quantitative RT-PCR. Supervised clustering of microarray data identified 2 gene clusters whose increased expression was associated with eRMS and non-ERMS, respectively (Figure 6B; Table S3). Supervised clustering that differentiated tumors with and without rhabdomyoblasts showed a pattern similar to clustering of eRMS vs. non-eRMS, respectively (data not shown). A third gene cluster was shared between skeletal muscle (SKM) and eRMS. Principal component analysis of both mouse tumors (Figure 6C) and human fusion negative soft tissue sarcomas including eRMS (Figure 6D) supports the assertion that eRMS and UPS lie in a continuum.
Five genes differentiates skeletal muscle, eRMS, UPS and aRMS
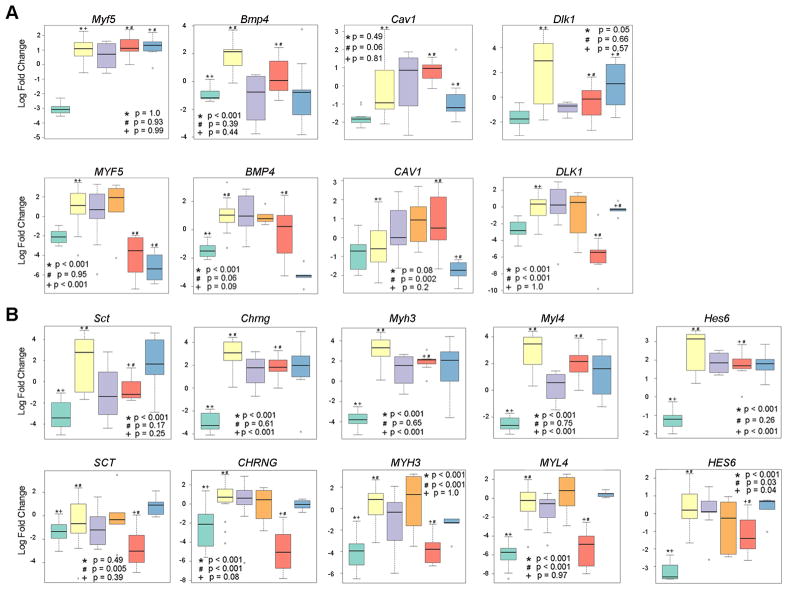
To determine whether a profile of a limited number of mRNA levels could be used diagnostically to differentiate human eRMS, UPS and aRMS, we used selected RT-PCR validations of gene expression data from results of microarray studies in Figure 6B to test possible tumor classification profiles. Using a K-nearest neighbors and Support Vector Machines methods to identify a reduced number of gene expression classifiers and a profile of 5 genes was developed (BMP4, MYF5, DLK1, PAX7, CAV1)(Figure 7A). Using Leave-One-Out cross validation, the 5 gene profile agreed with Pediatric CHTN pathologist diagnoses for 12 out of 12 human SKM, 23 out of 24 eRMS, 7 out of 9 UPS and 5 out of 5 aRMS (Table S4). This profile, however, classified 7 out of 10 spindle cell variant eRMS as classical eRMS, and predicted 4 out of 6 spindle cell rhabdomyosarcoma as eRMS. Overall sensitivities for these tumor types were 78–100% and specificities were 71–98% (Table S5).
Figure 7. Gene Expression Classifiers and Phenotypes in Mouse and Human Sarcomas.
(A) Expression of genes that as a profile differentiate SKM, eRMS, USCS/UPS and aRMS in humans, with corresponding graphs from mouse models. Pax7 is included in this five gene profile, but Pax7 expression data is presented in Figure 6A.
(B) Markers of activated satellite cells found to be highly expressed in mouse and human eRMS. Mouse genes have the first letter capitalized, whereas human genes have all letters capitalized. See also Figure S2.
Error bars represent SEM. See also Tables S4 and S5.
eRMS have a unique phenotype resembling an activated satellite cell program
Results of the preceding microarray analysis, RT-PCR validations and classification schemas not only reflected that eRMS expressed genes associated with myogenesis as well as previously reported RMS-related genes, but that eRMS harbored an activated satellite cell program. In general, genes whose increased expression was associated specifically with eRMS includes Secretin (Sct; Figure 7B), a gastrointestinal peptide expressed in the inner circular intestinal wall muscle of the developing mouse fetus(Siu et al., 2005), and the gamma (fetal) nicotinic cholinergic receptor (Chrng; Figure 7B), which was previously reported to be expressed in a high percentage of eRMS and aRMS(Gattenloehner et al., 1999). Other eRMS specific genes include embryonic myosin heavy and light chains (Myh3 and Myl4, respectively; Figure 7B), which are in keeping with an embryonic muscle development phenotype. Most interesting, however, is the number of genes shared between eRMS and activated satellite cells(Fukada et al., 2007). These shared genes include Pax7 (Figure 6A), Sct, Myh3, Chrng, cardiac troponin T2 (Tnnt2), hairy and enhancer of split 6 (Hes6; Figure 7B), RIKEN cDNA 1110002H13 gene, Otogelin (Otog), low density lipoprotein receptor-related protein 4 (Lrp4), guanine nucleotide binding protein alpha inhibiting 1 (Gnai1), unc-5 homolog B (Unc5b; Figure S2), MyoD, distal-less homeobox 2 (Dlx2), and Rho GDP dissociation inhibitor gamma (Arhgdig). Overall, eRMS-specific gene expression show great similarity to activated satellite cell gene expression.
Human eRMS and fusion-negative sarcomas have altered p53, Shh/Ptch1 and Rb1 signatures
To determine the relevance of our mouse sarcoma models to human disease, we examined the gene expression profiles of 111 primary human fusion-negative rhabdomyosarcomas and related cell lines for which microarray datasets were available(Davicioni et al., 2006; Lae et al., 2007).
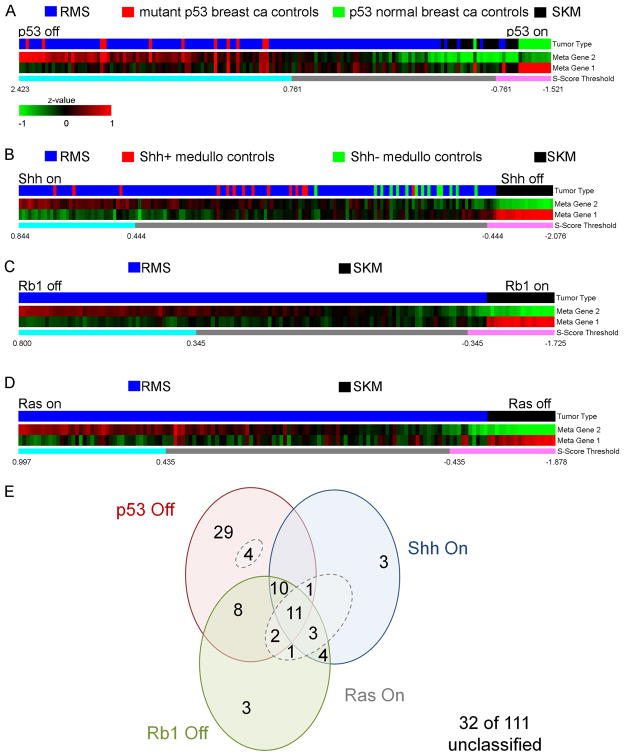
Individual p53 off, Shh on, Rb1 off and Ras on signatures are presented in Figures 8A–D, Tables S6–S9 and Figure S3. A global examination is given in Figure 8E. From this limited cohort of 111 fusion-negative soft tissue sarcoma samples (mostly diagnosed as eRMS by Children's Oncology Group central review or the Triche laboratory), the predominant feature is p53 loss in 59% of samples. Of that 59%, nearly half exhibited only the p53 off signature. Both Shh on and Rb1 off signatures were rare and often concurrent with one or more other signatures. Nineteen percent of tumors exhibited all three signatures. Surprisingly, a Ras signature was not seen alone in any tumor but existed together with at least one other signature in 21% of samples (Figure 8E; Table S10 and Figure S3). Clinical covariates by signature status are presented in Table S10. Too few samples were present to draw any conclusion on signature versus disease stage at diagnosis or survival; however, an interesting finding is that 3 of 4 undifferentiated spindle cell sarcoma (i.e., UPS) samples exhibited an Rb1 off signature. Also of note, 32 of 111 samples (29%) exhibited no p53, Shh, or Rb1 signature.
Figure 8. Heatmaps of human fusion-negative rhabdomyosarcomas for gene signatures representing the p53, Shh and Rb1 pathways.
(A) The p53 pathway heatmap represented as two metagenes was generated based on 320 significant genes (Benjamini and Hochberg adjusted p-value <0.05 & fold change > 2) with 183 genes were in metagene 1, whereas the other 136 genes were in metagene 2. In total, 65 of 111 fusion-negative RMS primary tumor samples (59%) exhibited a gene expression signature consistent with the p53 off state for which the S-score was greater than 0.761 (see Supplemental Experimental Procedures). All human breast cancer control samples with known p53 mutations also exhibited S-scores greater than 0.761.
(B) For the Shh signaling pathway heatmap, 111 genes were employed, 56 of which were in metagene 1, the other 55 genes were in metagene 2. Overall, 32 of 111 (29%) of tumors exhibited a gene expression signature consistent with a Shh on overdrive state for which S-score was greater than 0.444. The p-value for comparison of the S-score for Shh+ medulloblastoma controls and for Shh- medulloblastoma controls was 2.62x10−5 as calculated by the Wilcoxon rank sum test.
(C) For the Rb1 pathway, 381 genes were used to construct the heatmap, with 157 in metagene 1, and 224 genes in metagene 2. For the Rb1 pathway, 42 of 111 (38%) of samples demonstrated an Rb1 off state with an S-score greater than 0.345.
(D) To evaluate the Ras pathway, 87 genes common to zebrafish eRMS and human Ras-driven pancreatic cancer were used to construct the heatmap, with 24 genes in metagene 1, and 63 genes in metagene 2. For the Ras pathway, 25 of 111 of samples demonstrated a Ras on state with an S-score greater than 0.477, but after exclusion of 2 samples that did not also contain a Ras signature common to Ras-activated mammary epithelial cells, only 23 of 111 samples (21%) were felt to have a strict Ras signature. None of the Ras signature samples (breast similar or pancreatic similar) had a Ras activation signature in the absence of mutant p53, Shh or Rb1 signatures.
(E) Venn diagram showing the intersection of signatures amongst human fusion-negative soft tissue sarcomas. ca, cancer. Ras On samples are encircled with dotted lines.
See also Figure S3 and Tables S6–S10.
DISCUSSION
A great deal of research on rhabdomyosarcomas is given to aRMS, which is associated with especially poor prognosis when metastatic (<20% long term survival)(Breneman et al., 2003), whereas comparatively less attention is given to eRMS, for which long term survival when metastatic is still merely 40%(Breneman et al., 2003). Even lower attention has been given to UPS, although a few promising studies are emerging(Nakayama et al., 2007). In the current study we have examined several key aspects of eRMS and UPS biology. We have demonstrated that embryonal rhabdomyosarcoma, spindle cell rhabdomyosarcoma, and UPS lie in a continuum. Interestingly, the spindle cell subtype of embryonal rhabdomyosarcoma was originally identified as a well-differentiated subtype that occurred predominantly in children and had a favorable prognosis(Leuschner et al., 1993). However, subsequent studies have shown that spindle cell rhabdomyosarcomas can be very aggressive, especially in adults. The murine spindle cell rhabdomyosarcomas identified in our study appear to have more akin to spindle cell rhabdomyosarcomas in adults. While rhabdomyosarcoma can arise from various stages of myogenic differentiation ranging from satellite cells to maturing myoblasts, the proportion of rhabdomyosarcomas versus undifferentiated spindle cell sarcomas (i.e., UPS) varies both by cell/lineage of origin and mutational spectrum. We have shown that p53 and/or Ptch1 mouse models give rise to histologically and immunohistochemically authentic eRMS tumors with the same molecular expression profile as human eRMS, whereas Rb1 loss reduces the myodifferentiation potential of these tumor models and increases proliferative capacity to the level seen in aRMS. By global gene expression analysis, we uncoupled the gene expression signature for eRMS from that of UPS in the mouse, demonstrating that the former is consistent with an activated satellite cell phenotype. By examining human Pax:Fkhr fusion negative rhabdomyosarcomas, we have established that p53 loss of function, Shh gain of function, and Rb1 loss of function gene expression signatures are highly prevalent in human primary eRMS tumors and often are present concurrently. Thus, our mouse models are pertinent to specific subsets of human fusion negative soft tissue sarcomas.
A continuum exists between eRMS and UPS
Features of myogenic differentiation distinguish eRMS from other forms of sarcoma, although common disagreement about classification may be based more in biology than previously realized. Historical discordance rates between institutional and centralized Intergroup Rhabdomyosarcoma Study Group (IRSG) diagnoses of eRMS NOS, spindle cell eRMS, and undifferentiated sarcoma have been reported at 19, 97 and 37%, respectively(Qualman et al., 1998), although the diagnosis of undifferentiated sarcoma has become much more refined than in this past era due to the ability to identify even focal differentiation by improved immunostains. The relatedness of this family of sarcomas is also reflected in the fact that eRMS and Pax:Fkhr fusion-negative aRMS are difficult to discern from one another using gene expression and comparative genomic hybridization tools (Davicioni et al., 2009; Wexler and Ladanyi; Williamson et al.). Conversely, ultrastructural studies of UPS suggest features in common with myofibroblastic differentiation(Suh et al., 2000). The experimental results of our current study suggest that at least a subset of UPS and eRMS may share a common myogenic cell (or cells) of origin, but that UPS are more likely to evolve from Pax7-expressing satellite cells (muscle stem cells). From this common muscle stem cell of origin, divergence of maturation possibly accounts for the continuum of tumor cell phenotypes that was observed in this study. Recognizing the continuum from UPS to eRMS, one might prospectively be able to use the molecular signatures for UPS and eRMS obtained from this study to enhance the classification and to improve the ability to predict prognosis and response to molecularly targeted therapies. Our first attempt at this effort generated a 5 gene expression profile (BMP4, MYF5, DLK1, PAX7, CAV1) that correctly classified human eRMS vs. UPS vs. aRMS with high sensitivity and good specificity; however, to prove this profile s clinical value, prospective validation studies must be performed.
Rhabdomyosarcomas can arise from myogenic precursors, but not a single cell type or lineage of origin
An important question in the field is, “what is the cell of origin of rhabdomyosarcoma?”(Hettmer and Wagers). Epidemiology indicates that the incidence of eRMS increases from toddler (classical eRMS) to adolescent ages (spindle cell variant) (Ognjanovic et al., 2009; Pastore et al., 2006), periods when muscle growth accelerates and satellite cell activity may be highest. By contrast, aRMS incidence does not vary by age or sex(Ognjanovic et al., 2009; Pastore et al., 2006).
It is reasonable to expect that rhabdomyosarcoma tumor cell phenotypes might imply cell of origin. Indeed, some have suggested that human eRMS arise from satellite cells because these tumors express the satellite cell marker Pax7(Tiffin et al., 2003). Based upon expression patterns in mouse tumors, other investigators have similarly postulated a (neural crest) cell of origin for double mutant NF1 and p53 associated rhabdomyosarcoma (Vogel et al., 1999). However, a satellite cell phenotype for some forms of rhabdomyosarcomas is supported experimentally for tumor models driven by the Ras pathway(Langenau et al., 2007).
Further evidence that eRMS may arise from satellite cells is supported by the observation that 40% of eRMS arise in the orbit(Gurney et al., 1999), and that cranial muscle, including the orbit, has a higher density of satellite cells than most non-cranial hypaxial or epaxial muscle tissue(McLoon et al., 2004). Interestingly, the extra-ocular muscles have a very high density of muscle stem cells (satellite cells) that are continuously undergoing proliferation and renewal(McLoon et al., 2004) and are known to be more resilient and less prone to the effects of muscle dystrophy than non-cranial muscles(Porter et al., 2003). Head & neck tumors, but not orbital ones, exhibit the bimodal increased age incidences described above, probably because of the increase in spindle cell variant eRMS during adolescence(Pastore et al., 2006). Genitourinary sites exhibit a strong age distribution in 0–3 year old children, which is puzzling at first because these sites are generally not associated with either skeletal muscle or satellite cells. However, revealing contemporary reports of the presence of satellite cells in the urethral rhabdosphincter are emerging (Sumino et al., 2007), and the presence of similar cells in other regions of the bladder, the prostate and biliary tract are intriguing.
Our results show that Pax7-expressing murine satellite cells indeed can be the cell of origin of eRMS, although more differentiated, Myf6-expressing myoblasts are also capable of forming eRMS with approximately the same latency. Given the clinical presentations of rhabdomyosarcoma arising from satellite cell-rich muscles at corresponding age periods of muscle growth, one can conclude that satellite cells are a contributor to eRMS in humans - but maybe not the major (or only) cell of origin. Indeed, other cells of origin may not (initially) be myogenic at all. In an example inferred for alveolar rhabdomyosarcoma, Lagha and colleagues reported that in the mouse embryonic dermamyotome, the ratio of Pax3 to Foxc2 balances fate between myogenic and vascular cell fates, but that Pax3:Fkhr suppresses Foxc2 (Lagha et al., 2009). This latter result suggests that, in the susceptible cell, an angiogenic precursor might be converted to a myogenic state, a result which has been shown experimentally for embryonic aortic mesangioblasts (Messina et al., 2009). Therefore, our demonstration of susceptibility of myogenic precursors to embryonal rhabdomyosarcomagenesis may be only the beginning of a very interesting biological story about the plasticity of different cell lineages.
Mutational profile determines tumor phenotype
Many pathways lead to eRMS, but the most well known pathway is the germline disruption of p53 in Li-Fraumeni syndrome (LFS). Originally described as familial RMS(Li and Fraumeni, 1969), LFS is commonly associated with eRMS, although histologically diagnosed alveolar rhabdomyosarcoma (for which Pax:Fkhr fusion status is often unknown) can also occur in LFS(Diller et al., 1995). Our mouse model studies suggest that p53 loss can give rise to pure eRMS if derived from a Myf6-expressing maturing myoblast cell of origin, but not from a satellite cell of origin. However, our studies of human tumors suggest that a p53 loss of function signature is extremely prevalent in fusion negative- sarcomas and eRMS (as much as 59% of tumors). One of the potentially most clinically relevant findings from our mouse model studies is that mouse tumors with a p53 off signature only and a Myf6 lineage origin may have the highest potential for “differentiation” therapy using agents other than chemotherapy or radiation. Prospective mutational profiling and ex vivo myodifferentiation assays may identify patients who are candidates for this approach. An important caveat is that Myf6 expression itself would not be appropriate for identifying tumors derived from a Myf6 expressing cell of origin, because as we show from our RT-PCR assays (Figure S2), tumor phenotype does not reflect cell of origin.
A role for the Shh/Ptch1 signaling pathway in eRMS is implicated by the association of human congenital Ptch1 haploinsufficiency with the occurrence of cardiac rhabdomyoma and embryonal rhabdomyosarcoma(Beddis et al., 1983; Gorlin, 1987; Tostar et al., 2006). Additional supporting evidence for Shh playing a role in a subset of eRMS is provided by observational studies using human tumors wherein expression of the Shh signaling cascade are altered (Oue et al.; Tostar et al., 2006). The initial experimental demonstration of Ptch1 haploinsufficiency being linked to eRMS tumor initiation was reported by Hahn and colleagues(Hahn et al., 1998). A persistent role for Shh overdrive in tumor progression of Ptch1 haploinsufficient mice is suggested by Ptch1 reporter expression in murine rhabdomyosarcoma(Gerber et al., 2007). The results of our paper further indicate that Ptch1 haploinsufficiency can contribute to tumor initiation from myogenic cells of origin at every level of differentiation, which is not the case p53 alone is lost in Pax7-expressing satellite cells or Myf5expressing myoblasts. In our analysis of human fusion-negative sarcomas and eRMS, evidence of Shh overdrive was present in 29% of tumors, but almost always in association with p53 or Rb1 gene signatures. Thus, Ptch1 does play an important role in tumor initiation and neoplastic myogenesis; however, in sporadic tumors Ptch1 does not often appear to act in isolation. For this reason, the Pax7CreER-Ptch1-p53 mouse model may be a very good preclinical tool for studying the spindle cell variant of eRMS.
While Ras has been implicated in pediatric eRMS (Chen et al., 2006; Martinelli et al., 2009; Stratton et al., 1989), the surprising result that an activated Ras signature was only present in combination with other mutant signatures is consistent with the demonstrated role of Ras at initiation of adult pleomorphic rhabdomyosarcoma(Doyle et al., 2010; Tsumura et al., 2006) but suggests that Ras may only be a modifier in pediatric eRMS. We were struck by the contrast between the 10% of human tumors that expressed all 4 mutant signatures versus the 28% tumors that only coveted a p53 off signature. Therein may exist an opportunity for molecular stratification (and intervention) in eRMS.
Perhaps the most interesting observation is that we could not identify a signature for 29% of human fusion-negative soft tissue sarcoma, underlining the possibility that mutations independent of p53 have yet to be discovered and modeled.
Rb1 is a modifier in eRMS
Early studies suggested that Rb1 abnormalities rarely occur in eRMS or aRMS(De Chiara et al., 1993), but a recent study reports that Rb1 allelic imbalance occurred in 13 of 27 eRMS tested (but much less frequently than in aRMS(Kohashi et al., 2008)). In a second contemporary series, 6 of 36 eRMS lacked pRb1 staining on IHC(Takahashi et al., 2004). Other supporting evidence for pRb family proteins being implicated in eRMS comes from the observation of SV40 Tag expression from a LFS eRMS(Malkin et al., 2001). However, among patients with hereditary Rb1 mutations, rhabdomyosarcomas reported to have occurred were more often categorized as RMS “Not Otherwise Specified (NOS)” than eRMS (note that NOS may imply inadequate tissue rather than diagnostic uncertainty) (Kleinerman et al., 2007). Our studies in mouse models suggest that while Rb1 loss of function alone does not lead to tumor initiation, Rb1 loss of function in combination with other oncogenic factors is strongly associated with an undifferentiated phenotype, wherein myogenic marker expression is reduced or absent. Tumor cell proliferation (Ki67 positivity) and myodifferentiation capacity under low serum conditions) were also severely altered. Therefore, Rb1 is best characterized as a modifier of phenotype in eRMS.
eRMS share features of activated satellite cells
Perhaps the most fascinating aspect of this study is the high concordance of gene expression between murine eRMS and murine activated satellite cells–regardless of whether cell of origin is a satellite cell or a maturing myoblast. The data suggests that eRMS may represent a myogenic precursor state (as opposed to an opportunistic expression of early myogenic markers), and that treatment might be guided by this principle. Past clinical studies indirectly support this assertion, that in eRMS (but not aRMS) myodifferentiation of tumor cells correlates with response to treatment(Smith et al., 2002), and differentiated tumor cells (rhabdomyoblasts) do not cause relapse even if present many years(Arndt et al., 2006). Thus, we speculate that specific subsets of eRMS that are phenotypically similar to activated satellite cells are poised for differentiation.
EXPERIMENTAL PROCEDURES
Mice
All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC) at the Univ. of Texas Health Science Center at San Antonio. Detailed descriptions of mouse lines and repository status are given in Supplemental Experimental Procedures.
Human Tissue Samples
De-identified human samples were obtained with approval from the UTHSCSA institutional review board via the Pediatric Cooperative Human Tissue Network, Columbus, OH. Fusion status of aRMS samples was performed as previously described (Barr et al., 2006).
Pathology
Methods for histopathological analysis are given in the Supplemental Materials. Slides were sequentially reviewed by three pathologists (BR, ZY, and DP) and concurrence reached in all cases regarding diagnosis of eRMS. Diagnostic criteria of eRMS as well as the procedures for primary cell cultures are given in Supplemental Materials.
Microarray Gene Expression Analysis
Mouse gene expression microarray data were generated using Illumina Mouse Ref-8 BeadChip v1.1 (Illumina, San Diego, CA). Datasets were deposited in the GEO database (GSE22520). Detailed analysis procedures and methods for RT-PCR validation assays are given in the Supplemental Materials.
SIGNIFICANCE.
Histology-directed risk stratification is the basis for treating rhabdomyosarcoma in children, yet a spectrum of histopathological phenotypes exist for the embryonal subtype of this tumor, some of which can only be discerned from a broad family of undifferentiated pleomorphic sarcoma by an incremental increase in myogenic marker expression. Here, we demonstrate that embryonal rhabdomyosarcoma and some undifferentiated pleomorphic sarcomas are a continuum of disease, often with similar mutational profiles, but generally arising from different cell(s) of origin. Despite embryonal rhabdomyosarcomas having an activated satellite cell phenotype, these tumors are most likely to arise from differentiating myoblasts. Importantly, we highlight a subset of p53 deficient eRMS arising from Myf6-expressing myoblasts that are most likely to respond to therapeutic myodifferentiation approaches
HIGHLIGHTS.
Embryonal rhabdomyosarcoma & undifferentiated pleomorphic sarcoma form a continuum
Mutant p53, Ptch1 or Rb1 in satellite cells gives rise to undifferentiated sarcoma
Embryonal rhabdomyosarcoma arise from myoblasts but express satellite cell markers
Rb1 loss acts as an apparent modifier for sarcomas by inducing de-differentiation
Supplementary Material
Acknowledgments
Trainee and research support were provided by the Scott Carter Foundation as well K99NS064171-02 and 5R01CA133229-04. Human tissue samples were provided by the NCI-supported Cooperative Human Tissue Network. The NICHD-supported DSHB is maintained by The Univ. of Iowa. Authors thank Maricela Castillo for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arndt CA, Hammond S, Rodeberg D, Qualman S. Significance of persistent mature rhabdomyoblasts in bladder/prostate rhabdomyosarcoma: Results from IRS IV. J Pediatr Hematol Oncol. 2006;28:563–567. doi: 10.1097/01.mph.0000212978.21372.97. [DOI] [PubMed] [Google Scholar]
- Barr FG, Smith LM, Lynch JC, Strzelecki D, Parham DM, Qualman SJ, Breitfeld PP. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: a report from the Children's Oncology Group. J Mol Diagn. 2006;8:202–208. doi: 10.2353/jmoldx.2006.050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddis IR, Mott MG, Bullimore J. Case report: nasopharyngeal rhabdomyosarcoma and Gorlin's naevoid basal cell carcinoma syndrome. Med Pediatr Oncol. 1983;11:178–179. doi: 10.1002/mpo.2950110309. [DOI] [PubMed] [Google Scholar]
- Breneman JC, Lyden E, Pappo AS, Link MP, Anderson JR, Parham DM, Qualman SJ, Wharam MD, Donaldson SS, Maurer HM, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma--a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21:78–84. doi: 10.1200/JCO.2003.06.129. [DOI] [PubMed] [Google Scholar]
- Brown CB, Engleka KA, Wenning J, Min Lu M, Epstein JA. Identification of a hypaxial somite enhancer element regulating Pax3 expression in migrating myoblasts and characterization of hypaxial muscle Cre transgenic mice. Genesis. 2005;41:202–209. doi: 10.1002/gene.20116. [DOI] [PubMed] [Google Scholar]
- Chen Y, Takita J, Hiwatari M, Igarashi T, Hanada R, Kikuchi A, Hongo T, Taki T, Ogasawara M, Shimada A, Hayashi Y. Mutations of the PTPN11 and RAS genes in rhabdomyosarcoma and pediatric hematological malignancies. Genes Chromosomes Cancer. 2006;45:583–591. doi: 10.1002/gcc.20322. [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, Schofield DE, Buckley JD, Meyer WH, Sorensen PH, Triche TJ. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol. 2009;174:550–564. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- De Chiara A, T'Ang A, Triche TJ. Expression of the retinoblastoma susceptibility gene in childhood rhabdomyosarcomas. J Natl Cancer Inst. 1993;85:152–157. doi: 10.1093/jnci/85.2.152. [DOI] [PubMed] [Google Scholar]
- Diller L, Sexsmith E, Gottlieb A, Li FP, Malkin D. Germline p53 mutations are frequently detected in young children with rhabdomyosarcoma. J Clin Invest. 1995;95:1606–1611. doi: 10.1172/JCI117834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J Pathol. 2010 doi: 10.1002/path.2748. [DOI] [PubMed] [Google Scholar]
- Fletcher CDMFKU, Mertens F. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2002. Pleomorphic malignant fibrous histiocytoma/undifferentiated high grade pleomorphic sarcoma. In World Health Organization Classification of Tumours. [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gattenloehner S, Dockhorn-Dworniczak B, Leuschner I, Vincent A, Muller-Hermelink HK, Marx A. A comparison of MyoD1 and fetal acetylcholine receptor expression in childhood tumors and normal tissues: implications for the molecular diagnosis of minimal disease in rhabdomyosarcomas. J Mol Diagn. 1999;1:23–31. doi: 10.1016/S1525-1578(10)60605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26:1122–1136. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ. Nevoid basal-cell carcinoma syndrome. Medicine (Baltimore) 1987;66:98–113. doi: 10.1097/00005792-198703000-00002. [DOI] [PubMed] [Google Scholar]
- Grass B, Wachtel M, Behnke S, Leuschner I, Niggli FK, Schafer BW. Immunohistochemical detection of EGFR, fibrillin-2, P-cadherin and AP2beta as biomarkers for rhabdomyosarcoma diagnostics. Histopathology. 2009;54:873–879. doi: 10.1111/j.1365-2559.2009.03303.x. [DOI] [PubMed] [Google Scholar]
- Gurney JG, Young JL, Jr, Roffers SD, Smith MA, Bunin GR. In: Soft Tissue Sarcomas. In Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995, National Cancer Institute, SEER Program. NIH Pub. No. 99–4649. Ries LAG SM, Gurney JG, Linet M, Tamra T, Young JL, Bunin GR, editors. Bethesda, MD: National Cancer Institute; 1999. [Google Scholar]
- Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5- independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmer S, Wagers AJ. Muscling in: Uncovering the origins of rhabdomyosarcoma. Nat Med. 16:171–173. doi: 10.1038/nm0210-171. [DOI] [PubMed] [Google Scholar]
- Hosoyama T, Nishijo K, Garcia MM, Schaffer BS, Ohshima-Hosoyama S, Prajapati SI, Davis MD, Grant WF, Scheithauer BW, Marks DL, et al. A Postnatal Pax7+ Progenitor Gives Rise to Pituitary Adenomas. Genes & Cancer. 2010;1:388–402. doi: 10.1177/1947601910370979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. The Journal of cell biology. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinerman RA, Tucker MA, Abramson DH, Seddon JM, Tarone RE, Fraumeni JF., Jr Risk of soft tissue sarcomas by individual subtype in survivors of hereditary retinoblastoma. J Natl Cancer Inst. 2007;99:24–31. doi: 10.1093/jnci/djk002. [DOI] [PubMed] [Google Scholar]
- Kohashi K, Oda Y, Yamamoto H, Tamiya S, Takahira T, Takahashi Y, Tajiri T, Taguchi T, Suita S, Tsuneyoshi M. Alterations of RB1 gene in embryonal and alveolar rhabdomyosarcoma: special reference to utility of pRB immunoreactivity in differential diagnosis of rhabdomyosarcoma subtype. J Cancer Res Clin Oncol. 2008;134:1097–1103. doi: 10.1007/s00432-008-0385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lae M, Ahn EH, Mercado GE, Chuai S, Edgar M, Pawel BR, Olshen A, Barr FG, Ladanyi M. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX- FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212:143–151. doi: 10.1002/path.2170. [DOI] [PubMed] [Google Scholar]
- Lagha M, Brunelli S, Messina G, Cumano A, Kume T, Relaix F, Buckingham ME. Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev Cell. 2009;17:892–899. doi: 10.1016/j.devcel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner I, Newton WA, Jr, Schmidt D, Sachs N, Asmar L, Hamoudi A, Harms D, Maurer HM. Spindle cell variants of embryonal rhabdomyosarcoma in the paratesticular region. A report of the Intergroup Rhabdomyosarcoma Study. Am J Surg Pathol. 1993;17:221–230. doi: 10.1097/00000478-199303000-00002. [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF., Jr Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst. 1969;43:1365–1373. [PubMed] [Google Scholar]
- Malkin D, Chilton-MacNeill S, Meister LA, Sexsmith E, Diller L, Garcea RL. Tissue-specific expression of SV40 in tumors associated with the Li-Fraumeni syndrome. Oncogene. 2001;20:4441–4449. doi: 10.1038/sj.onc.1204583. [DOI] [PubMed] [Google Scholar]
- Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M, Dominici C. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer. 2009;48:975–982. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- McLoon LK, Rowe J, Wirtschafter J, McCormick KM. Continuous myofiber remodeling in uninjured extraocular myofibers: myonuclear turnover and evidence for apoptosis. Muscle Nerve. 2004;29:707–715. doi: 10.1002/mus.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina G, Sirabella D, Monteverde S, Galvez BG, Tonlorenzi R, Schnapp E, De Angelis L, Brunelli S, Relaix F, Buckingham M, Cossu G. Skeletal muscle differentiation of embryonic mesoangioblasts requires pax3 activity. Stem Cells. 2009;27:157–164. doi: 10.1634/stemcells.2008-0503. [DOI] [PubMed] [Google Scholar]
- Morotti RA, Nicol KK, Parham DM, Teot LA, Moore J, Hayes J, Meyer W, Qualman SJ. An immunohistochemical algorithm to facilitate diagnosis and subtyping of rhabdomyosarcoma: the Children's Oncology Group experience. Am J Surg Pathol. 2006;30:962–968. doi: 10.1097/00000478-200608000-00005. [DOI] [PubMed] [Google Scholar]
- Nakayama R, Nemoto T, Takahashi H, Ohta T, Kawai A, Seki K, Yoshida T, Toyama Y, Ichikawa H, Hasegawa T. Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma. Mod Pathol. 2007;20:749–759. doi: 10.1038/modpathol.3800794. [DOI] [PubMed] [Google Scholar]
- Nishijo K, Hosoyama T, Bjornson CR, Schaffer BS, Prajapati SI, Bahadur AN, Hansen MS, Blandford MC, McCleish AT, Rubin BP, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. Faseb J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer. 2009;115:4218–4226. doi: 10.1002/cncr.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oue T, Yoneda A, Uehara S, Yamanaka H, Fukuzawa M. Increased expression of the hedgehog signaling pathway in pediatric solid malignancies. J Pediatr Surg. 45:387–392. doi: 10.1016/j.jpedsurg.2009.10.081. [DOI] [PubMed] [Google Scholar]
- Pastore G, Peris-Bonet R, Carli M, Martinez-Garcia C, Sanchez de Toledo J, Steliarova-Foucher E. Childhood soft tissue sarcomas incidence and survival in European children (1978– 1997): report from the Automated Childhood Cancer Information System project. Eur J Cancer. 2006;42:2136–2149. doi: 10.1016/j.ejca.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Porter JD, Merriam AP, Khanna S, Andrade FH, Richmonds CR, Leahy P, Cheng G, Karathanasis P, Zhou X, Kusner LL, et al. Constitutive properties, not molecular adaptations, mediate extraocular muscle sparing in dystrophic mdx mice. FASEB J. 2003;17:893–895. doi: 10.1096/fj.02-0810fje. [DOI] [PubMed] [Google Scholar]
- Qualman S, Lynch J, Bridge J, Parham D, Teot L, Meyer W, Pappo A. Prevalence and clinical impact of anaplasia in childhood rhabdomyosarcoma : a report from the Soft Tissue Sarcoma Committee of the Children's Oncology Group. Cancer. 2008;113:3242–3247. doi: 10.1002/cncr.23929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualman SJ, Coffin CM, Newton WA, Hojo H, Triche TJ, Parham DM, Crist WM. Intergroup Rhabdomyosarcoma Study: update for pathologists. Pediatr Dev Pathol. 1998;1:550–561. doi: 10.1007/s100249900076. [DOI] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu FK, Sham MH, Chow BK. Secretin, a known gastrointestinal peptide, is widely expressed during mouse embryonic development. Gene Expr Patterns. 2005;5:445–451. doi: 10.1016/j.modgep.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Smith LM, Anderson JR, Coffin CM. Cytodifferentiation and clinical outcome after chemotherapy and radiation therapy for rhabdomyosarcoma (RMS) Med Pediatr Oncol. 2002;38:398–404. doi: 10.1002/mpo.10060. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49:6324–6327. [PubMed] [Google Scholar]
- Suh CH, Ordonez NG, Mackay B. Malignant fibrous histiocytoma: an ultrastructural perspective. Ultrastruct Pathol. 2000;24:243–250. doi: 10.1080/01913120050176699. [DOI] [PubMed] [Google Scholar]
- Sumino Y, Hirata Y, Sato F, Mimata H. Growth mechanism of satellite cells in human urethral rhabdosphincter. Neurourol Urodyn. 2007;26:552–561. doi: 10.1002/nau.20369. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Oda Y, Kawaguchi K, Tamiya S, Yamamoto H, Suita S, Tsuneyoshi M. Altered expression and molecular abnormalities of cell-cycle-regulatory proteins in rhabdomyosarcoma. Mod Pathol. 2004;17:660–669. doi: 10.1038/modpathol.3800101. [DOI] [PubMed] [Google Scholar]
- Tiffin N, Williams RD, Shipley J, Pritchard-Jones K. PAX7 expression in embryonal rhabdomyosarcoma suggests an origin in muscle satellite cells. Br J Cancer. 2003;89:327–332. doi: 10.1038/sj.bjc.6601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208:17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- Tsumura H, Yoshida T, Saito H, Imanaka-Yoshida K, Suzuki N. Cooperation of oncogenic K-ras and p53 deficiency in pleomorphic rhabdomyosarcoma development in adult mice. Oncogene. 2006;25:7673–7679. doi: 10.1038/sj.onc.1209749. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SWG, John R. Malignant fibrous histiocytoma. In: Weiss SWG, John R, editors. Enzinger and Weiss s Soft Tissue Tumors. St Louis: Mosby; 2007. [Google Scholar]
- Wexler LH, Ladanyi M. Diagnosing Alveolar Rhabdomyosarcoma: Morphology Must Be Coupled With Fusion Confirmation. J Clin Oncol. doi: 10.1200/JCO.2009.27.5339. [DOI] [PubMed] [Google Scholar]
- Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, Thway K, Orbach D, Lae M, Freneaux P, et al. Fusion Gene-Negative Alveolar Rhabdomyosarcoma Is Clinically and Molecularly Indistinguishable From Embryonal Rhabdomyosarcoma. J Clin Oncol. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.