Abstract
Uracil residues are eliminated from cellular DNA by uracil-DNA glycosylase, which cleaves the N-glycosylic bond between the uracil base and deoxyribose to initiate the uracil-DNA base excision repair pathway. Co-crystal structures of the core catalytic domain of human uracil-DNA glycosylase in complex with uracil-containing DNA suggested that arginine 276 in the highly conserved leucine intercalation loop may be important to enzyme interactions with DNA. To investigate further the role of Arg276 in enzyme-DNA interactions, PCR-based codon-specific random mutagenesis, and site-specific mutagenesis were performed to construct a library of 18 amino acid changes at Arg276. All of the R276X mutant proteins formed a stable complex with the uracil-DNA glycosylase inhibitor protein in vitro, indicating that the active site structure of the mutant enzymes was not perturbed. The catalytic activity of the R276X preparations was reduced; the least active mutant, R276E, exhibited 0.6% of wild-type activity, whereas the most active mutant, R276H, exhibited 43%. Equilibrium binding studies utilizing a 2-aminopurine deoxypseudouridine DNA substrate showed that all R276X mutants displayed greatly reduced base flipping/DNA binding. However, the efficiency of UV-catalyzed cross-linking of the R276X mutants to single-stranded DNA was much less compromised. Using a concatemeric [32P]U·A DNA polynucleotide substrate to assess enzyme processivity, human uracil-DNA glycosylase was shown to use a processive search mechanism to locate successive uracil residues, and Arg276 mutations did not alter this attribute.
The uracil-DNA glycosylase family-1 enzymes (UDGs)1 are named for their homology to the first uracil-DNA glycosylase activity observed, Escherichia coli uracil-DNA glycosylase (Ung), described by Lindahl et al. (1). UDGs are highly conserved among different species and are thought to have evolved from a common ancestor (2). Ung family UDGs remove uracil residues from DNA that arise from misincorporation of dUMP in place of dTMP during DNA replication or from deamination of existing cytosine residues in DNA, and are active on single-as well as double-stranded DNA (1, 3). These enzymes catalyze the hydrolysis of the N-glycosylic bond joining the uracil base to the deoxyribose phosphate backbone of DNA and produce two reaction products: the free uracil base and AP-site DNA (1). Removal of the uracil base constitutes the first step in the multistep DNA repair pathway known as base excision repair (4). Drawing on x-ray crystallographic data (Fig. 1) generated by Tainer and co-workers (5, 6), a recent kinetic study of the Ung reaction mechanism by Wong et al. (7) proposed that UDGs excise uracil from DNA by means of a “pinch-pull-push” catalytic mechanism. The temporal sequence of events associated with uracil excision was postulated to be: 1) UDG binds, kinks, and compresses the duplex DNA backbone via the action of the 4 Pro-Ser loop and the Gly-Ser loop while scanning the minor groove for a uracil residue; 2) upon locating the uracil target in DNA, UDG extracts the uracil base from the DNA helix and displaces the helical space of the flipped-out uracil residue by the insertion of a conserved leucine side chain into the minor groove of the DNA; and 3) the flipped-out uracil base is captured and stabilized by the UDG active center, where the glycosylic bond is cleaved (7). The E. coli enzyme, Ung, was shown by Bennett et al. (8) to utilize a processive search mechanism for locating sequential uracil resides on the same DNA strand.
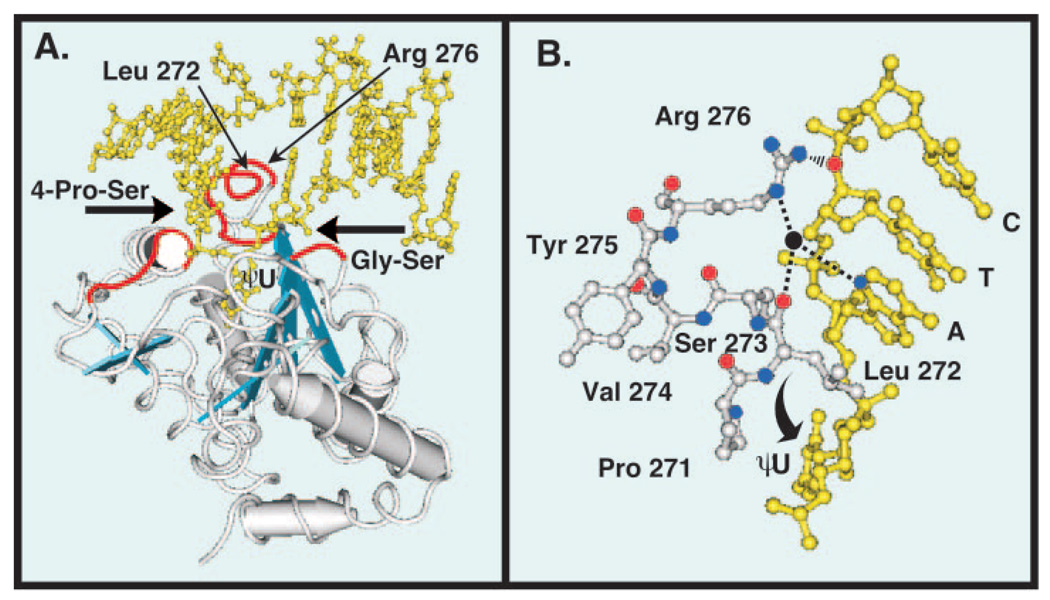
Fig. 1. Tertiary structure of human uracil-DNA glycosylase bound to DNA.
Co-crystal structure of the core catalytic domain UNG* bound to DNA containing the uncleaved uracil analogue dψU (5). The DNA is shown in yellow and the view is looking into the major groove. A, three distinct amino acid sequences (red tubes) of the UNG* polypeptide backbone (silver tubes) critical to the proposed pinch-pull-push catalytic mechanism (7) are shown. The 4 Pro-Ser loop (165PPPPS169) and the Gly-Ser loop (246GS247) compress (“pinch”) the deoxyribose phosphate backbone from the 5′ and 3′ directions, respectively (6). The Leu272 loop (268HPSPLSVYR276), which contains Arg276, penetrates the DNA base stack (“push”) and occupies the helical space of the flipped-out ψU residue (6). Conserved amino acid residues (Gln144, Asn204, and His268) in the UNG* binding pocket capture (“pull”) and stabilize the expelled extrahelical dψU. α-Helices are depicted as silver cylinders and β-sheets are illustrated as blue strands. B, ball-and-stick diagram of the UNG Leu272 loop (271PLSVYR276) shown in silver and a portion of the oligonucleotide sequence 3′-CTA dψU-5′ shown in yellow. The ηN of the Arg276 guanidinium side chain (nitrogen atoms, blue balls) is shown as interacting (black rippled lines) with the 5′-phosphate of the cytosine residue (oxygen atom, red ball), as stated for cleaved U·G DNA by Slupphaug et al. (17). The εN participates in water-bridged (water, black ball) hydrogen bonding (dashed lines) with the N3 of adenine (blue ball) and the carbonyl group (red ball) of Leu272 as shown in Parikh et al. (6). Structures were drawn with the Cn3D 4.0 software program using Protein Data Bank code 1EMH (MMDB 13471) deposited by Parikh et al. (5) in the Molecular Modeling Data base of the National Center for Biotechnology Information.
Several enzymes capable of uracil excision have been identified and cloned in human cells, including uracil-DNA glycosylase (UNG) (9), thymine-DNA glycosylase (10), single-strand selective monofunctional uracil-DNA glycosylase (SMUG1) (11), and methyl-CpG-binding protein 4 (12). However, recent evidence suggests that UNG is the major repair enzyme for removal of uracil from U·A and U·G mispairs, and U in single-stranded DNA (2, 13). Two forms of human uracil-DNA glycosylase, nuclear (UNG2) and mitochondrial (UNG1), are generated from the UNG gene using two promoters, followed by splicing of the nuclear form exon 1A transcript into a consensus splice at codon 35 of the mitochondrial form exon 1B; the remaining sequence is the same for both transcripts (14). UNG1 consists of 304 amino acids, the first 35 of which are unique to this form, whereas UNG2 comprises 313 amino acids, the first 44 of which are unique to UNG2. The unique N-terminal sequences in UNG1 and UNG2 are required for mitochondrial and nuclear localization, respectively, but not for catalytic activity (14, 15). The 269-amino acid core catalytic domain shared by UNG1 and UNG2, designated UNG* in this report (or UNGΔ84 in the literature), has been extensively studied, and its structure, substrate specificity, and molecular mechanism of catalysis characterized (5, 13, 16–19). In vitro, UNG* removes uracil residues in single-stranded DNA or duplex DNA containing U·G mispairs or U·A base pair in the order single-stranded U > U·G > U·A (16). UNG* was also reported to excise the uracil analogues 5-hydroxyuracil, isodialuric acid, and alloxan at a relatively low rate (20).
Mutational and structural studies of UNG* have implicated amino acids involved in enzyme activity and uracil recognition. Mutational analysis demonstrated that Asn204, Gln144, Asp145, His268, Ser169, and Ser270 are critical to UNG* activity; Gln144, Asp145, and His268 appeared to be involved in stabilization of the uracilate anion reaction intermediate (18, 21). Substitution of Leu272 with alanine (L272A) severely impaired enzyme activity on U·G- and U·A-containing DNA (6). Replacement of Asn204 by Asp and Tyr147 by Ala, Cys, or Ser resulted in enzymes that have cytosine-DNA glycosylase activity or thymine-DNA glycosylase activity, respectively (19). Additional insight into the relationship between structure and function evolved following the solution of the co-crystal structures of UNG* in complex with DNA (5, 6, 17). Parikh et al. (6) reported that of the five arginines and eight lysines localized to the positively charged active site face of UNG*, only Arg276 contacted the DNA. As illustrated in Fig. 1B, the εN of Arg276 participates in water-bridged hydrogen bonds with the N3 of adenine and the carbonyl group of Leu272. Also, the ηN of the Arg276 guanidinium side chain is shown as interacting with the dCMP 5′ phosphate, as stated by Slupphaug et al. (17) for cleaved U·G DNA. However, the precise role of the Arg276 residue in DNA binding and uracil recognition has not been studied, nor has the role of Arg276 in the UNG* processive search mechanism been investigated.
In this study, PCR-based codon-specific random mutagenesis was performed on the UNG* gene and 18 R276X mutants were isolated. The mutant proteins were overproduced, purified, and characterized with respect to inhibition by Ugi, catalytic efficiency, DNA binding/base flipping, UV cross-linking to single-stranded DNA, and enzyme processivity using a polymeric uracil-containing [32P]DNA substrate.
EXPERIMENTAL PROCEDURES
Materials
Chloramphenicol and kanamycin were purchased from Sigma, and ampicillin was from Fisher. Nickel-nitrilotriacetic acid-agarose was obtained from Qiagen. Hydroxyapatite and P-4 Bio-Gel were purchased from Bio-Rad, and poly(U)-Sepharose, CM-Sephadex, and Sephacryl S-500 were from Amersham Biosciences. DEAE-cellulose (DE-52) and phosphocellulose (P-11) were obtained from Whatman. Single-stranded DNA-agarose was prepared as previously described (22). [γ-32P]ATP was purchased from PerkinElmer Life Sciences and [3H]dUTP was from Amersham Biosciences. Restriction endonucleases DpnI, EcoRI, HindIII, SacI, and NdeI were obtained from New England Biolabs, as were E. coli exonuclease III and Deep Vent DNA polymerase. Pfu Turbo DNA polymerase was purchased from Stratagene. T4 polynucleotide kinase and T4 DNA ligase were obtained from Fermentas. Bacteriophage PBS-2 uracil-DNA glycosylase inhibitor (Ugi) protein (fraction IV) and E. coli Ung (fraction V) were purified as previously described by Sanderson and Mosbaugh (23).
Plasmid, pUNG15, was obtained from American Type Culture Collection (number 65269), pTrc99A from Amersham Biosciences, pET-22b from Novagen, and pRP from Stratagene. Plasmid DNA was isolated using a Qiagen miniprep kit following the manufacturer’s instructions. Oligonucleotides were 5′-end phosphorylated as previously described by Bennett et al. (8).
E. coli strain BW1067 (ung-153::kan thi-1 relA1 spoT1) was a gift from B. Weiss (Emory University) and E. coli JM105 and phage P1vira were provided by W. Ream (Oregon State University). E. coli JM109 was purchased from Stratagene, and E. coli BLR from Novagen. Using E. coli BW1067 as the donor strain and JM105 as recipient, JM105 (ung::kan), renamed as CY10, was created using standard phage P1 transduction methods as described by Miller (24). Similarly, using E. coli BH157 as the donor strain and JM105 as recipient, JM105 (dug::tet), renamed as CY01, was created by phage P1 transduction. E. coli CY10 was transduced to recA (Δ(srl-recA)306::Tn10(TcR)) using phage P1 lysate from the donor strain BLR; this strain was called CY10rec. E. coli CY11 (ung dug) was created by transduction of CY10 with phage P1 lysate from the donor strain BH157. E. coli CY10rec and CY11 were transformed with pRP (Cmr) that contained the arginine AGA/AGG tRNA and proline CCC tRNA genes (Stratagene).
Preparation of Competent Cells
Terrific Broth (25) (50 ml) containing kanamycin (25 µg/ml) and chloramphenicol (34 µg/ml) was inoculated with an E. coli CY10rec/pRP colony and incubated with shaking (250 rpm) overnight at 25 °C. Cells were harvested in early log phase (A600 nm = 0.4), incubated on ice for 10 min, harvested by centrifugation, and processed according to the standard protocol of the Z-Competent Transformation Kit (Zymo Research). Aliquots (100 µl) of the competent cell preparation were stored at −80 °C.
Construction of Plasmids for Overproduction of Human UNG* and UNG
The nucleotide sequence corresponding to the core catalytic domain of human uracil-DNA glycosylase (UNG*) was amplified in a polymerase chain reaction (100 µl) containing pUNG15 as template (0.5 µg), primers (1 µm each) RI: 5′-GCGAATTCTTTGGAGAGAGCTGGAAG-3′, and H3: 5′-GCAAGCTTTCACAGCTCCTTCCAGTC-3′, as forward and reverse primers, respectively, 1× ThermoPol (New England Biolabs) buffer, 200 µm each dATP, dTTP, dCTP, and dGTP, and 2 units of Deep Vent DNA polymerase. Following digestion with EcoRI and HindIII, the UNG* fragment (700 bp) was gel-purified and ligated to pTrc99A DNA that was similarly digested and purified. E. coli CY10rec/pRP was then transformed with the ligation mixture and plated on medium containing 100 µg/ml ampicillin and 25 µg/ml chloramphenicol. A His6-tagged UNG* construct (named UNG in this report) was generated by PCR using UNG*/pTrc99A as a template, the forward primer 5′-GCGAATTCCATCACCATCACCATCACTTTGGAGAGAGCTGGAAG-3′, and the H3 reverse primer as described above. The UNG gene construct was ligated into pTrc99A as described for UNG*, and subsequently subcloned into pET-22b at NdeI and HindIII sites. DNA sequence analysis verified that the nucleotide sequence of the UNG* and UNG constructs was identical to that reported by Slupphaug et al. (16).
Codon-specific Oligonucleotide-directed Random Mutagenesis
Codon-specific PCR-based random mutagenesis of the UNG Arg276 (AGA) codon was carried out following the QuikChange procedure (Stratagene). Primers, 5′-CTTTGTCAGTGTATNNNGGGTTCTTTGGATG-3′ (FP-31-mer) and 5′-CATCCAAAGAACCCNNNATACACTGACAAAGG-3′ (RP-32-mer), were synthesized that contained an equal mixture of the four phosphoramidite nucleoside monomers (NNN) at the AGA codon corresponding to Arg276. PCR mixtures (50 µl) contained 50 ng of UNG/pET-22b DNA template, 14 pmol each of the forward (FP-31-mer) and reverse (RP-32-mer) primers, 250 µm each of dATP, dTTP, dCTP, and dGTP, 2.5 units of Pfu Turbo DNA polymerase, and 1× Pfu reaction buffer (Stratagene). PCR was carried out in a Hybaid PCR Express thermocycler according to the following parameters: 1 min denaturation at 94 °C, 18 cycles of 94 °C (30 s), 55 °C (1 min), 68 °C (12 min), and 72 °C (12 min). The PCR-DNA product was then digested with DpnI (25 units) for 2 h at 37 °C. E. coli CY10rec/pRP Z-competent cells (1–2 × 1010 cells) were thawed on ice, gently mixed with 5 µl of DpnI-digested DNA, incubated on ice for 1 h, spread on pre-warmed 2× YT plates containing 1% glucose, 100 µg/ml ampicillin, and 34 µg/ml chloramphenicol, and incubated overnight at 37 °C.
Site-specific Oligonucleotide-directed Mutagenesis
Site-specific PCR-based mutagenesis of the Arg276 codon of UNG was carried out for five amino acid substitutions as described above except that the forward primers (31-mers) were 5′-CTTTGTCAGTGTATXXXGGGTTCTTTGGATG-3′, where XXX was TGC for cysteine, CAC for histidine, ATG for methionine, GTA for valine, and TGG for tryptophan amino acid substitutions. The corresponding reverse primers (31-mers) were 5′-CATCCAAAGAACCCYYYATACACTGACAAAG-3′, where YYY was GCA, GTG, CAT, TAC, and CCA for the cysteine, histidine, methionine, valine, and tryptophan amino acid substitutions, respectively.
Preparation of Cell Extracts for Uracil-DNA Glycosylase Activity Assay
Overnight cultures of ampicillin-resistant E. coli CY10rec/pRP/UNG-pET-22b were used to inoculate (1/25 dilution) flasks containing Terrific Broth (30 ml) supplemented with 100 µg/ml ampicillin, and incubation was continued for 3 h at 37 °C. Cells were harvested by centrifugation, resuspended in 1 ml of ice-cold sonification buffer (50 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride), placed on ice, and subjected to a 2-min sonic pulse (Branson Sonifier 450, microtip, 30% duty cycle, power setting 3). The cell lysates were clarified by centrifugation at 14,000 × g for 10 min at 4 °C, and the protein concentration of the supernatant fraction was determined using the Bio-Rad Protein assay using bovine serum albumin as the standard. Extracts were diluted 1/10 and 1/100 in UDB buffer (50 mm Hepes-KOH (pH 7.9), 1 mm EDTA, 1 mm DTT, and 50 µg/ml acetylated bovine serum albumin) and uracil-DNA glycosylase activity was measured as described below. Growth of cultures whose cell extract exhibited lower than average uracil-DNA glycosylase specific activity (<10 units/mg of protein) was continued, and the UNG/pET-22b plasmids were isolated using a Qiagen miniprep kit for DNA sequence analysis.
Overproduction and Purification of UNG*
E. coli CY11/pRP/UNG*-pTrc99A was grown at 37 °C with shaking (250 rpm) in 1.5 liters of 2× YT medium containing 1% glucose and 100 µg/ml ampicillin. Upon reaching mid-log phase growth (A600 nm = ~0.6), the culture temperature was reduced to 30 °C and isopropyl 1-thio-β-d-galactopyranoside was added to 1 mm final concentration. Following incubation for 5 h at 30 °C, the cells were harvested by centrifugation and resuspended in 30 ml of buffer SB (50 mm Tris acetate (pH 7.0), 10 mm NaCl, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm DTT, and 10% (w/v) glycerol), and adjusted to 500 µg/ml lysozyme (Sigma) prior to storage at −80 °C. The cell suspension was thawed and sonification was conducted as described above. All operations were carried out at 4 °C unless otherwise indicated. After centrifugation at 20,000 × g for 15 min, the supernatant fraction was reserved, and the cell pellet was resuspended in 30 ml of buffer SB. Both the sonification and centrifugation steps were repeated and the supernatant fractions were combined to produce the cell-free extract (Fraction I). An equal volume of 1.6% streptomycin sulfate was slowly added to the crude extract while mixing on ice for 1 h. Precipitates were then removed by centrifugation at 15,000 × g for 15 min and the supernatant fraction (~90 ml) was designated Fraction II.
Fraction II was loaded onto a DEAE-cellulose column (4.9 cm2 × 20 cm) equilibrated in buffer SB and the column was washed with 150 ml of the equilibration buffer. Fractions containing uracil-DNA glycosylase activity were pooled (Fraction III) and applied to a CM-Sephadex column (4.9 cm2 × 10 cm) equilibrated in buffer SB. After washing the column with 500 ml of equilibration buffer, a 150-ml linear gradient of 10–400 mm NaCl in buffer SB was applied. The peak of UNG* activity eluted at ~150 mm NaCl; active fractions were pooled, dialyzed against buffer SB, and designated Fraction IV. Fraction IV was loaded onto a phosphocellulose column (4.9 cm2 × 10 cm) equilibrated in the same buffer. The column was washed with 100 ml of buffer SB and eluted with a 150-ml linear gradient of 10–400 mm NaCl in buffer SB. The enzyme activity peak that eluted at ~250 mm NaCl was pooled and dialyzed against buffer SB (Fraction V). Fraction V was loaded onto a single-stranded DNA-agarose column (19.6 cm2 × 5 cm) equilibrated in buffer SB. The column was washed with 500 ml of equilibration buffer, and a 500-ml linear gradient of 10–800 mm NaCl in buffer SB was applied. Eluted fractions were assayed for UNG* activity, and a single peak of activity was detected at ~175 mm NaCl. The enzyme was pooled as Fraction VI, dialyzed against buffer SB containing 40 mm NaCl, and applied to a poly(U) column (1.8 cm2 × 3 cm) equilibrated in the same buffer. After washing the column with 50 ml of equilibration buffer, the enzyme was eluted using a 50-ml linear gradient of 40–600 mm NaCl in buffer SB. Fractions containing UNG* activity were analyzed by 12.5% SDS-polyacrylamide gel electrophoresis and pooled, constituting fraction VII (peak activity eluted at ~275 mm NaCl). After dialyzing fraction VII against buffer KDP (10 mm potassium phosphate (pH 7.4), 1 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride) containing 50 mm KCl, the sample was loaded onto a hydroxyapatite column (1.6 cm2 × 2 cm) equilibrated in the same buffer. The column was washed with 50 ml of equilibration buffer and proteins were eluted using a 30 ml of linear gradient of KCl (50–600 mm) in buffer KDP. Uracil-DNA glycosylase activity was eluted as a single symmetrical peak (~450 mm KCl), pooled, and stored at −80 °C as Fraction VIII.
Overproduction and Purification of UNG
E. coli CY10rec/pRP/ UNG-pET-22b was grown in 1.0 liter of 2× YT medium containing 1% glucose, 100 µg/ml ampicillin, and 34 µg/ml chloramphenicol at 37 °C with shaking. When the A600 of the culture reached 0.5, the temperature was reduced to 25 °C and incubation was continued for 20 min before the addition of isopropyl 1-thio-β-d-galactopyranoside to 1 mm. After incubation for 6 h, the cells were harvested by centrifugation, resuspended in 25 ml of buffer A (50 mm sodium phosphate (pH 8.0), 500 mm NaCl, 1 mm β-mercaptoethanol, 10 mm imidazole, 5% (w/v) glycerol) and lysozyme was added to 500 µg/ml prior to storage at −80 °C. The cell suspension was thawed on ice, supplemented with phenylmethylsulfonyl fluoride to 1 mm, and subjected to sonification as described above. The cell lysate was similarly processed except using buffer A. Following the sonification, the cell lysate (Fraction I) was diluted to a protein concentration of 1.5 mg/ml with buffer A and applied to a nickel-nitrilotriacetic acid-agarose column (0.8 cm2 × 2.5 cm) equilibrated in buffer A. The column was washed with 20 ml of equilibration buffer and step eluted with buffer A containing 300 mm imidazole. Fractions containing UNG activity were pooled and dialyzed against buffer B (10 mm sodium phosphate (pH 8.0), 20 mm NaCl, 1 mm DTT, 5% (w/v) glycerol) constituting Fraction II. Fraction II was applied to a hydroxyapatite column (1.8 cm2 × 1.1 cm) equilibrated in buffer B. The column was washed with 20 ml of buffer B containing 200 mm NaCl and step eluted with 20 ml of buffer B containing 300 mm NaCl. Fractions containing uracil-DNA glycosylase activity were pooled, dialyzed against buffer C (30 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1 mm DTT, 5% (w/v) glycerol), and designated Fraction III. Fraction III was applied to a poly(U) column (1.8 cm2 × 1.2 cm) equilibrated in buffer C. After washing the column with 4 ml of buffer C and then with 20 ml of buffer C containing 150 mm NaCl, the enzyme was eluted with 20 ml of buffer C containing 500 mm NaCl. Active fractions were pooled and dialyzed against buffer C (Fraction IV), and stored at −80 °C. The Arg276 mutant protein preparations were purified using the same procedure.
Uracil-DNA Glycosylase Activity Assay
Uracil-DNA glycosylase activity was measured in standard reaction mixtures as previously described by Bennett and Mosbaugh (26). Enzyme preparations were diluted in 50 mm Hepes-KOH (pH 7.9), 1 mm EDTA, 1 mm DTT, and 100 µg/ml acetylated bovine serum albumin. One unit of uracil-DNA glycosylase is defined as the amount of enzyme that releases 1 nmol of uracil per hour under standard conditions.
Nuclease Assay
Reaction mixtures (100 µl) contained 8–200 ng of wild-type or mutant UNG (Fraction IV), 100 µg/ml acetylated bovine serum albumin, buffer C, and 12.5 ng of single-stranded carboxyfluorescein (FAM) 5′-end labeled oligonucleotide: 5′-FAM-T-25-mer (5′-FAMGGGGCGCGTATAAGGAATTCGTACC-3′) or 25 ng of double-stranded oligonucleotide: 5′-FAM-T-25-mer annealed to A-25-mer (5′-GGTACGAATTCCTTATACGAGCCCC-3′). Following incubation at 37 °C for 30 min, the reaction products were resolved by 15% polyacrylamide, 8.3 m urea gel electrophoresis. The gels were scanned using an FMBioII fluorescence imaging system (Hitachi Genetic Systems) and the fluorescent bands were analyzed with ImageQuant software (Amersham Biosciences).
UNG·Ugi Binding Assay
Ung (fraction V), UNG, or R276X (Fraction IV) mutant protein (40 pmol) was combined with Ugi (100 pmol) in a reaction mixture (15 µl) containing buffer C and incubated at 20 °C for 10 min and then at 4 °C for 20 min. Following complex formation, 120 µl of 40 mm CAPS-NaOH (pH 10.5) and 30 µl of buffer LB (300 mm CAPS-NaOH (pH 10.5), 25% sucrose, and 0.04% bromphenol blue) were added to each sample. Non-denaturing polyacrylamide slab gel electrophoresis was performed by a modification of the procedure described by Bennett and Mosbaugh (27). Briefly, samples (100 µl) were loaded onto a non-denaturing 10% polyacrylamide gel, polymerized using 0.3% ammonium persulfate, which contained 150 mm CAPS-NaOH (pH 10.5) in place of Tris-HCl (pH 8.8) buffer. Electrophoresis was conducted at 4 °C and 250 V with 40 mm CAPS-NaOH (pH 10.5) running buffer until the tracking dye migrated ~8 cm. Protein bands were detected after staining the gel for 1 h in dye solution containing 0.04% Coomassie Brilliant Blue G-250 in 2.5% HClO4. Gels were destained with 5% acetic acid and then imaged with a UVP ImageStore 7500.
DNA Binding Assay
Oligonucleotides 2′-deoxypseudouridine-25-mer (dψU-25-mer, 5′-GGGGCTCGTAψUAAGGAATTCGTACC-3′) and 2-aminopurine-25-mer (2AP-25-mer, 5′-GGTACGAATTCCTT2APTACGAGCCCC-3′) were hybridized as described by Wong et al. (7) with some modifications. The annealing reaction was carried out by heating the oligonucleotides (5 nmol) in buffer C (500 µl) to 85 °C for 5 min followed by slow cooling to room temperature. DNA binding assays were performed in reaction mixtures (250 µl) that contained buffer C, 50 nm double-stranded dψU·2AP-25-mer, and various amounts of UNG (0–1.5 µm) or R276X mutant protein. Fluorescence intensity was measured using a SLM-Aminco model 8100 series 2 spectrometer equipped with a quartz mini-cell cuvette (0.5 × 0.5 cm). The excitation wavelength was 310 nm; emission was monitored at 370 nm and integrated at 1-s intervals for 60 s. DNA binding was detected by measuring the enhancement of 2AP fluorescence intensity that followed enzyme addition and was determined in triplicate.
Photochemical Cross-linking Reactions
Three oligonucleotides were 5′-end 32P-labeled in reaction mixtures containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 5 mm DTT, 10 mm MgCl2, 330 µCi of [γ-32P]ATP (6000 Ci/mmol), 200 units of T4 polynucleotide kinase, and 35 nmol of dψU-25-mer, T-25-mer (5′-GGGGCTCGTATAAGGAATTCGTACC-3′), or U-25-mer (5′-GGGGCTCGTAUAAGGAATTCGTACC-3′). After incubation for 20 min at 37 °C, ATP was added to 1mm and incubation was continued for 30 min. Unreacted [γ-32P]ATP and ATP were removed using a P-4 Bio-Gel (Bio-Rad) spun column as described previously (27). The 32P-labeled oligonucleotides were concentrated, buffer exchanged into TE buffer using a Centricon 3 centrifugal filter unit (Millipore), and adjusted to 20 µm in TE buffer. Samples (12 µl) containing 20 pmol of 32P-labeled oligonucleotide (18,000 cpm/pmol), 40 pmol of UNG (Fraction IV), and buffer C, were placed on ice and UV-irradiated (λmax = 254 nm) in a Stratalinker 1800 (Stratagene) for various times as indicated in the figure legends. Following UV irradiation, 3 µl of buffer S (45 mm Tris base, 445 mm boric acid, 1 mm EDTA, 25% sucrose, 0.04% bromphenol blue) was added and samples (6 µl) were analyzed by non-denaturing 10% polyacrylamide gel electrophoresis. The gels were dried and the amount of 32P radioactivity detected in each band was determined using a PhosphorImager and ImageQuant software (Amersham Biosciences).
Preparation of Double-stranded Uracil-containing Concatemeric [32-P]DNA Substrate
Oligonucleotides, U-30-mer (5′-GCGTGACGCACTGAUAAGTGAATTCGACCG-3′) and A-30-mer (5′-CGTCACGCCGGTCGAATTCACTTATCAGTG-3′) were 5′-end phosphorylated as described above, except that 50 mm NaCl was included in each reaction. [γ-32P]ATP was added to the U-30-mer reaction, whereas nonradioactive ATP was included in the A-30-mer reaction. Oligonucleotides [32P]U-30-mer and A-30-mer were annealed in a Hybaid PCR Express Thermocycler using the following parameters: 2 min each at 85, 75, 65, 60, 55, 50, 45, 40, 35, 25, 15, and 5 °C. Duplex monomer (30-mer) units were joined by ligation in a reaction mixture (4.4 ml) containing buffer K (50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 5 mm DTT, 10 mm MgCl2) supplemented with 1 mm ATP, 440 units of T4 DNA ligase, and 58 nmol of [γ-32P]U·A-30-mer. The ligation reaction mixture was divided among 12 microcentrifuge tubes (367 µl each) and incubated in a Hybaid thermocycler programmed to carry out 198 cycles of 1 min at 10 °C followed by 1 min at 22 °C. After thermocycling, incubation was continued at 16 °C for 6 h, and the reaction mixtures were pooled and adjusted to 5% (w/v) glycerol. The reaction mixture was loaded onto a Sephacryl S-500 column (1.8 cm2 × 70 cm) equilibrated in TE buffer containing 150 mm NaCl and eluted with equilibration buffer. Fractions (1 ml) were collected and analyzed using non-denaturing 4% polyacrylamide gel electrophoresis followed by autoradiography. Fractions that contained high molecular weight (>600 bp) concatenated [32P]DNA were pooled, concentrated, and buffer-exchanged into TE buffer using Centricon 30 centrifugal filters (Millipore). The concentration of the [32P]DNA substrate was determined spectrophotometrically (1 A260 nm = 50 µg/ml).
Restriction Endonuclease Digestion of the Uracil-containing Concatemeric Polynucleotide Substrate
Restriction endonuclease reaction mixtures (10 µl) contained concatenated [32P]U·A-DNA (10 pmol), EcoRI (10 units), or HpaII (10 units), and either 1× EcoRI buffer or NEB buffer 1 (both from New England Biolabs), respectively. After incubation for 60 min at 37 °C, the reactions were terminated by the addition of 3 m K2HPO4 (pH 13.7) to 0.3 m and heating at 95 °C for 30 min. The samples were then neutralized by adding 1.5 m KH2PO4 to 0.3 m followed by 13.3 µl of TE buffer. The concatenated [32P]DNA (10 pmol) was also digested with UNG (70 units) for 60 min at 37 °C in two reaction mixtures (10 µl each) containing 70 mm Hepes-KOH (pH 7.9), 1 mm EDTA, and 1 mm DTT; reactions were terminated with Ugi (1 µg). One reaction was subjected to alkaline hydrolysis followed by neutralization as described above; the other reaction was divided into two aliquots (5.5 µl). One aliquot was supplemented with TE buffer (4.5 µl) and the other was adjusted to 1× EcoRI buffer for incubation with EcoRI (10 units) for 60 min at 37 °C. The reaction was terminated, and both samples were processed as described above. An equal volume (26.7 µl) of denaturing sample buffer (95% deionized form-amide, 1 mm EDTA, 0.04% bromphenol blue) was added to each sample. Portions (6 µl) of each sample were subjected to denaturing 12% polyacrylamide, 8.3 m urea gel electrophoresis until the tracking dye migrated 17 cm. Gels were dried and used to expose PhosphorImager screens that were subsequently scanned with a PSI PhosphorImager (Amersham Biosciences). The scanned images were quantified using the ImageQuant program.
Uracil-DNA Glycosylase Processivity Assay
The processivity of UNG and R276X mutants was determined using the concatemeric [32P]U·A polynucleotide DNA substrate described above. Standard reaction mixtures (100 µl) contained 80 pmol of [32P]U·A-DNA (48.6 × 104 cpm/pmol) and 0.06–1.0 unit of enzyme. Incubation was performed at 37 °C and samples (10 µl) were removed at various times (0–80 min). Reactions were stopped with the addition of Ugi and placed on ice. Each sample was divided into two (5.5 µl) aliquots. TE buffer (4.5 µl) was added to the first aliquot and the sample was processed using the alkaline hydrolysis/neutralization treatment as described above. The second aliquot was adjusted to 1× EcoRI buffer, digested with EcoRI (10 units) for 60 min at 37 °C, and then similarly processed. Portions (6 µl) of each reaction were subjected to analysis by 12% polyacrylamide, 8.3 m urea gel electrophoresis as described above.
RESULTS
Overproduction and Purification of UNG and R276X Proteins
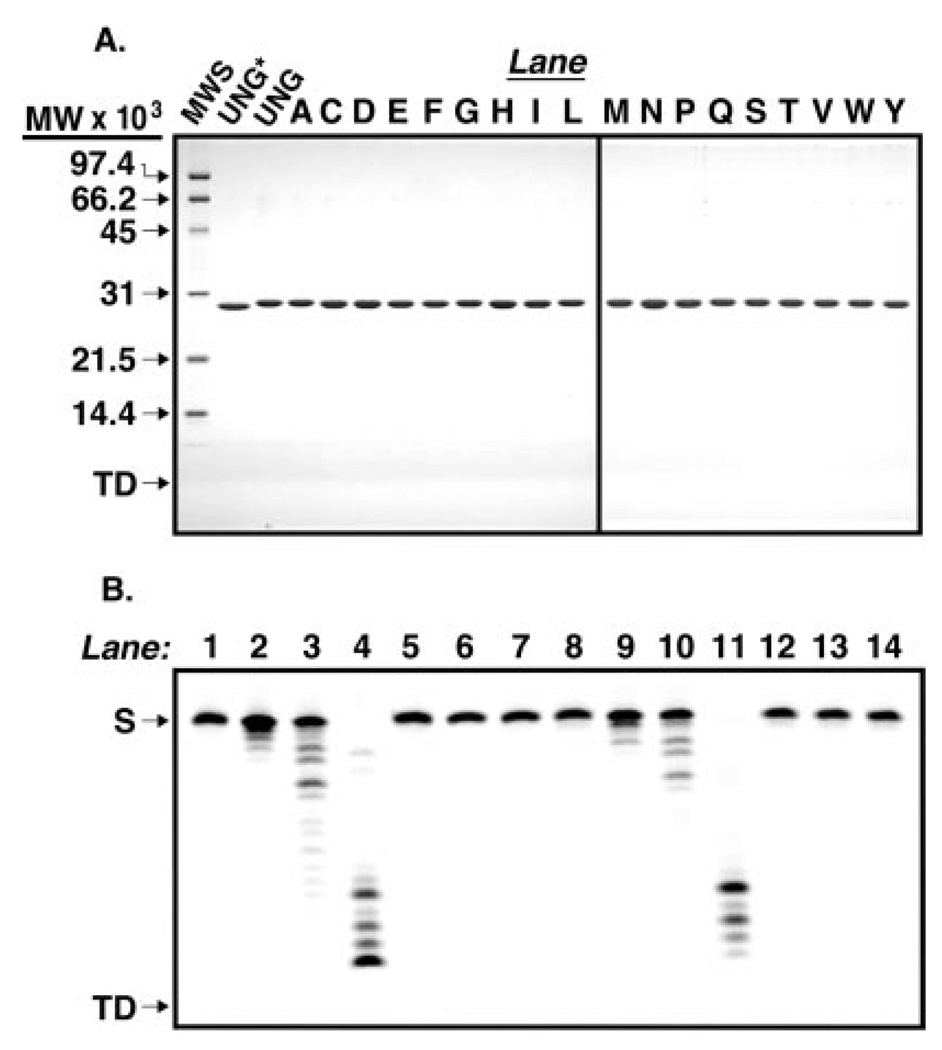
Codon-specific random mutagenesis was performed on arginine 276 of the human UNG gene and 18 R276X (R276X) mutants were isolated by activity screening as described under “Experimental Procedures.” To facilitate the mutational analysis of human uracil-DNA glycosylase, the recombinant core catalytic domain of human uracil-DNA glycosylase (UNG*), consisting of residues 85–304 (numbering according to mitochondrial pre-form UNG1 (16)), UNG* containing six N-terminal histidine residues, designated UNG, and the R276X mutant UNG proteins were overproduced in E. coli and purified as described under “Experimental Procedures.” To examine the purity and relative gel mobility of each protein preparation, 4 µg of UNG*, UNG, and R276X mutant proteins (Fraction IV) were subjected to 12.5% SDS-polyacrylamide gel electrophoresis. Analysis of the Coomassie-stained gel showed that UNG*, UNG, and R276X mutant proteins were purified (>95%) to apparent homogeneity and exhibited mobility consistent with the predicted molecular weight (Fig. 2A). The purified protein preparations were found to be free of contaminating nuclease activity in assays utilizing 5′-end carboxyfluorescein-labeled single- and double-stranded oligonucleotides (Fig. 2B and data not shown).
Fig. 2. Purity of the enzymes used in this study.
A, UNG* (fraction VIII), UNG, and various Arg276 mutant proteins (fraction IV) were purified as described under “Experimental Procedures.” Samples (4 µg) of UNG*, UNG, and each Arg276 mutant protein were subjected to 12.5% SDS-polyacrylamide gel electrophoresis, and protein bands were visualized after staining with Coomassie Brilliant Blue G-250. The mobility of molecular weight standards (SDS-PAGE, low-range standards, Bio-Rad) as well as the tracking dye (TD) are indicated by arrows from top to bottom, respectively (lane MWS). Lanes containing R276X mutant proteins are represented by conventional single letter amino acid abbreviations. B, UNG (fraction IV) was assayed for nuclease activity using 5′-end carboxyfluorescein-labeled single- and double-stranded oligonucleotide substrates, 5′-FAM-T-25-mer and 5′- FAM-T·A-25-mer, respectively, as described under “Experimental Procedures.” Mock reaction mixtures contained 5′-FAM-T-25-mer (12.5 ng) or 5′-FAM-T·A-25-mer (25 ng) in reaction buffer (lanes 1 and 8, respectively), control reactions contained 0.02, 0.2, or 2 units of E. coli exonuclease III and either 5′-FAM-T-25-mer or 5′-FAM-T·A-25-mer (lanes 2–4 and 9–11, respectively), and reactions containing UNG (8, 40, and 200 ng) and 5′-FAM-T-25-mer or 5′-FAM-T·A-25-mer (lanes 5–7, and 12–14, respectively) were incubated at 37 °C for 30 min. The reaction products were resolved by 15% polyacrylamide, 8.3 m urea, gel electrophoresis, and the gel was analyzed with a FMBioII fluorescence imaging system. Arrows indicate the locations of oligonucleotide substrate (S) and tracking dye (TD).
Ability of R276X Mutant Proteins to Bind Ugi
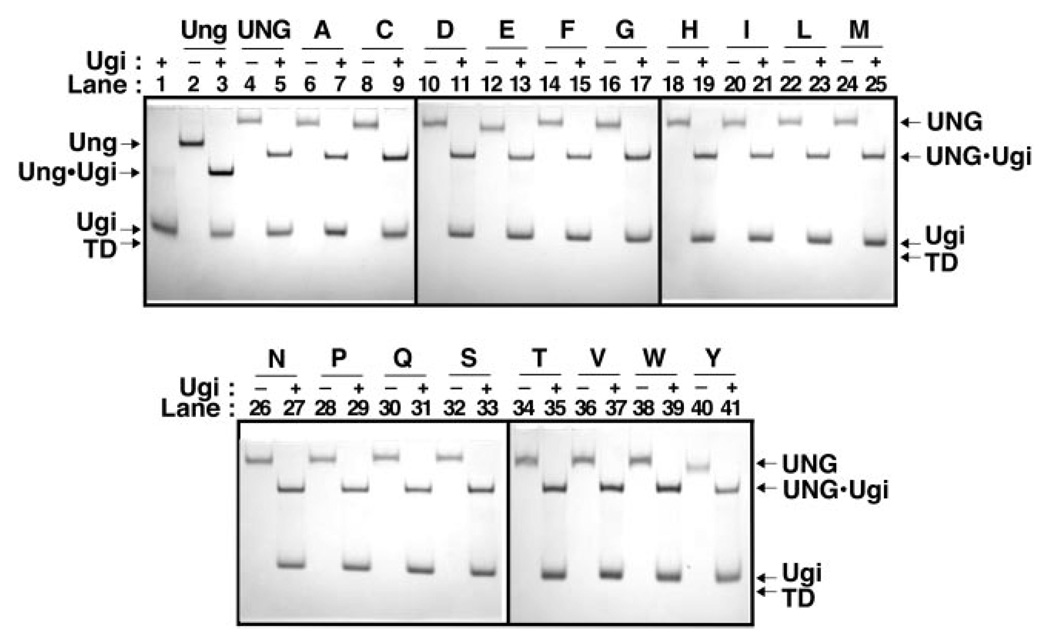
The uracil-DNA glycosylase inhibitor protein (Ugi), encoded by the Bacillus subtilis bacteriophage PBS-2, inactivates Ung-family uracil-DNA glycosylases by forming an essentially irreversible protein-protein complex with 1:1 stoichiometry (26). The x-ray crystal structures of the core catalytic domain of human uracil-DNA glycosylase and E. coli uracil-DNA glycosylase in complex with Ugi show that Ugi binds tightly to the sequence-conserved DNA-binding groove of the enzymes in a manner described as protein mimicry of DNA (28, 29). Therefore, we decided to use Ugi as a probe to determine whether the R276X mutant proteins in this study were properly folded, reasoning that Ugi would only bind to enzymes with a structurally intact DNA-binding region. Each mutant protein was reacted with a small molar excess (2.5-fold) of Ugi and the mixture was resolved by non-denaturing polyacrylamide gel electrophoresis (Fig. 3). Inspection of the gel shows that each R276X mutant protein analyzed formed a stable complex with Ugi, and that the appearance of the UNG·Ugi complex band was concurrent with the disappearance of the UNG band (Fig. 3, +Ugi lanes). These results demonstrate that mutagenesis of Arg276 did not perturb the active site structure of the mutant proteins.
Fig. 3. Ability of UNG and R276X mutant proteins to bind Ugi.
Reaction mixtures (15 µl) containing 40 pmol of E. coli Ung (lanes 2 and 3), UNG (lanes 4 and 5), or R276X mutant protein (lanes 6–41), with or without (+ or −) Ugi (100 pmol) were incubated as described under “Experimental Procedures.” A control reaction containing Ugi (100 pmol) alone was similarly processed (lane 1). Samples were analyzed at 4 °C by non-denaturing 10% polyacrylamide gel electrophoresis, proteins were visualized with Coomassie Brilliant Blue G-250 stain, and the gel was imaged as described under “Experimental Procedures.” Arrows indicate the location of Ung, UNG, Ugi, Ung·Ugi, UNG·Ugi, and the tracking dye front (TD). Arg276 mutant protein lane assignments are indicated by single letter amino acid abbreviations.
Uracil-DNA Glycosylase Activity of R276X Mutants
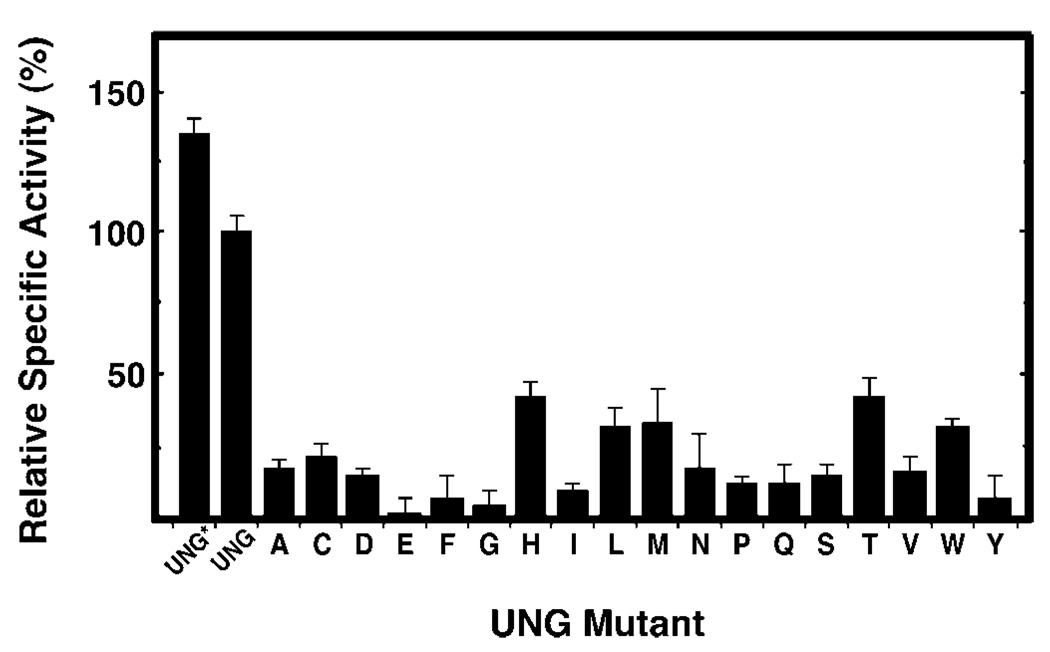
To determine the specific activity of UNG*, UNG, and R276X mutant proteins, standard uracil-DNA glycosylase reactions were conducted with 0.04–0.13 units of each protein preparation as described under “Experimental Procedures.” The specific activity (units/mg) of each preparation was then calculated and expressed as a percentage of the specific activity of UNG, which was set to 100% (Fig. 4). All 18 R276X mutant proteins showed a reduction in specific activity. The most active mutant was histidine (43%), whereas the least active was glutamic acid (0.6%) (Fig. 4, bars A–Y). The activity of the remaining 16 mutant proteins was intermediate, ranging from 4 (R276G) to 33% (R276M) of wild-type UNG activity (Fig. 4). UNG* was more active (135%) than UNG (100%).
Fig. 4. Specific uracil-DNA glycosylase activity of R276X mutant proteins.
Uracil-DNA glycosylase activity was measured under standard reaction conditions using 0.04–0.13 units of UNG*, UNG, or the indicated R276X mutant protein, as described under “Experimental Procedures.” Protein concentrations were determined using the Bradford method (36) and the Protein Assay reagent (Bio-Rad). The specific activity (units/mg) of UNG* and the R276X mutant enzymes was normalized to that of UNG (3.76 × 105 units/mg), which was defined as 100%. Arg276 mutant proteins are denoted by single letter amino acid abbreviations. Error bars represent the standard deviation of four experimental determinations.
Effect of R276X Mutations on 2-Aminopurine Fluorescent Intensity
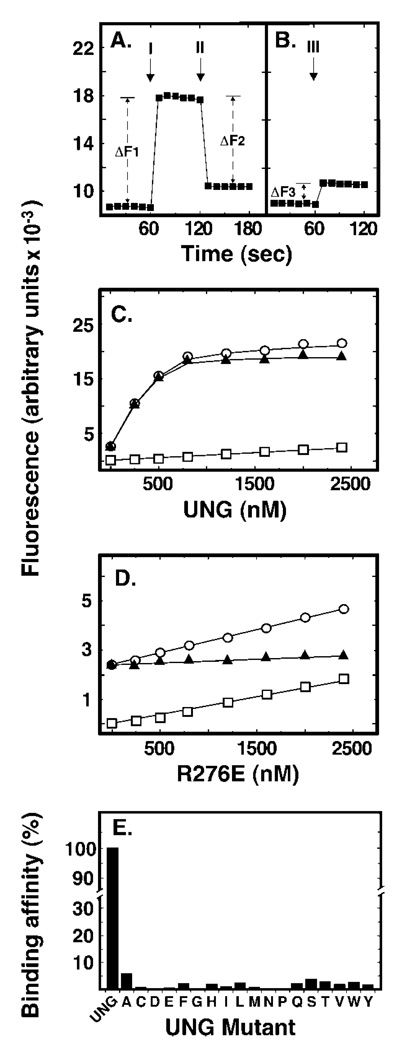
Crystallographic studies by Parikh and co-workers (5) demonstrated that UNG* was unable to cleave the C1-C1′ bond between the deoxypseudouracil base (ψU) and its cognate 2′-deoxyribose. However, it was reported that substitution of ψU for uracil did not affect the deoxyribose pucker, aromaticity, or hydrogen-bonding interactions of the pseudo-substrate with the enzyme, and that the ψU base was flipped of the double helix and into the UNG* active site pocket (5). Kinetic studies by Stivers and co-workers (30) showed an increase (2–8-fold) in 2AP fluorescence when E. coli Ung bound uracil substrate analogues were positioned adjacent or opposite to 2AP (30). Because 2AP fluorescence is quenched when the base analogue is stacked within the DNA helix, the increase in 2AP fluorescence indicated that the uracil base adjacent to 2AP was flipped out (30). Based on these reports, we reasoned that ψU was positioned opposite 2AP (dψU·2AP-25-mer), when flipped of the DNA helix by UNG, would also result in quantitative 2AP fluorescence enhancement. An increase in 2AP fluorescence concomitant with UNG addition was indeed observed, as is shown in the Fig. 5A (labeled I). To determine whether the enhancement of 2AP fluorescence was dependent on UNG interactions with dψU·2AP-25-mer, an excess of uracil-DNA glycosylase inhibitor protein (Ugi) was added to the reaction to abolish UNG DNA binding (26). Following supplementation with Ugi (Fig. 5A, II), 2AP fluorescence was quenched (ΔF2) to the level (ΔF3) observed for Ugi alone (Fig. 5B, III). Thus, the increase in 2AP fluorescence intensity upon UNG addition (ΔF1) was equal to the decrease in fluorescence intensity caused by Ugi (ΔF2) plus the Ugi/DNA fluorescence background (ΔF3): ΔF1 = ΔF2 + ΔF3. This result showed that the observed 2AP fluorescence enhancement was the result of UNG binding to dψU·2AP-25-mer. The dependence of the 2AP fluorescence enhancement on UNG concentration was determined using reaction mixtures containing 0, 100, 250, 500, 1000, or 1500 nm UNG and 50 nm dψU·2AP-25-mer (Fig. 5C). The average fluorescence at each UNG concentration was acquired and the data were fit to a non-linear regression curve. As shown in Fig. 5, 2AP fluorescence increased as a linear function of UNG concentration until the binding/flipping reaction was saturated between 500 and 1000 nm UNG. The net increase in 2AP fluorescence was obtained by subtracting the fluorescence of the enzyme acquired in the absence of dψU·2AP-25-mer DNA from the total fluorescence intensity (Fig. 5, legend). Interestingly, we did not observe saturation of 2AP fluorescence in binding reactions containing the R276X mutant enzymes. However, the 2AP fluorescence intensity observed at mutant enzyme concentrations from 0 to 500 nm did increase in a linear manner; therefore, we decided to use the initial slope of the fluorescence binding curve as a measure of enzyme affinity for the dψU-containing substrate. In the wild-type UNG reaction, the slope of the curve was determined from the 0, 100, 250, and 500 nm UNG data points and found to be 22.7 (Fig. 5C). In contrast, the defining slope of the 2AP fluorescence curve obtained for the R276E mutant was 0.134, 169-fold lower compared with that of UNG (Fig. 5D). Using this approach, the relative binding affinities of the 18 R276X mutant proteins for the dψU-containing double-stranded DNA substrate were determined; all R276X mutations negatively affected DNA binding/dψU flipping (Fig. 5E).
Fig. 5. Effect of Arg276 mutations on DNA binding and base flipping as measured by 2-aminopurine fluorescence.
Reaction mixtures (250 µl) contained 50 nm double-stranded dψU·2AP-25-mer and various amounts (0–1500 nm) of UNG or Arg276 mutant proteins. Fluorescence intensity measurements were performed as described under “Experimental Procedures.” A, the baseline fluorescent intensity of the 2-aminopurine-containing (dψU·2AP-25-mer) DNA substrate was measured at 1-s intervals for 1 min. UNG (500 nm) was then added and additional fluorescent intensity measurements were continued for 1 min. Arrow I marks the time of UNG addition; ΔF1 is the average fluorescence intensity enhancement caused by UNG addition. Lastly, Ugi (5 µm) was added to the reaction and the fluorescence intensity was monitored for another minute. Arrow II marks the time of Ugi addition and ΔF2 is the average 2AP fluorescent intensity quench that resulted from Ugi addition. The rectangular plot symbols represent the average intensity of 10 consecutive 1-s measurements. B, the fluorescent intensity of the dψU·2AP-25-mer was monitored for 1 min as in A prior to the addition of Ugi (5 µm). Arrow III marks the time of Ugi addition; ΔF3 corresponds to the average increase in 2AP fluorescent intensity caused by Ugi addition. C, reaction mixtures containing dψU·2AP-25-mer (50 nm) and 0, 100, 250, 500, 1000, or 1500 nm UNG were prepared and the fluorescent intensity was determined at 1-s intervals for 1 min. Open circles represent total observed fluorescence, open squares represent the fluorescence of the protein solution alone, and filled triangles represent the net fluorescence (open circles minus open squares). D, samples were prepared as described in C, except that the R276E mutant enzyme replaced UNG. The linear dependence of fluorescent intensity as a function of the enzyme concentration was calculated and termed the initial “slope” of the binding curve. Plot symbols are as described in C. E, sample preparation, fluorescent intensity measurements, and slope calculations were carried out as described in D for each of the Arg276 mutant enzyme preparations and compared with the initial slope of UNG, which was set to 100%. Error bars indicating the standard deviation of the 60 1-s fluorescent intensity measurements in C–E are obscured by the plot symbols.
Photochemical Cross-linking of UNG and R276X Mutant Proteins to Single-stranded dψU-25-mer
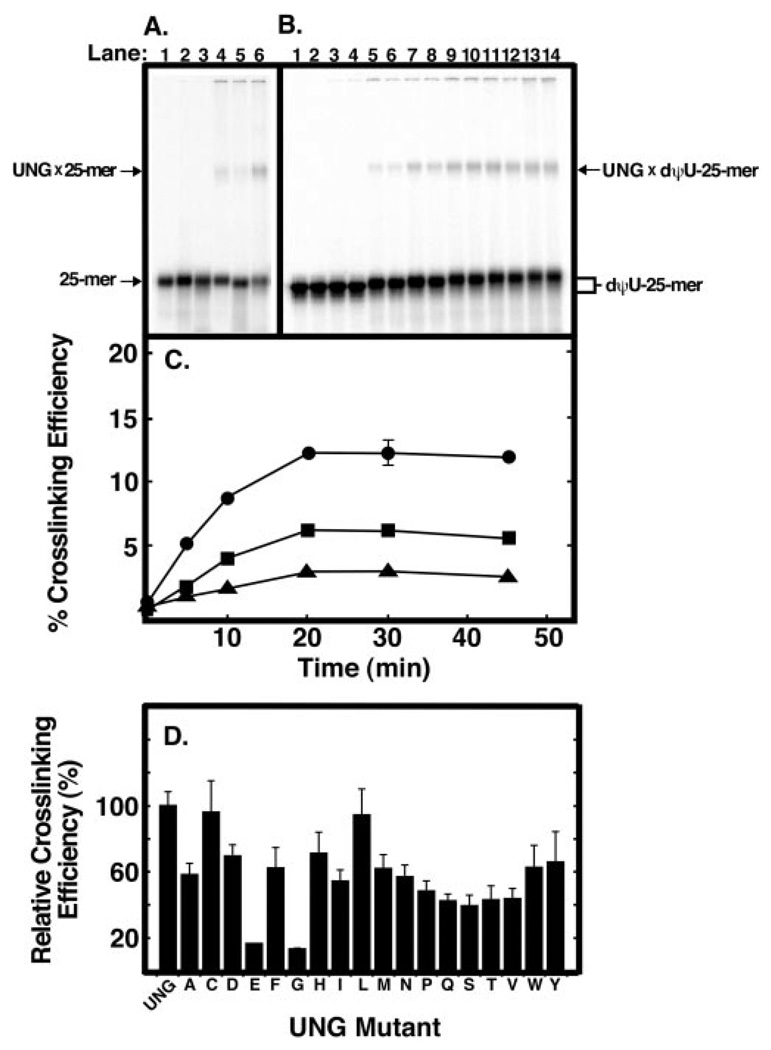
To examine the effect of Arg276 mutations on UNG interaction with single-stranded DNA, we performed UV cross-linking experiments with the 5′-end 32P-labeled single-stranded 25-mer oligonucleotide that contained site-specific T, U, or dψU nucleotides. The reaction mixtures were UV-irradiated for 30 min, subjected to non-denaturing polyacrylamide gel electrophoresis, and analyzed by PhosphorImager as described under “Experimental Procedures.” As shown in Fig. 6, two radioactive bands were identified: one band migrated rapidly and corresponded to free [32P]25-mer, whereas the other migrated more slowly and represented the UNG × [32P]25-mer protein-DNA cross-linked complex. The cross-linking efficiency of UNG to each oligonucleotide was: dψU-25-mer (12%) > T-25-mer (6%) > U-25-mer (3%)(Fig. 6A, lanes 6, 4, and 5, respectively). To determine the UV dose dependence of UNG·[32P]25-mer cross-link formation, a time course of UV irradiation was carried out using [32P]dψU-25-mer DNA (Fig. 6B). Inspection of the autoradiogram revealed the time-dependent emergence of the mobility retarded UNG·[32P]dψU-25-mer cross-linked band (Fig. 6B). Similar UV irradiation time course experiments were conducted with [32P]T-25-mer and [32P]U-25-mer DNA. Quantification of the 32P radioactivity (Fig. 6C) showed that, as before (Fig. 6A), UV cross-linking was more efficient with [32P]dψU-25-mer DNA than either [32P]T- or [32P]U-25-mer DNA. Moreover, the accumulation of UV cross-linked UNG × [32P]25-mer complex increased in a linear manner until ~20 min, at which point no further increase in cross-linked product was observed for any of the DNA substrates (Fig. 6C). Based on these results, the [32P]dψU-25-mer DNA substrate and the 10-min UV exposure time were selected for subsequent experiments.
Fig. 6. Ability of UNG and Arg276 mutant proteins to form UV-catalyzed cross-links to [32P]25-mer DNA.
A, samples (12 µl) containing 20 pmol of 5′-end 32P-labeled oligonucleotide T-25-mer, U-25-mer, or dψU-25-mer without UNG (lanes 1–3, respectively) or with 40 pmol of UNG (lanes 4–6, respectively) were UV-irradiated for 30 min as described under “Experimental Procedures.” Following irradiation, samples were subjected to non-denaturing polyacrylamide gel electrophoresis; the gels were dried and analyzed using a PhosphorImager. The positions of the UNG × [32P]DNA-25-mer cross-linked complex bands and free [32P]DNA-25-mer bands are indicated by arrows. B, reaction mixtures (12 µl) were prepared in duplicate that contained 20 pmol of [32P]dψU-25-mer and 40 pmol of UNG. Following UV irradiation for 0, 5, 10, 20, 30, and 45 min (lanes 3–14, respectively), reactions were analyzed as in A. Control reactions (lanes 1 and 2), containing 20 pmol of [32P]dψU-25-mer, were not irradiated. C, reaction mixtures (12 µl) were prepared in duplicate that contained 20 pmol of [32P]dψU-25-mer (closed circles), [32P]T-25-mer (closed squares), or [32P]U-25-mer (closed triangles), and 40 pmol of UNG. Following UV irradiation for 0, 5, 10, 20, 30, and 45 min, reactions were analyzed as in A, and the PhosphorImager data were quantified using the ImageQuant program. The cross-linking efficiency (%) was calculated by dividing the intensity of the UNG × [32P]25-mer band by the sum of the [32P]25-mer and UNG × [32P]25-mer bands and multiplying by 100. D, reaction mixtures (12 µl) were prepared that contained 20 pmol of [32P]dψU-25-mer and 40 pmol of each Arg276 mutant enzyme. The reactions were UV-irradiated for 10 min and analyzed as described in C. The cross-linking efficiency of each mutant preparation, indicated by the corresponding single letter amino acid abbreviation, is compared with that of UNG. Error bars represent the standard deviation of three experiments.
To ascertain the effect of the R276X mutations on DNA binding as reflected by UV cross-linking efficiency, each of the 18 R276X mutant enzymes were combined with [32P]dψU-25-mer DNA, irradiated for 10 min, and the cross-linking efficiency relative to UNG was determined as described above (Fig. 6D). Mutations that resulted in the lowest cross-linking efficiency were R276E and R276G, whereas the R276C and R276L were essentially same as UNG (Fig. 6D). The cross-linking efficiency of the remaining 14 R276X mutant proteins was intermediate and did not appear to correlate with side chain class (e.g. aliphatic, acidic). All R276X mutations, except R276C and R276L, showed a decrease in UV cross-linking efficiency.
Processivity of UNG on Concatemeric Uracil-containing [32P]DNA
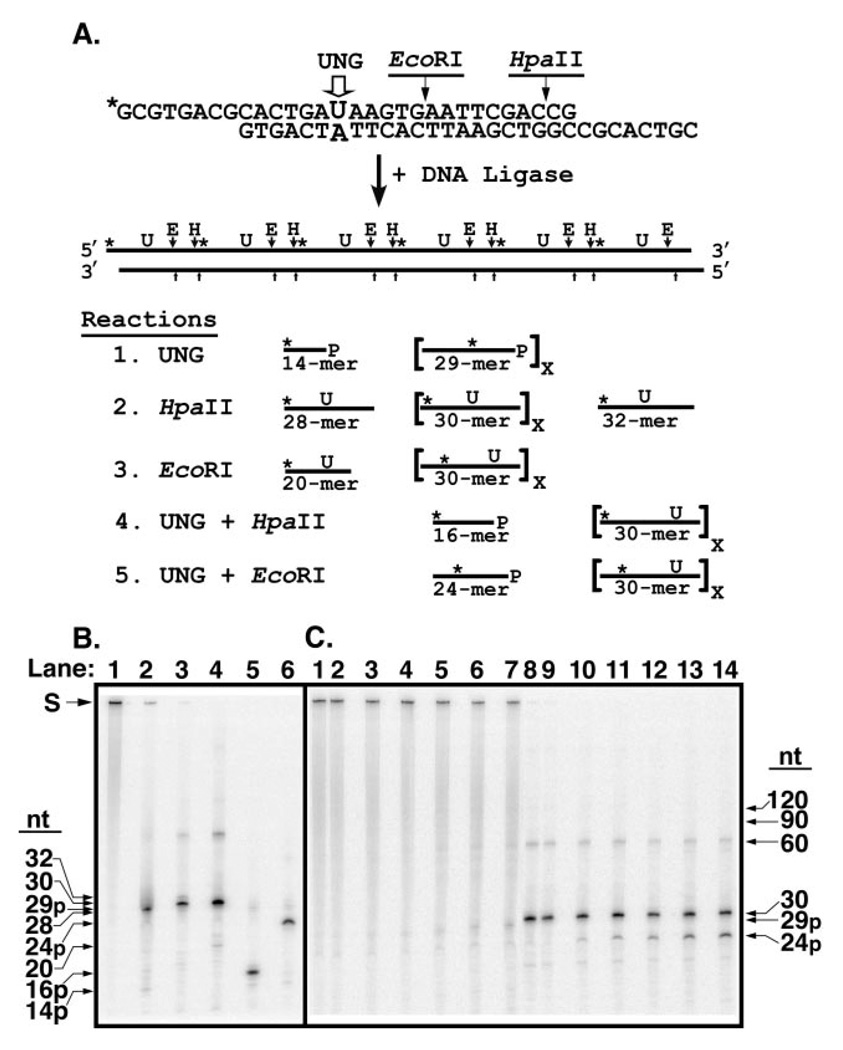
Proteins that recognize a particular site or sequence in DNA can locate their targets by random three-dimensional diffusion or by facilitated diffusion (31), processes are often referred to, respectively, as distributive or processive search mechanisms. To determine the search mechanism used by UNG to locate sequential uracil residues on the same DNA strand, a defined concatemeric DNA substrate was constructed from repeating 30-nucleotide monomer units (Fig. 7A), as described under “Experimental Procedures.” Each monomer unit contained a U·A base pair in a defined sequence context; the uracil-containing strand was 5′-32P-end labeled. Thus, a [32P]29-mer bearing a 3′-phosphate ([32P]29p-mer) would be released if adjacent uracil residues were successively excised (processive excision) and the apyrimidinic sites cleaved by alkali treatment. A [32P]29p-mer would be released in the event of successive uracil cleavage rather than a [32P]30-mer, because hot alkali catalyzes β-elimination of the 3′-phosphodiester bond of the AP-site followed largely by δ-elimination of the 5′-phosphodiester bond of the β-elimination product, resulting in the loss of the baseless sugar. However, if uracil residues were excised at random, little [32P]29p-mer would be produced. By comparing the amount of [32P]29p-mer released relative to the total amount of uracil excised during the early stages of the reaction, the degree of processive excision (processivity) of each enzyme could be evaluated. This approach was successfully used previously to determine the impact of ionic strength on the processive search mechanism of E. coli uracil-DNA glycosylase and rat mitochondrial uracil-DNA glycosylase (8). In the present report, the 5′ and 3′ ends of the [32P]U·A-30-mer monomer unit contained an eight-nucleotide complementary overlapping sequence that formed an HpaII restriction endonuclease recognition site upon hybridization of two or more monomer units (Fig. 7A). Each [32P]U·A-30-mer monomer also contained an internal EcoRI cleavage site. Concatemeric [32P]DNA substrates with uracil residues at regular intervals (30 nucleotides) along one strand of the duplex polymer were produced by DNA ligation. Analysis of the ligation products by non-denaturing polyacrylamide gel electrophoresis revealed that greater than 90% of the polymeric [32P]U·A DNA was ≥300 bp (data not shown).
Fig. 7. Analysis of reaction products generated by UNG from a concatenated uracil-containing [32P]DNA substrate.
A, the concatenated uracil-containing [32P]DNA substrate was prepared by ligation of double-stranded 30-mer units as described under “Experimental Procedures.” Each 30-mer unit contained a uracil residue at position 15 in the upper strand opposite adenine in the lower, complementary, DNA strand; only the uracil-containing strand was 5′-32P-end labeled (marked with an asterisk, *). An EcoRI restriction endonuclease recognition site was situated 3′ to nucleotide 20 of the uracil-containing DNA strand. The 30-mer unit was designed with self-complementary overhanging 5′-ends of eight nucleotides that formed a HpaII endonuclease recognition site upon ligation. Ligation of the 30-mer units created a concatenated uracil-containing [32P]DNA substrate in which each uracil residue as well as the EcoRI and HpaII recognition sites were separated by 30 nucleotides. Reaction 1: sequential uracil excision by UNG followed by alkaline hydrolysis of the resultant AP-site produces 29-mers containing an internal 32P label and a 3′-phosphate (29p-mer). Excision of the 5′-terminal uracil produces a 5′-32P-end labeled 14-mer with a 3′-phosphate (14p-mer). Reaction 2: HpaII digestion of the concatenated [32P]DNA substrate and subsequent alkaline hydrolysis generates 30-mers with an internal 32P label. Cleavage by HpaII at the 5′- and 3′-ends yields a 28-mer containing a 5′-32P-end label and a 32-mer containing an internal 32P label, respectively. Reaction 3: EcoRI digestion produces 30-mers except at the 5′ terminus where a [32P]DNA-20-mer is generated. Reaction 4: concatenated [32P]DNA digested with UNG, then exhaustively digested with HpaII followed by alkaline hydrolysis, yields 16-mers containing a 3′-phosphate (16p-mer) where the uracil has been excised, but [32P]30-mers from intact uracil-containing 30-mer units, as shown in Reaction 2. Only 32P-labeled reaction products are shown. Reaction 5: concatenated [32P]DNA digested with UNG, then exhaustively digested with EcoRI followed by alkaline hydrolysis, yields [32P]24-mers containing a 3′-phosphate (24p-mer) where the uracil has been excised, but [32P]30-mers from uracil-containing 30-mer units, as shown in Reaction 3. Only 32P-labeled reaction products are shown. B, characterization of the concatenated uracil-containing [32P]DNA substrate. Reaction mixtures (10 µl) containing 10 pmol of concatenated [32P]DNA and no addition (lane 1), 70 units of UNG (lane 2), 10 units of EcoRI (lane 3), 10 units of HpaII (lane 4), 70 units of UNG and 10 units of HpaII (lane 5), or 70 units of UNG and 10 units of EcoRI (lane 6), were incubated at 37 °C, subjected to alkaline hydrolysis, and analyzed by 12% denaturing polyacrylamide gel electrophoresis as described under “Experimental Procedures.” The mobility of the unreacted concatenated [32P]DNA substrate (S) and various oligonucleotide reaction products (32-, 30-, 29p-, 28-, 24p-, 20-, 16p-, and 14p-mer) is indicated by arrows. C, time course of UNG digestion. A reaction mixture (100 µl) containing 0.06 units of UNG and 80 pmol of concatenated uracil-containing [32P]DNA was incubated at 37 °C. Aliquots (10 µl) were removed at 0, 5, 10, 20, 40, and 80 min and terminated by the addition of Ugi as described under “Experimental Procedures.” A control reaction contained 10 pmol of the [32P]DNA substrate only. Following reaction termination, each time point was divided into two aliquots. One, designated the UNG reaction, was subjected to alkaline hydrolysis. The other, designated as the EcoRI reaction, underwent EcoRI digestion and subsequent alkaline hydrolysis. Reactions were analyzed by denaturing 12% polyacrylamide, 8.3 m urea gel electrophoresis as described under “Experimental Procedures.” UNG reactions corresponding to the control reaction, and 0-, 5-, 10-, 20-, 40-, and 80-min time points were loaded in lanes 1–7, respectively, whereas the corresponding EcoRI reactions were loaded in lanes 8–14. Arrows indicate the positions of the various oligonucleotide reaction products. The extent of uracil excision after each incubation time was determined by dividing the amount of [32P]24p-mer oligonucleotide released by EcoRI digestion by the sum of [32P]24p-mer + [32P]30-mer, and multiplying the quotient by 100; this percentage was called the Extent of Digestion.
To isolate high molecular weight [32P]U·A concatemeric DNA, the polymeric [32P]U·A ligation products were subjected to gel filtration chromatography, and fractions that contained [32P]U·A concatemeric DNA greater than 600 bp as judged by non-denaturing polyacrylamide gel electrophoresis were pooled and used as substrate for processivity reactions The average length of the concatenated [32P]U·A DNA substrate was determined by three additional methods, all of which involved enzymatic “collapse” of the [32P]U·A concatemeric substrate to its monomer components (Fig. 7A). First, the concatemeric substrate was subjected to excess UNG and alkali treatment, and the percentage of [32P]14p-mer determined following denaturing polyacrylamide gel electrophoresis (Fig. 7B, lane 2). The ratio of [32P]14p-mer to the sum of [32P]29p-mer plus [32P]14p-mer was dictated by the length of the concatemeric substrate, because digestion of every uracil residue released 32P-containing 29p-mers except at the 5′-and 3′-ends. The 5′-end digestion fragment was a [32P]14p-mer; however, the 15-mer 3′-end fragment did not contain [32P] and therefore was undetectable. UNG digestion and subsequent alkaline treatment was found to produce 5.6% [32P]14p-mer. In the second and third methods, the length of the average [32P]concatemer was determined by restriction endonuclease digestion (Fig. 7A). Digestion of the [32P]U·A concatemer with HpaII produced [32P]30-mers except at the 5′- and 3′-ends, which released [32P]28-mer and [32P]32-mer, respectively (Fig. 7B, lane 3). The amount of [32P]28-mer was 9.2%, whereas the amount of [32P]32-mer was 9.3%. Digestion of the [32P]U·A concatemer with EcoRI yielded [32P]30-mer except for the 5′-end, which released [32P]20-mer; the percentage of [32P]20-mer was 6.2% (Fig. 7B, lane 4). Whereas the amounts of 5′-end 32P-fragments generated by the UNG and EcoRI digestions were in good agreement (5.6 and 6.2%, respectively), the amount of the 5′-end [32P]28-mer generated by HpaII digestion of the concatemeric [32P]DNA substrate was somewhat higher (9.2%). Therefore, we used the data from the UNG and EcoRI digestions to estimate the length of the average concatemeric [32P]DNA substrate, which was found to be ~510 bp. As shown in Fig. 7B (lanes 2–4, at the S arrow) it appeared that a minor percentage of the concatemeric DNA substrate was intractable to UNG (<7.4%), HpaII (<0.8%), or EcoRI (<0.2%) cleavage. We also observed a minor band at the 60-nucleotide position that was refractory to cleavage. However, subsequent processivity experiments were not materially affected by the minor refractory DNA substrates. We speculate that the estimate of the average size of the [32P]concatemeric DNA substrate as determined by non-denaturing polyacrylamide gel electrophoresis was higher than the estimates provided by the method of enzymatic collapse because annealed but unligated junctions between monomer units would not be detected during non-denaturing polyacrylamide gel electrophoresis.
To measure the processivity of UNG, it was critical to relate the amount of processive excision product ([32P]29p-mer) detected to the total amount of uracil excised. The [32P]concatemeric DNA substrate was designed such that following uracil excision by UNG, exhaustive digestion with HpaII or EcoRI, and hot alkali treatment, would produce [32P]30-mer if the uracil residue within the monomer unit was not excised, and [32P]16p-mer and [32P]24p-mer, respectively, if the uracil residue was excised (Fig. 7B, lanes 5 and 6). Thus, the extent of UNG digestion (the overall amount of uracil excised during the reaction) could be determined, using EcoRI, for example, as the ratio of [32P]24p-mer detected to the sum of [32P]24p-mer +[32P]30-mer detected (Fig. 7A, Reaction 5). Processivity could then be assessed by plotting the amount of processive product produced ([32P]29p-mer) as a function of the extent of digestion. Because processive excision generates more [32P]29p-mer, the ratio of [32P]24p-mer to [32P]24p-mer + [32P]30-mer would be greater than that generated by distributive excision, which would produce little [32P]29p-mer, especially during the early stages of the reaction. In the case of distributive excision, the accumulation of [32P]29p-mer products in large amounts would appear only late in the reaction time course when a significant percentage of the uracil residues on a particular [32P]concatemer had already been excised, raising the likelihood that a single distributive excision event would release a [32P]29p-mer.
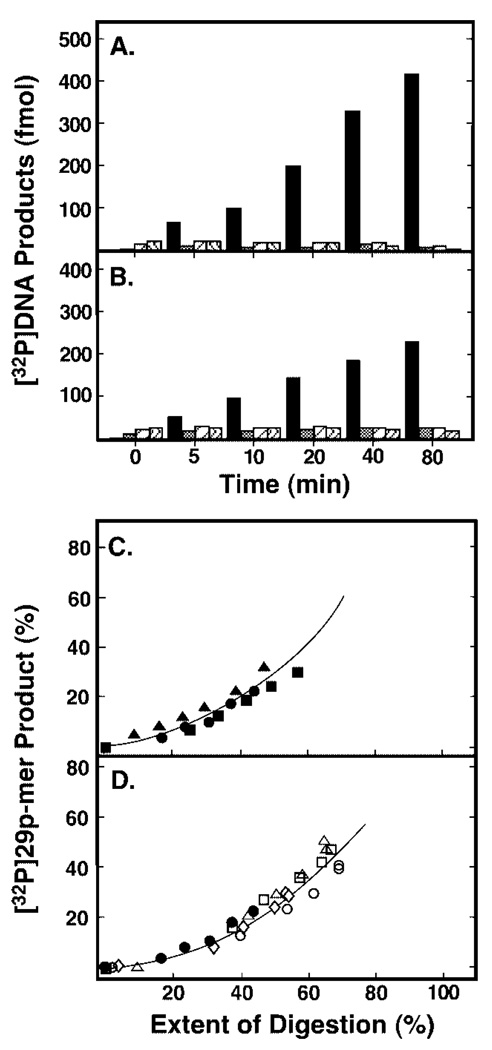
To assess UNG processivity, a high molar ratio of DNA substrate to enzyme was used to ensure that, on average, no more than one UNG molecule was bound initially to a particular concatemeric-DNA molecule. Standard processivity assays were performed in reactions containing 10 pmol of concatemeric [32P]U·A DNA substrate and 0.06 units of UNG. A time course reaction of uracil excision was conducted, and the products of each reaction time point were divided into two aliquots. One aliquot was subjected to AP-site hydrolysis and used to determine the percentage of total [32P]DNA substrate that appeared as [32P]29p-mer units that were indicative of a processive excision search mechanism. The other aliquot was used to determine the extent of digestion, i.e. the amount of uracil released from polymeric [32P]DNA substrate regardless of position. The extent of digestion was measured by treating the UNG reaction products with excess EcoRI restriction endonuclease, as described above. Both aliquots were then analyzed together by 12% denaturing polyacrylamide gel electrophoresis (Fig. 7C). As the reaction progressed, only the [32P]29p-mer products were visible and partially digested [32P]oligonucleotides appeared at the relatively low level in the later time points (Fig. 7C, lanes 1–7). The extent of UNG digestion at each point in the reaction time course was evaluated by EcoRI digestion (Fig. 7C, lanes 8–14). Quantification of the gel results revealed that the emergence of the [32P]24p-mer bands appeared commensurate with the production of the [32P]29p-mer bands in the UNG reaction product, as shown in Fig. 8C. Similar results were found in reactions containing either 0.04 units of Ung or 0.06 units of UNG* and the same 32P-labeled concatemeric DNA substrate (Fig. 8, A and B, respectively). The molar quantities of [32P]29p-mer excision products generated by Ung and UNG* on the concatenated uracil-containing [32P]DNA were dominant throughout the reaction time course. Analysis of the Ung and UNG* gels also showed that the emergence of the [32P]24p-mer band was proportional to the production of the [32P]29p-mer band in both Ung and UNG* reactions (Fig. 8C). To arrive at a more quantitative assessment of processivity, the amount of processive excision product ([32P]29p-mer, y axis) was divided by the extent of digestion (x axis) at each early time point to determine the initial slope of the “processivity” plot. The slope of the E. coli Ung processivity plot was determined to be 0.51, which was in good agreement with an earlier measurement (0.56) of Ung processivity reported by Bennett et al. (8). The slope of the UNG* processivity plot was determined to be 0.44, consistent with an earlier processivity measurement (0.45) reported for rat liver mitochondrial uracil-DNA glycosylase (8), and the slope of the UNG processivity plot was 0.39. These results suggested that the core catalytic domain of human uracil-DNA glycosylase (UNG*), like E. coli Ung and rat liver mitochondrial uracil-DNA glycosylase, located uracil residues using a processive search mechanism. In addition, the results indicated that the N-terminal His6 tag of UNG did not significantly affect the processive character of the enzyme, as the processivity analysis of the UNG* and UNG reaction products were very similar (Fig. 8C).
Fig. 8. Analysis of reaction products generated by Ung, UNG*, UNG, or its Arg276 mutant proteins from concatenated uracil-containing [32P]DNA.
Two processivity reaction mixtures (100 µl) containing either 0.04 units of E. coli Ung (A) or 0.06 units of human UNG* (B), and 80 pmol of concatenated uracil-containing [32P]DNA substrate were prepared as described under “Experimental Procedures.” Samples (10 µl) were withdrawn after 0, 5, 10, 20, 40, and 80 min of incubation at 37 °C, and divided into two aliquots as described in the legend to Fig. 7. From the reactions subjected to alkaline hydrolysis only, the molar quantities of the four detectable [32P]oligonucleotide UNG digestion products were determined and are presented graphically as: 29p-mer (black bar), 59p-mer (cross-hatched bar), 89p-mer (striped bar), and 119p-mer (wavy bar). C, the amount of [32P]29p-mer product generated at each incubation time (A and B, black bars) was determined relative to the total [32P]DNA detected, and the extent of digestion at various incubation times was determined after EcoRI treatment. A time course reaction identical to that described for Ung and UNG* but containing 0.06 unit of UNG was also conducted and analyzed as described above. The processivity of Ung (0.04 unit, filled circles), UNG* (0.06 unit, filled squares), or UNG (0.06 unit, filled triangles) was analyzed by graphing the amount (%) of processive [32P]29p-mer product detected as a function of the extent of digestion (%). D, processivity analysis of uracil-DNA glycosylase reactions containing R276D (0.3 unit, open diamonds), R276L (0.3 unit, open circles), R276T (1.0 unit, open triangles), and R276W (1.0 unit, open squares) is presented and compared with UNG (0.06 unit, filled circles).
Processivity of R276X Mutants on Concatemeric Uracil-containing [32P]DNA
To assess the effect of Arg276 mutations on the processive search mechanism of UNG, we selected the following four mutants for analysis: (i) R276L, chosen because the efficiency of UV cross-linking to [32P]ψ U-25-mer was unaffected; (ii) R276W, because it had highest activity among aromatic side chain mutants; (iii) R276D and (iv) R276T were chosen because they exhibited similar moderate defects in uracil-DNA glycosylase activity and UV cross-linking efficiency. The standard processivity assay was conducted in reactions containing 10 pmol of concatemeric [32P]U·A DNA substrate and various amounts of R276D, R276L, R276T, and R276D as described in the legend to Fig. 8. The reaction products of each time point were analyzed for processive [32P]29p-mer product release and the extent of digestion as described above. Next, the processivity of the mutant proteins was assessed by graphing the percentage of processive [32P]29p-mer product released as a function of the extent of digestion, and analysis of the results showed that the release of [32P]29p-mer was proportional to the overall amount of uracil excised (Fig. 8D). From the processivity plots the initial slopes were determined for R276D, R276L, R276T, and R276W, and found to be 0.39, 0.41, 0.55, and 0.54, respectively. These results were in general agreement with those obtained for UNG, UNG*, and Ung (Fig. 8C). Moreover, similar results were observed for processivity reactions containing R276H, R276C, and R276S (data not shown). Taken together, these experiments indicated that mutations at Arg276 did not significantly affect the processive search mechanism of UNG.
DISCUSSION
We have used PCR-based codon-specific random mutagenesis and site-specific mutagenesis to construct a library of 18 amino acid changes at arginine 276 of the core catalytic domain of human uracil-DNA glycosylase. The R276X mutations were characterized as to their effect on Ugi complex formation, uracil-DNA glycosylase catalytic activity, DNA binding/base flip-ping, and UV cross-linking efficiency. The results show that, without exception, mutations at arginine 276 resulted in significantly decreased UNG catalytic activity on uracil-containing double-stranded DNA and lower affinity to double-stranded dψU·2AP-containing oligonucleotide. R276X mutations had a less pronounced effect of UV cross-linking efficiency to single-stranded DNA. In addition, UNG was demonstrated to utilize a processive search mechanism to locate successive uracil residues site-specifically situated along on one strand of a concatemeric uracil-containing [32P]DNA substrate, and mutations at arginine 276 did not alter the processive character of the UNG search mechanism.
The negative effect of R276X mutations on UNG catalytic activity may have resulted from perturbation of the enzyme-DNA interaction during the formation of the enzyme-DNA complex prior to glycosylic bond cleavage. Wong and co-workers (7) used pre-steady-state kinetic analysis of a single catalytic turnover by E. coli Ung to predict three sequential steps in the enzyme mechanism that occurred prior to cleavage of the glycosylic bond: 1) rapid equilibrium binding of Ung to DNA (nonspecific); 2) uracil flipping; and 3) a protein conformation change. The possibility that mutation of Arg276 affects the initial binding step of UNG to double-stranded DNA will be discussed in light of three experimental observations. First, structural data from the double L272R/D145N UNG mutant in complex with (cleaved) U·G DNA indicated that the η-NH2 of the Arg276 guanidinium side chain formed a hydrogen bond with the oxygen residue of the third phosphate residue 3′ to the site of the cleaved uracil residue (17). Also, in the co-crystal structure of wild-type UNG in complex with either cleaved U·G or cleaved U·A DNA, the Arg276 side chain ε-nitrogen was reported to form a water-bridged hydrogen bond with the N3 site of an adenine residue 3′ to the uracil residue as UNG “read” the DNA minor groove (5). Thus, mutations at Arg276 may disrupt protein-DNA interactions, effectively lowering the affinity of the enzyme for DNA. However, it is not at all clear that disruption of one positively charged amino acid side chain of 13 would strongly affect DNA binding, because it appears that electrostatic interactions, not direct or water-mediated DNA phosphate interactions, orient the enzyme on DNA for initial damage detection (6). It should also be noted that the Arg276/DNA phosphate interaction observed by Slupphaug et al. (17) was not reported by Parikh et al. (6). To determine the mechanism of catalysis by UNG, Dinner and co-workers (32) used a hybrid quantum mechanical/molecular mechanical approach based on the UNG-DNA co-crystal structure solved by Parikh et al. (5). They calculated that the primary contribution to lowering the activation energy of uracil excision came not from the enzyme but rather from the burial and positioning of four 5′-phosphate groups in the substrate (T4, dU5, dA6, and dT7) that were thought to stabilize the cationic sugar in the transition state. According to this model, an interaction between Arg276 NH2 and the 5′-phosphate of dC8 or lack thereof would not significantly affect the activation energy of uracil excision.
Second, the fluorescence equilibrium binding experiments reported in this article demonstrated that binding of R276X mutant proteins to a dψU·2AP-containing duplex oligonucleotide was not saturable. This result suggested that the KD of the R276X mutant proteins for duplex DNA was significantly elevated relative to wild-type UNG. However, in these experiments it was not possible to distinguish between binding to DNA and binding the flipped-out uracil. The fluorescence signal in these experiments came from DNA containing 2AP opposite ψU; flipping of the dψU residue led to increased 2AP fluorescence. Therefore, it may be argued that the R276X mutations affected the efficiency of dψU flipping rather than DNA binding. Parikh et al. (6) observed that mutation of Leu272 to Ala did not affect the rate of binding (kass) to 4′-thio-U·G DNA relative to wild-type UNG, but the rate of dissociation (kdiss) was approximately three times slower for the wild-type enzyme, presumably because insertion of the Leu272 side chain in the DNA minor groove slowed binding of the DNA phosphate backbone by the Leu272 loop. In the model of Wong et al. (7), insertion of the Leu272 loop acts as a “door stop” to prevent facile return of the flipped out uracil to the DNA base stack. Structural data from Parikh et al. (6) indicated that the εN of the Arg276 guanidinium side chain also participated in a water-mediated hydrogen bond with the carbonyl oxygen of Leu272. Therefore, ablation of the Arg276 side chain by mutation could destabilize the Leu272 intercalating loop and reduce its ability to prevent the uracil residue from flipping back into the DNA base stack. In addition, mutation of Arg276 may interfere with the ability of Tyr275 to widen the DNA minor grove and facilitate uracil flipping (6).
Third, the effect of mutations at Arg276 on UV-catalyzed cross-linking to single-stranded dψU-containing oligonucleotide was much less severe than the effects observed for experiments involving double-stranded DNA. In fact, the UV cross-linking efficiency of two mutant proteins, R276C and R276L, was comparable (~95%) to that of wild-type. This result indicated that whereas Arg276 was required for efficient recognition and base flipping of uracil in double-stranded DNA, its role in binding the more flexible single-stranded 25-mer DNA was much less important. The efficiency of UV cross-linking UNG to dψU-25-mer was ~4-fold greater than to U-25-mer and 2-fold greater than to T-25-mer. This result is consistent with the report by Shoyer et al. (33), who observed that UV cross-linking of E. coli Ung to single-stranded DNA 20-mer containing one uracil residue amid 19 dTMP residues was less efficient (~0.5) than to dT20. We hypothesize that UV cross-linking of UNG to dψU-25-mer was more efficient because of the non-cleavable nature of the C1-C1′ glycosylic bond linking the pseudouracil base to deoxyribose. Thus, the residence time of flipped dψU in the active site may be longer, and the association of UNG active site residues with dψU and adjacent nucleotides prolonged. If the duration of the protein-DNA contact were longer, then the efficiency of UV cross-linking would be expected to increase, as was observed. Whereas every amino acid side chain has the potential to form UV-catalyzed cross-links with DNA, the aromatic side chains of Phe, Trp, Tyr, and His were reported to form DNA cross-links more efficiently (34). However, substitution of these aromatic amino acids for Arg276 did not lead to greater UV cross-linking efficiency; instead, substitution with Cys or Leu resulted in near wild-type cross-link formation. Whereas a straightforward explanation for these observations is not obvious, we speculate that the differential UV cross-linking efficiencies of the R276X mutant proteins may have to do with the relative effectiveness of the leucine loop in maintaining the dψU base in a flipped conformation and the single-stranded DNA in the DNA binding groove of the enzyme.
Using a concatemeric [32P]U·A polynucleotide substrate with uracil residues and an EcoRI endonuclease recognition site at non-overlapping 30-nucleotide intervals, we have demonstrated that the catalytic domain of human uracil-DNA glycosylase utilized a processive mechanism to excise successive uracil residues. This result was consistent with previously reported processivity measurements of E. coli Ung and rat liver mitochondrial uracil-DNA glycosylase (8). We interpret a processivity plot slope measurement of 0.5 to mean that, on average, two uracil residues are excised per DNA encounter, where, for a slope p, the expected number of excisions is 1/(1-p).2 Thus, uracil-DNA glycosylase from different sources exhibit modest processivity, consistent with short sliding or hopping facilitated diffusion (35). Interestingly, the processivity of the R276X mutant proteins studied did not differ significantly from that of UNG, although the Arg276 mutations resulted in uracil-DNA glycosylase enzymes with notably decreased catalytic activity. Thus, aggregate electrostatic interactions between the DNA binding groove of the enzyme and nonspecific DNA may be the primary determinant of the search mechanism.
Acknowledgments
We acknowledge Dr. Michael Schimerlik (Oregon State University) for assistance with fluorescence-based kinetic studies and the Center for Gene Research and Biotechnology for performing DNA sequencing.
Footnotes
This work was supported by National Institutes of Health Grants GM66245 (to S. E. B.) and GM32823 (to D. W. M.) and NIEHS National Institutes of Health Grant P30 ES00210.
The abbreviations used are: UDG, family-1 of uracil-DNA glycosylase; Ung, E. coli uracil-DNA glycosylase; UNG*, core catalytic domain of human uracil-DNA glycosylase; UNG, His6-tagged UNG*; Ugi, uracil-DNA glycosylase inhibitor protein; Arg276, arginine 276 of UNG; dψU, 2′-deoxypseudouridine; AP site, apyrimidinic/apurinic site; CAPS, 3-(cyclohexylamino)-1-propanesulfonic acid; FAM, carboxyfluorescein; DTT, dithiothreitol.
R. T. Smythe, personal communication.
REFERENCES
- 1.Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 2.Aravind L, Koonin EV. Genome Biol. 2000;1:0007.0001–0007.0008. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearl LH. Mutat. Res. 2000;460:165–191. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 4.Mosbaugh DW, Bennett SE. Prog. Nucleic Acids Res. Mol. Biol. 1994;48:315–370. doi: 10.1016/s0079-6603(08)60859-4. [DOI] [PubMed] [Google Scholar]
- 5.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parikh SP, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. J. Biol. Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 8.Bennett SE, Sanderson RJ, Mosbaugh DW. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- 9.Slupphaug G, Markussen F-H, Olsen LC, Aasland R, Aarsaether N, Bakke O, Krokan HE, Helland DE. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. J. Biol. Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 11.Haushalter KA, Stukenberg PT, Kirschner MW, Verdine GL. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 12.Hendrich B, Bird A. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 14.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 17.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 18.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 19.Kavli B, Slupphaug G, Mol CD, Arvai AS, Petersen SB, Tainer JA, Krokan HE. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 20.Dizdaroglu M, Karakaya A, Jaruga P, Slupphaug G, Krokan HE. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianchet MA, Seiple LA, Jiang YL, Ichikawa Y, Amzel LM, Stivers JT. Bichemistry. 2003;42:12455–12460. doi: 10.1021/bi035372+. [DOI] [PubMed] [Google Scholar]
- 22.Bennett SE. Characterization of the Escherichia coli Uracil-DNA Glycosylase-Inhibitor Complex. Ph.D. thesis. Corvallis, OR: Oregon State University; 1995. [Google Scholar]
- 23.Sanderson RJ, Mosbaugh DW. J. Biol. Chem. 1996;271:29170–29181. doi: 10.1074/jbc.271.46.29170. [DOI] [PubMed] [Google Scholar]
- 24.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Bennett SE, Mosbaugh DW. J. Biol. Chem. 1992;267:22512–22521. [PubMed] [Google Scholar]
- 27.Bennett SE, Jensen ON, Barofsky DF, Mosbaugh DW. J. Biol. Chem. 1994;269:21870–21879. [PubMed] [Google Scholar]
- 28.Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 29.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 30.Stivers JT, Pankeiwicz KW, Watanabe KA. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 31.Berg OG, Winter RB, von Hippel PH. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 32.Dinner AR, Blackburn GM, Karplus M. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 33.Shroyer MJ, Bennett SE, Putnam CD, Tainer JA, Mosbaugh DW. Biochemistry. 1999;38:4834–4845. doi: 10.1021/bi982986j. [DOI] [PubMed] [Google Scholar]
- 34.Shetlar MD. Photochem. Photobiol. Rev. 1980;5:105–197. [Google Scholar]
- 35.Gerland U, Moroz JD, Hwa T. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12015–12020. doi: 10.1073/pnas.192693599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]